Abstract
Porphyromonas gingivalis is one of the well-characterized periodontal pathogens involved in periodontitis. The invasive and proteolytic activities of P. gingivalis clinical isolates have been shown to be associated with heterogenic virulence, as determined in a mouse abscess model. The aims of the present study were to identify a P. gingivalis strain with a low virulence among clinical isolates, based on its invasive ability and cytokine proteolytic activities, and to explore the preferential degradation of a certain cytokine by P. gingivalis. P. gingivalis ATCC 33277, W50, and 10 clinical isolates were used. After incubating bacteria with IL-4, IL-6, IL-10, IL-17A, TNFα, IFNγ, and IL-1α, the amounts of remaining cytokines were determined by ELISA. Invasion ability was measured by a flow cytometric invasion assay. There was inter-strain variability both in the cytokine proteolytic activities and invasion ability. In addition, differential degradation of cytokines by P. gingivalis was observed: while IFNγ and IL-17A were almost completely degraded, inflammatory cytokines TNFα and IL-1α were less susceptible to degradation. Interestingly, the invasion index, but not cytokine proteolytic activities, of P. gingivalis had strong positive correlations with clinical parameters of subjects who harbored the isolates. Therefore, the invasive ability of P. gingivalis is an important virulence factor, and the bacterial invasion step may be a good target for new therapeutics of periodontitis.
Introduction
Periodontitis is a chronic inflammatory disease caused by the interactions between the subgingival biofilm and host immune responses, which lead to the destruction of tooth-supporting tissues. Periodontitis is characterized as a polymicrobial disease that involves dysbiosis of the indigenous flora in the subgingival biofilm.Citation1 Among the more than 500 bacterial species harbored in the subgingival biofilm, Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia are well-characterized periodontal pathogens.Citation2
P. gingivalis, a gram-negative obligate anaerobe, has a number of virulence factors such as cysteine proteases (gingipains), major fimbriae, lipopolysaccharide, and capsule.Citation3 Among the virulence factors of P. gingivalis, gingipains, which consist of Arg-gingipain (Rgp) and Lys-gingipain (Kgp), can degrade a number of host proteins, including cytokines/chemokines, immunoglobulins, complement proteins, and host cell receptors.Citation4,5 The degradation of various cytokines, including IL-1β, IL-4, IL-5, IL-6, IFNγ, and TNFα, by purified gingipains or live P. gingivalisCitation6-10 suggests a possibility that P. gingivalis may be able to modulate the immune response through preferential inactivation of a certain cytokine. However, the degradation of these cytokines has not been simultaneously studied.
P. gingivalis fimbriae are filamentous appendages that mediate the bacterial adherence to various host cells, other bacteria, and host macromolecules.Citation11 The fimA gene, which encodes FimA (a subunit of major fimbriae), has been classified into 6 types (I-V and Ib) based on the nucleotide sequences.Citation11 While type II, IV, and Ib genotypes are associated with periodontitis, the type I genotype is associated with periodontal health.Citation12,13 In particular, P. gingivalis isolated from patients mainly contain the fimA genotype II.Citation12,14
P. gingivalis has been shown to invade various host cells in vitro, including gingival epithelial cells, endothelial cells, and gingival fibroblasts.Citation15-17 In addition, P. gingivalis has been detected within gingival tissues obtained from patients with periodontitis, suggesting an important role of tissue invasion in the pathogenesis of periodontitis.Citation18,19
Inaba et al.Citation20 reported that the invasive and proteolytic activities of P. gingivalis clinical isolates are associated with heterogenic virulence, which has been studied in the mouse abscess model. Although probiotics have only included commensal bacteria and shown their usefulness as an adjunct therapy in chronic periodontitis,Citation21,22 P. gingivalis strains with a low virulence may be also used for the prevention of colonization or replacement of highly virulent P. gingivalis. The aims of the present study were to identify a P. gingivalis strain with a low virulence among clinical isolates based on its invasive ability and cytokine proteolytic activities, and to explore the preferential degradation of a certain cytokine by P. gingivalis.
Results
Ten P. gingivalis clinical isolates were isolated: one (KUMC-H1) from a periodontally healthy subject and 9 (KUMC-P1∼KUMC-P8 and KUMC-P10) from periodontitis patients. The demographic and clinical characteristics of the study subjects are summarized in . Patient P6 had a past history of tooth extraction (#16, #17, and #27) due to root caries below the crowns in 2008. Although the cause for the loss of the other teeth was not known, the levels of remaining alveolar bones were not substantially reduced compared to those of the adjacent tooth, suggesting a cause other than periodontitis. The causes for the tooth loss in patients P1, P7, and P8 were not available.
Table 1. The demographic and clinical characteristics of study subjects.
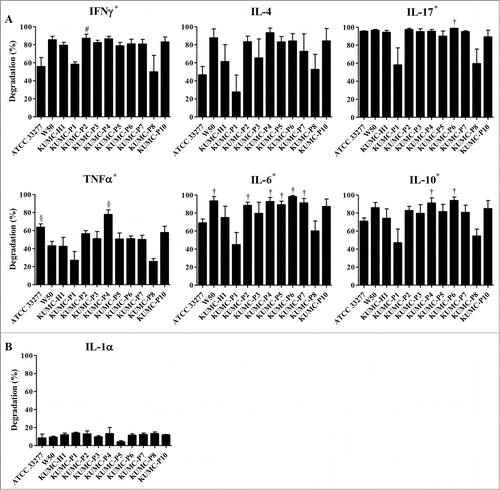
CD4+ T-helper (Th) cell subsets, such as Th1, Th2, Th17, and Tregs, play a critical role in modulating the immune responses.Citation23,24 To investigate the possibility that P. gingivalis clinical isolates differentially modulate the immune response through the preferential degradation of a certain cytokine, a mixture of IFNγ, IL-4, IL-17A, TNFα, IL-6, and IL-10 (Th1, Th2, Th17, two inflammatory, and an anti-inflammatory cytokine, respectively) was incubated with viable P. gingivalis. The amounts of residual recombinant cytokines determined by ELISA revealed a substantial inter-strain difference in the degradation of IFNγ (p = 0.007), IL-17A (p = 0.007), TNFα (p = 0.000), IL-6 (p = 0.004), and IL-10 (p = 0.018). In the case of the laboratory strains, W50 tended to have a higher degradation ability than ATCC 33277 for 4 cytokines. In general, KUMC-P1 and KUMC-P8 strains showed reduced degradation of all tested cytokines compared with other strains (). Interestingly, TNFα was less susceptible to degradation by P. gingivalis. In particular, W50, KUMC-P2, KUMC-P5, and KUMC-P6 strains showed significant differences in the degradation rate between TNFα and all other cytokines, suggesting the preferential protection of an inflammatory cytokine. Therefore, the ability of P. gingivalis to degrade IL-1α, another inflammatory cytokine, was examined. Interestingly, unlike other cytokines, IL-1α was resistant to degradation by P. gingivalis, which showed only 3 to 13% degradation rate with no significant inter-strain difference (). Taken together, there was inter-strain variability in the ability to degrade cytokines and the inflammatory cytokines TNFα and IL-1α were less susceptible to degradation by all strains of P. gingivalis.
Figure 1. Various proteolytic activity of P. gingivalis strains. (A) A mixture of recombinant cytokines (IL-4, IL-6, IL-10, IL-17A, IFNγ, and TNFα) or (B) IL-1α were prepared in KGM medium, and incubated with viable P. gingivalis at 37°C for 1 h. After incubation, the supernatants were collected and the amount of remaining cytokines was determined by ELISA. The results indicate degradation percentage compared with control samples incubated without P. gingivalis. *P < 0.05 significant difference among strains, †P < 0.05 versus KUMC-P1, #P < 0.05 vs. KUMC-P8, §P < 0.05 versus both KUMC-P1 and KUMC-P8.
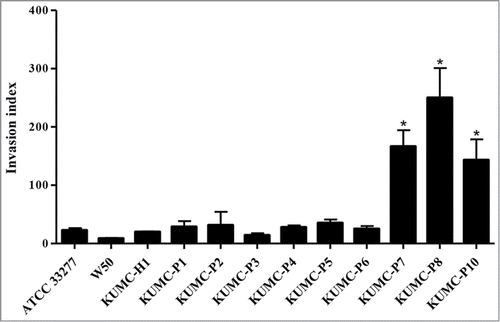
The invasion of P. gingivalis clinical isolates into HOK-16B cells was examined by a flow cytometric invasion assay. P. gingivalis ATCC 33277 and W50 strains were used as controls. In the case of the laboratory strains, the invasive ability of P. gingivalis ATCC 33277 was greater than that of W50, as was reported by another groupCitation25; this difference, however, was not significant. All 10 clinical isolates were able to invade HOK-16B cells. Among them, the invasion indexes of KUMC-P7, KUMC-P8, and KUMC-P10 strains were substantially higher than those of other strains, which resulted in an approximately 20-fold difference between the lowest and the highest strains ().
Figure 2. Various invasive ability of P. gingivalis strains. HOK-16B cells (6 × 10Citation4 cells/well) were seeded into 24-well plates and incubated with viable CFSE-labeled P. gingivalis at the MOI of 1000 for 4 h. After quenching the fluorescence of bacteria bound on the cell surface with trypan blue, the fluorescence of HOK-16B cells containing intracellular bacteria was analyzed by flow cytometry. *P < 0.05 KUMC-P7, KUMC-P8, and KUMC-P10 have a significant difference compared with rest of strains.
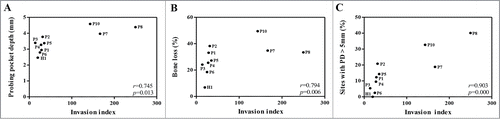
We examined whether the invasive abilities or proteolytic activities of the P. gingivalis clinical isolates have a relationship to any clinical parameters. Unexpectedly, the invasion index of P. gingivalis had strong positive correlations with the mean probing depth (PD, r = 0.745, p = 0.013), mean marginal bone loss (r = 0.794, p = 0.006), and % of sites with PD greater than 5 mm (r = 0.903, P < 0.0005) (), but not with those of the site from which P. gingivalis was isolated. These clinical parameters were not associated with degradation of any cytokine.
Figure 3. Strong positive correlations between clinical parameters of 10 subjects and the invasive ability of P. gingivalis strains. Two-tailed Spearman's rho correlations of the invasive ability of P. gingivalis with 3 clinical parameters (PD, marginal bone loss, and sites with PD > 5 mm) of subjects are shown.
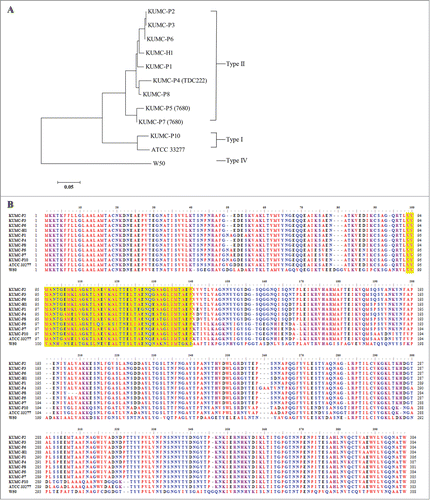
Because fimA–disrupted mutants have been shown to lose almost all of their invasive ability,Citation26 we analyzed the entire coding sequence of the fimA gene to determine an underlying mechanism for the heterogenic invasive ability of P. gingivalis. A sequence similarity search revealed that KUMC-P10 has a type I genotype, while all the other strains have type II. The translated sequences of KUMC-P4 and KUMC-P7 fimA genes were identical to those of TDC222 and 7680 strains in the database, respectively. KUMC-P2 and KUMC-P3 had identical FimA sequences. Far from our expectations, the strains with high invasive ability (KUMC-P7, KUMC-P8, and KUMC-P10) were not clustered together in the phylogenetic tree (). In the aligned FimA sequences, we could not identify any area where the amino acid sequences were shared by KUMC-P7, KUMC-P8, and KUMC-P10 but not by the others ().
Figure 4. Analysis of P. gingivalis fimA sequences. Using the genomic DNA from 10 P. gingivalis clinical isolates, the entire coding area of fimA gene was amplified, sequenced, and translated into amino acid sequences. (A) A phylogenetic tree was constructed using the translated FimA sequences of 10 clinical isolates and 2 laboratory strains from the database by the neighbor-joining method. The scale bar represents genetic distance. (B) The translated amino acid sequences were aligned and yellow shaded area represents the epithelial binding domain of P. gingivalis fimbriae.Citation40
Discussion
In this study, we investigated the invasion ability and cytokine degradation by various P. gingivalis clinical isolates in order to identify a strain with a low virulence. Substantial inter-strain variability was observed both in the cytokine degradation and invasiveness, as reported by other groups.Citation20,25,27-29 Although a positive correlation of invasive efficiency with inflammatory parameters determined in the mouse abscess model has been reported,Citation20 a strong correlation between the invasion efficiency and the clinical indexes of patients who harbor the P. gingivalis strain is a novel and unexpected finding. Considering the ecological complexity of subgingival biofilm, the fact that a single virulence parameter of one species reflects disease severity is unexpected. It has been reported that P. gingivalis degrades paxillin and focal adhesion kinase after invasion into epithelial cells, which inhibits cellular migration and proliferation.Citation30 Therefore, one possible explanation is that microulceration and delayed wound healing caused by invasive P. gingivalis may contribute to the induction of changes in subgingival microbial composition, which in turn may contribute to the progression of periodontitis. This explanation is in line with the recent concept of the ‘keystone pathogen’ that low abundance pathogens such as P. gingivalis can induce dysbiosis of normally benign microbiota.Citation31
Another controversial issue is in regards to the clonality of P. gingivalis within an individual. The detection of intra-individual clonal diversity by multilocus sequence typing changed the general concept that patients are colonized with a single strain of P. gingivalis.Citation32 A sensitive culture-independent method has detected P. gingivalis in the healthy sites of patients as well as in the diseased sites. It has been proposed that the virulence of P. gingivalis clones found in the healthy sites is different from those found in the diseased sites.Citation32 In our study, however, the invasive ability of P. gingivalis did not correlate with the clinical indexes of the sites from which P. gingivalis were isolated, but did correlate with the mean indexes. This contradicts the hypothesis that the disease development is determined by the heterogenic pathogenicity of P. gingivalis within a patient. There is a possibility that multiple clones detected within an individual are genetically related to each other in terms of pathogenicity. Otherwise, the multiple clones may be genetically diverse, but functionally similar; the Human Microbiome Project identified ubiquitous functional pathways among individuals despite the high degree of inter-individual variability in microbial taxa.Citation33 The variable amounts of subgingival biofilm and P. gingivalis, rather than heterogenic pathogenicity of P. gingivalis, may be an important determinant of periodontal status within a patient. The fact that P. gingivalis was sampled from a single site was a clear limitation of the study to extrapolate results on a patient level. Further studies using isolates from the multiple sites of each individual are warranted to clarify this issue.
Aside from the invasive ability, the ability of P. gingivalis to degrade cytokines did not show an association with any clinical parameters. However, the pathogenic heterogeneity of P. gingivalis determined in the mouse abscess model had a positive correlation with the activity of secreted Kgp gingipains.Citation20 This discrepancy may be attributed to the fact that the bacteria were subcutaneously injected in the mouse abscess model, bypassing the need of tissue invasion. In addition, proteases other than Kgp gingipains may be involved in cytokine degradation. The degradation of TNFα, IFNγ, and IL-4 by Rgp gingipain has been reported.Citation6,10,34 Another interesting finding was the nearly complete degradation of IFNγ and IL-17A along with the relative resistance of TNFα and IL-1α to degradation by P. gingivalis. The lower rate of IL-1β degradation compared to that of IL-6 has been also reported.Citation8 The degradation of Th1 and Th17 cytokines may contribute to the persistency of P. gingivalis infections, while the protection of inflammatory cytokines may contribute to the induction of chronic inflammation in gingival tissue. It is noteworthy similar resistance of IL-1α to degradation by T. denticola.Citation35
The analysis of the fimA gene sequences revealed that the majority of P. gingivalis isolated from patients have type II fimA genotype, which coincides with a previous report.Citation12 However, the type II fimA genotype was not associated with a high invasive ability of P. gingivalis. This fact also coincides with a previous report that the fimA genotypes of P. gingivalis are not related to the adhesion and invasion abilities.Citation29 We failed to identify a determinant of the high invasion ability within the primary sequence of FimA; this suggests that the invasion ability of P. gingivalis might be determined by the 3-D structure of FimA. Gingipains are also known to be involved in the adhesion/invasion of P. gingivalis through maturation of fimbriae and direct interaction with epithelial cells via the adhesin domain of gingipains.Citation36,37
KUMC-H1, a clone that was isolated from the healthy subject, had the second lowest invasion ability. However, the relatively young age (41 year) of H1 among the subjects in the study may have contributed to the healthy status. KUMC-P1 is an interesting clone in terms of its both low invasion and low proteolytic abilities. The P1 patient, who was 64 years old, showed relatively shallow PD and low percentage of PD greater than 5 mm. A limitation of the current study is a small sample size to adjust for other risk factors, such as age, smoking, and oral hygiene. Compared to the traditional invasion assay (based on colony-forming unit counts), the flow cytometric invasion assay is easy and fast. Future studies involving larger sample sizes and the adjustment of other risk factors are required to assess the true risk of P. gingivalis invasive ability in periodontal health status.
In conclusion, the invasion of epithelial cells but not the degradation of cytokines by P. gingivalis was associated with clinical parameters of periodontitis, which indicates that the invasive ability of P. gingivalis may be an important virulence factor. In addition, the bacterial invasion step may provide a good target for new therapeutics of periodontitis.
Materials and Methods
Study subjects
A total of 10 adult subjects who sought dental treatment at the Anam Hospital of Korea University were recruited for the study. The study protocol was approved by the Institutional Review Board for Human Subjects of the Korea University Anam Hospital (IRB No. ED10053). Written informed consent was obtained from all individuals. Exclusion criteria were pregnancy, diabetes, and other systemic conditions that could affect the periodontal status. PD was measured at 6 sites per each tooth. The amount of marginal alveolar bone loss was estimated as the percentage of the length from the cement-enamel junction to the alveolar crest relative to the length of the root on panoramic view. One periodontally healthy subject exhibited no sites with PD > 4 mm and no marginal bone loss > 20%. The nine subjects with periodontitis exhibited at least 2 or more sites with PD ≥ 5 mm and 2 or more teeth with marginal bone loss > 20% in one jaw.
Isolation and identification of P. gingivalis from clinical plaque samples
To isolate P. gingivalis, subgingival plaque samples were collected with a sterile paper point (Caulk-Dentsply) from the healthy site of a healthy subject and the diseased sites of periodontitis patients. The paper point was then placed in a sterilized dental transport medium. The plaque dispersed in the transport medium was streaked onto sheep blood agar with hemin and menadione (Sigma–Aldrich). The plates were incubated under an anaerobic atmosphere (10% CO2, 5% H2, and 85% N2). After incubation, black-pigmented colonies were selected and anaerobically grown in a brain heart infusion broth (BD Diagnostic Systems) that was supplemented with 5 μg/ml hemin and 5 μg/ml menadione. The genomic DNA from cultured bacteria was used for PCR with 16S rRNA-based P. gingivalis-specific primers.Citation38 After confirming the P. gingivalis 16S rRNA gene sequence, bacterial stocks were stored in liquid nitrogen until further use. P. gingivalis ATCC 33277 (American Type Culture Collection), W50 (American Type Culture Collection), and 10 clinical strains of P. gingivalis were used and cultured under the same conditions.
Degradation of cytokines by P. gingivalis and ELISA
To examine the degradation of cytokines by P. gingivalis, a cytokine mixture of IL-4, IL-6, IL-10, IL-17A, TNFα, and IFNγ (Peprotech) were prepared in keratinocyte growth medium containing supplementary factors (KGM; Clonetics) without antibiotics at 33 pM (equivalent to 500 pg/ml of IL-4) and added to 24-well plates. Viable P. gingivalis (1 × 107cells/well) strains were added to a mixture of recombinant cytokines and incubated at 37°C for 1 h. After centrifugation, the residual cytokine concentration in the supernatants were determined using ELISA kits (R&D Systems) according to the manufacturer's instructions and were compared to that of the basal levels of controls, which were incubated with KGM alone. The degradation of IL-1α recombinant protein (500 pg/ml) (R&D Systems) was performed separately in a similar manner.
Cell culture and flow cytometric invasion assay
Immortalized human gingival keratinocyte HOK-16B cells were maintained in KGM. A flow cytometric invasion assay was performed as described previously.Citation39 HOK-16B cells were infected with 5- (and 6-) carboxy-fluorescein diacetate succinimidyl ester (CFSE; Molecular Probe)-labeled bacteria at a multiplicity of infection (MOI) of 1000 in KGM without antibiotics for 4 h. Infected HOK-16B cells were washed with PBS and detached with trypsin-EDTA. After quenching the fluorescence of the bacteria bound on the surface with 0.4% trypan blue, the cells were analyzed using a FACSCalibur (BD Bioscience). For the negative controls, cells fixed with 3.7% formaldehyde were also exposed to the same amount of CFSE-labeled bacteria. In the flow cytometric invasion assay, the fluorescence intensity of host cells with invading bacteria is affected by the fluorescence intensity of labeled bacteria. The CFSE-labeling efficiency slightly varied from experiment to experiment, depending on the strains. To compare the invasive ability of different strains, the invasion index was calculated as follows: [mean fluorescence intensity (MFI) of infected cells-MFI of negative control cells]/MFI of CFSE-labeled bacteria*100.
fimA sequence analysis
To determine the fimA gene sequences of P. gingivalis clinical isolates, the fimA genes were amplified by PCR using fimA-universal primers as previously described,Citation38 and the products were then sequenced (Cosmo Genetech). The obtained sequences were BLAST searched (http://blast.ncbi.nlm.nih.gov/Blast.cgi) against the database to find the closest fimA gene; and were translated using public software (http://www.fr33.net/). In addition, the translated amino acid sequences were aligned using the CLUSTAL W algorithm in the MEGA 6.0 (http://www.megasoftware.net/) and subjected to phylogenetic analysis by applying neighbor-joining methods in the MEGA 6.0. The fimA gene sequences were registered in GenBank (GenBank accession numbers: KM987010∼987019).
Statistical analysis
All data were expressed as the mean plus standard error of the mean (SEM) of the 3 experiments. Because the measured values were not normally distributed, nonparametric statistical methods were used. Inter-strain differences were analyzed by Kruskal-Wallis with the post-hoc tests. The correlations between the clinical parameters of 10 subjects and the invasive ability or proteolytic activity of P. gingivalis clinical isolates were determined by Spearman's rank correlation coefficient. All analyses were performed by 2-tailed tests using SPSS18.0 (SPSS Inc.). Data were considered statistically significant at P value of <0 .05.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This study was supported by the National Research Foundation of Korea (Daejun, Korea) Grant 2014050477 funded by the Korean Government through the Oromaxillofacial Dysfunction Research Center for the Elderly at Seoul National University and a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare (HI13C0016), Republic of Korea.
References
- Lamont RJ, Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med 2014; pii: S1471-4914(14):00196-8; PMID:25498392; http://dx.doi.org/10.1016/j.molmed.2014.11.004
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. Microbial complexes in subgingival plaque. J Clin Periodontol 1998; 25: 134-44. PMID:9495612; http://dx.doi.org/10.1111/j.1600-051X.1998.tb02419.x
- Holt SC, Kesavalu L, Walker S, Genco CA. Virulence factors of Porphyromonas gingivalis. Periodontol 2000 1999; 20:168-238; PMID:10522227; http://dx.doi.org/10.1111/j.1600-0757.1999.tb00162.x
- Imamura T. The role of gingipains in the pathogenesis of periodontal disease. J Periodontol 2003; 74:111-8; PMID:12593605; http://dx.doi.org/10.1902/jop.2003.74.1.111
- Potempa J, Sroka A, Imamura T, Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr Protein Pept Sci 2003; 4:397-407; PMID:14683426; http://dx.doi.org/10.2174/1389203033487036
- Calkins CC, Platt K, Potempa J, Travis J. Inactivation of tumor necrosis factor-alpha by proteinases (gingipains) from the periodontal pathogen, Porphyromonas gingivalis. Implications of immune evasion. J Biol Chem 1998; 273:6611-4; PMID:9506956; http://dx.doi.org/10.1074/jbc.273.12.6611
- Mikolajczyk-Pawlinska J, Travis J, Potempa J. Modulation of interleukin-8 activity by gingipains from Porphyromonas gingivalis: implications for pathogenicity of periodontal disease. FEBS Lett 1998; 440:282-6; PMID:9872387; http://dx.doi.org/10.1016/S0014-5793(98)01461-6
- Stathopoulou PG, Benakanakere MR, Galicia JC, Kinane DF. The host cytokine response to Porphyromonas gingivalis is modified by gingipains. Oral Microbiol Immunol 2009; 24:11-7; PMID:19121064; http://dx.doi.org/10.1111/j.1399-302X.2008.00467.x
- Tam V, O'Brien-Simpson NM, Chen YY, Sanderson CJ, Kinnear B, Reynolds EC. The RgpA-Kgp proteinase-adhesin complexes of Porphyromonas gingivalis inactivate the Th2 cytokines interleukin-4 and interleukin-5. Infect Immun 2009; 77:1451-8; PMID:19168731; http://dx.doi.org/10.1128/IAI.01377-08
- Yun LW, Decarlo AA, Collyer C, Hunter N. Enhancement of Th2 pathways and direct activation of B cells by the gingipains of Porphyromonas gingivalis. Clin Exp Immunol 2003; 134:295-302; PMID:14616790; http://dx.doi.org/10.1046/j.1365-2249.2003.02287.x
- Enersen M, Nakano K, Amano A. Porphyromonas gingivalis fimbriae. J Oral Microbiol 2013; 5; PMID:23667717; http://dx.doi.org/10.3402/jom.v5i0.20265
- Amano A, Kuboniwa M, Nakagawa I, Akiyama S, Morisaki I, Hamada S. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J Dent Res 2000; 79:1664-8; PMID:11023261; http://dx.doi.org/10.1177/00220345000790090501
- Nakagawa I, Amano A, Ohara-Nemoto Y, Endoh N, Morisaki I, Kimura S, Kawabata S, Hamada S. Identification of a new variant of fimA gene of Porphyromonas gingivalis and its distribution in adults and disabled populations with periodontitis. J Periodontal Res 2002; 37:425-32; PMID:12472836; http://dx.doi.org/10.1034/j.1600-0765.2002.01637.x
- Enersen M, Olsen I, Kvalheim Ø, Caugant DA. fimA genotypes and multilocus sequence types of Porphyromonas gingivalis from patients with periodontitis. J Clin Microbiol 2008; 46:31-42; PMID:17977992; http://dx.doi.org/10.1128/JCM.00986-07
- Amornchat C, Rassameemasmaung S, Sripairojthikoon W, Swasdison S. Invasion of Porphyromonas gingivalis into human gingival fibroblasts in vitro. J Int Acad Periodontol 2003; 5:98-105; PMID:14604058
- Deshpande RG, Khan MB, Genco CA. Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis. Infect Immun 1998; 66:5337-43; PMID: 9784541
- Lamont RJ, Chan A, Belton CM, Izutsu KT, Vasel D, Weinberg A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun 1995; 63:3878-85; PMID:7558295
- Choi YS, Kim YC, Ji S, Choi Y. Increased bacterial invasion and differential expression of tight-junction proteins, growth factors, and growth factor receptors in periodontal lesions. J Periodontol 2014; 85:e313-22; PMID:24527855; http://dx.doi.org/10.1902/jop.2014.130740
- Kim YC, Ko Y, Hong SD, Kim KY, Lee YH, Chae C, Choi Y. Presence of Porphyromonas gingivalis and plasma cell dominance in gingival tissues with periodontitis. Oral Dis 2010; 16:375-81; PMID:20233323; http://dx.doi.org/10.1111/j.1601-0825.2009.01649.x
- Inaba H, Nakano K, Kato T, Nomura R, Kawai S, Kuboniwa M, Ishihara K, Ooshima T, Amano A. Heterogenic virulence and related factors among clinical isolates of Porphyromonas gingivalis with type II fimbriae. Oral Microbiol Immunol 2008; 23:29-35; PMID:18173795; http://dx.doi.org/10.1111/j.1399-302X.2007.00386.x
- Vivekananda MR, Vandana KL, Bhat KG. Effect of the probiotic Lactobacilli reuteri (Prodentis) in the management of periodontal disease: a preliminary randomized clinical trial. J Oral Microbiol 2010; 2:PMID:21523225; http://dx.doi.org/10.3402/jom.v2i0.5344
- Teughels W, Durukan A, Ozcelik O, Pauwels M, Quirynen M, Haytac MC. Clinical and microbiological effects of Lactobacillus reuteri probiotics in the treatment of chronic periodontitis: a randomized placebo-controlled study. J Clin Periodontol 2013; 40:1025-35; PMID:24164569; http://dx.doi.org/10.1111/jcpe.12155
- Wan YY, Flavell RA. How diverse–CD4 effector T cells and their functions. J Mol Cell Biol 2009; 1:20-36; PMID:19482777; http://dx.doi.org/10.1093/jmcb/mjp001
- Gaffen SL, Hajishengallis G. A new inflammatory cytokine on the block: re-thinking periodontal disease and the Th1/Th2 paradigm in the context of Th17 cells and IL-17. J Dent Res 2008; 87:817-28; PMID:18719207; http://dx.doi.org/10.1177/154405910808700908
- Dorn BR, Burks JN, Seifert KN, Progulske-Fox A. Invasion of endothelial and epithelial cells by strains of Porphyromonas gingivalis. FEMS Microbiol Lett 2000; 187:139-44; PMID:10856647
- Kato T, Kawai S, Nakano K, Inaba H, Kuboniwa M, Nakagawa I, Tsuda K, Omori H, Ooshima T, Yoshimori T, Amano A. Virulence of Porphyromonas gingivalis is altered by substitution of fimbria gene with different genotype. Cell Microbiol 2007; 9:753-65; PMID:17081195; http://dx.doi.org/10.1111/j.1462-5822.2006.00825.x
- Dolgilevich S, Rafferty B, Luchinskaya D, Kozarov E. Genomic comparison of invasive and rare non-invasive strains reveals Porphyromonas gingivalis genetic polymorphisms. J Oral Microbiol 2011; 3; PMID:21541093; http://dx.doi.org/10.3402/jom.v3i0.5764
- Eick S, Rödel J, Einax JW, Pfister W. Interaction of Porphyromonas gingivalis with KB cells: comparison of different clinical isolates. Oral Microbiol Immunol 2002; 17:201-8; PMID:12121469; http://dx.doi.org/10.1034/j.1399-302X.2002.170401.x
- Umeda JE, Missailidis C, Longo PL, Anzai D, Wikström M, Mayer MP. Adhesion and invasion to epithelial cells by fimA genotypes of Porphyromonas gingivalis. Oral Microbiol Immunol 2006; 21:415-9; PMID:17064402; http://dx.doi.org/10.1111/j.1399-302X.2006.00312.x
- Nakagawa I, Inaba H, Yamamura T, Kato T, Kawai S, Ooshima T, Amano A. Invasion of epithelial cells and proteolysis of cellular focal adhesion components by distinct types of Porphyromonas gingivalis fimbriae. Infect Immun 2006; 74:3773-82; PMID:16790749; http://dx.doi.org/10.1128/IAI.01902-05
- Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol 2012; 10:717-25; PMID: 22941505; http://dx.doi.org/10.1038/nrmicro2873
- Enersen M. Porphyromonas gingivalis: a clonal pathogen?: Diversities in housekeeping genes and the major fimbriae gene. J Oral Microbiol 2011; 3; PMID:22125739; http://dx.doi.org/10.3402/jom.v3i0.8487
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012; 486:207-14; PMID:22699609; http://dx.doi.org/10.1038/nature11234
- Yun PL, DeCarlo AA, Hunter N. Modulation of major histocompatibility complex protein expression by human gamma interferon mediated by cysteine proteinase-adhesin polyproteins of Porphyromonas gingivalis. Infect Immun 1999; 67:2986-95; PMID:10338509
- Jo AR, Baek KJ, Shin JE, Choi Y. Mechanisms of IL-8 suppression by Treponema denticola in gingival epithelial cells. Immunol Cell Biol 2014; 92:139-47; PMID:24296811; http://dx.doi.org/10.1038/icb.2013.80
- Chen T, Duncan MJ. Gingipain adhesin domains mediate Porphyromonas gingivalis adherence to epithelial cells. Microb Pathog 2004; 36:205-9; PMID:15001226; http://dx.doi.org/10.1016/j.micpath.2003.12.001
- Kato T, Tsuda T, Omori H, Kato T, Yoshimori T, Amano A. Maturation of fimbria precursor protein by exogenous gingipains in Porphyromonas gingivalis gingipain-null mutant. FEMS Microbiol Lett 2007; 273:96-102; PMID:17559394; http://dx.doi.org/10.1111/j.1574-6968.2007.00779.x
- Baek KJ, Choi Y, Ji S. Gingival fibroblasts from periodontitis patients exhibit inflammatory characteristics in vitro. Arch Oral Biol 2013; 58:1282-92; PMID:24011303; http://dx.doi.org/10.1016/j.archoralbio.2013.07.007
- Ji S, Shin JE, Kim YS, Oh JE, Min BM, Choi Y. Toll-like receptor 2 and NALP2 mediate induction of human beta-defensins by Fusobacterium nucleatum in gingival epithelial cells. Infect Immun 2009; 77:1044-52; PMID:19103770; http://dx.doi.org/10.1128/IAI.00449-08
- Sojar HT, Han Y, Hamada N, Sharma A, Genco RJ. Role of the amino-terminal region of Porphyromonas gingivalis fimbriae in adherence to epithelial cells. Infect Immun 1999; 67:6173-6; PMID:10531284
