Abstract
Invasive syndrome caused by Klebsiella pneumoniae (KP), including liver abscess, is mainly caused by community-acquired strains with characteristics of positive hypermucoviscosity (HV) phenotype and regulator of mucoid phenotype A (rmpA) and transcriptional activator (rmpA2) genes. Extended- spectrum β-lactamase-producing KP (ESBL-KP) is commonly nosocomial and rarely HV-positive. We aimed to explore the reasons of the rarer prevalence of HV phenotype, rmpA and rmpA2 as well as the virulence phenotype among the ESBL-KP isolates from clinical specimens than those non-ESBL isolates. The β-lactamase genes, rmpA, rmpA2 and genes for K capsule serotype of 440 KP isolates were analyzed. The virulence of the isolates was characterized by the mouse lethality experiments. The prevalence rates of HV phenotype (∼50% vs. < 10%) as well as rmpA and rmpA2 genes (∼50–60% vs. < 20–30%) were significantly higher in non-ESBL group than in the ESBL group (p < 0.0001). Expression of HV phenotype in the rmpA-positive KP isolates was significantly rarer in the ESBL group than in non-ESBL group (33.3% vs. 91.9%, p < 0.0001). The frameshift mutations of rmpA and/or rmpA2 corresponded to negative HV phenotype of KP isolates that harbored the rmpA and/or rmpA2, resulting in variable mouse lethality (LD50, ∼103 - >5 × 107 CFU). The mutation rates might significantly differ among KP isolates from various sources. Virulence was dependent on rmpA-related HV phenotype. In conclusion, ESBL-KP isolates were less hypermucoviscous and less virulent than non-ESBL KP isolates, mostly due to concurrently lower carriage and higher mutation rates of the rmpA and rmpA2 genes.
Introduction
Klebsiella pneumoniae (KP) can cause various infections, like septicemia, pneumonia, urinary tract infection, meningitis, and tissue abscesses.Citation1,2 In Taiwan, primary liver abscess is mainly caused by KP with diverse virulence determinants, like K1 or K2 capsular serotype, hypermucoviscosity (HV) phenotype, as well as the plasmid-borne regulator of mucoid phenotype A (rmpA) and rmpA2 (a transcriptional activator) genes.Citation1-7 The HV phenotype was correlated with high serum resistance of KP.Citation3 Isolates with rmpA and HV phenotype were highly virulent in the mouse lethality tests.Citation7
We previously discovered the prevalence rates of HV phenotype and rmpA of 151 blood KP isolates were 38% and 48%, respectively.Citation1 Among them, 52 (90%) of 58 HV-positive isolates possessed rmpA, whereas, 20 (21.5%) of 93 HV-negative isolates carried rmpA. That is, 52 (72%) of 72 rmpA-positive isolates expressed HV phenotype. The association between rmpA and HV phenotype of KP was highly significant (p < 0.0001), however, we could not exactly explain the reasons why some (28%) rmpA-positive isolates did not exhibit the HV phenotype.Citation1 Besides, Fang et al found that the rmpA was universally present in all tested strains, including invasive and non-invasive isolates.Citation3 Furthermore, Yeh et al. reported non-virulence of some rmpA-positive KP isolates from Singapore.Citation5 In similar, Lee et al. found 18% of 56 HV-negative KP isolates carrying rmpA or rmpA2.Citation8 From the above observation, it seems that some KP isolates did not fully express the function of the rmpA gene. Therefore, the relationship between the rmpA and rmpA2 genes and the exhibition of the HV phenotype of KP has remained a mystery.
Extended-spectrum β-lactamase-producing KP (ESBL-KP) is mainly nosocomial and has rarely been found to be associated with the HV phenotype. In Korea, ESBL-KP comprised 33.8% of hospital-acquired bacteremia and only 8.4% of community-acquired bacteremia.Citation9 Of 435 patients with community-onset KP bacteremia, 33 (7.6%) were infected with ESBL producers.Citation10 In Taiwan, ESBL is rarely identified in community-acquired KP infection and is not associated with primary KP liver abscess.Citation11 In addition, Lee et al. reported a negative association of the HV phenotype with ESBL-KP isolates (OR 0.042, p = 0.003).Citation8 In China, Li et al. found a significantly lower proportion of ESBL-KP in the HV-positive isolates than in the HV-negative isolates (17% vs. 56%, P = 0.0006). Li et al noticed an increasing trend of ESBL-KP among the hypermucoviscous isolates over time.Citation12 In a small series from Algiers, the virulence profiles of 54 KP isolates have been well elucidated.Citation13 The study comprised 74.1% isolates with multiresistance particularly to extended-spectrum cephalosporins, which can be due to ESBL production. Overall, 5 (9.2%) and 2 (3.7%) isolates had HV and rmpA, respectively. One rmpA-positive isolate did not have HV phenotype. From the above-mentioned studies, it remains unknown why the ESBL-KP isolates rarely exhibit the HV phenotype.
The goals of the study were to explain the difference in the prevalence of the HV-related virulence determinants between the KP isolates with and without the ESBL production and to explore the reason why some rmpA-positive KP strains were HV-negative. We hypothesized that low prevalence of rmpA and/or rmpA2 in ESBL-KP isolates contributed to their rare HV phenotype. Our additional hypothesis was that genetic mutation of rmpA and/or rmpA2 was associated with the negative HV phenotype in the rmpA- and/or rmpA2-positive KP isolates.
Results
Clinical isolates and microbiological characteristics
All investigated bacterial strains were recovered from clinical specimens of the patients hospitalized in Chi Mei Medical Center in Tainan City in Taiwan. These human samples were obtained as part of routine care on clinical indication. A total of 226 non-repetitive clinical isolates were collected from1 January 2004 to 31 December 2005. ESBL-KP was identified in 57 isolates (25.2%) from various specimens, including sputum (n = 24), urine (n = 18), blood (n = 10), ascites (n = 2), bile (n = 1), pleural fluid (n = 1), and bronchoalveolar lavage fluid (n = 1). The remaining 169 non-ESBL KP isolates included sputum (n = 105), blood (n = 39), urine (n = 8), abscess pus (n = 6), wound (n = 3), central venous catheter (CVC) tip (n = 2), ascites (n = 3), bile (n = 1), pleural effusion (n = 1) and pericardial effusion (n = 1). Furthermore, 166 ESBL-KP blood isolates were collected from 1 January 2007 to 31 July, 2010 and 48 non-ESBL KP blood isolates collected in 2010 (randomly 4 isolates per month) were also used for comparison as external validation.
The prevalence of HV phenotype among isolates
The HV phenotype was significantly lower in ESBL-KP isolates than that in non-ESBL KP isolates (8.8% vs. 53.8%. p < 0.0001,). Specifically, the HV-positive sputum isolates were more common in non-ESBL than that in ESBL group (59% vs 4.2%). However, the HV phenotype of urine isolates was significantly rarer than that of sputum and blood isolates within non-ESBL group. The HV phenotype of blood isolates (204–2005) was more common than non-blood isolates within ESBL-KP group (p = 0.033), but was not significantly different to non-ESBL blood isolates, albeit with a lower trend (30% vs 55%, p = 0.171). To reduce the selection bias from the small sample size of the ESBL-KP blood isolates (2004–2005, n = 10), therefore, we further collected 166 ESBL-KP blood isolates (2007–2010) and 48 non-ESBL KP blood isolates (2010). The difference in HV prevalence of the ESBL-KP blood isolates between 2 periods (2004–2005 vs 2007–2010) was not statistically significant. Again, the HV phenotype of ESBL-KP blood isolates (2007–2010) was more common than ESBL-KP non-blood isolates (2004–2005), but became significantly less common than non-ESBL KP blood isolates (2004–2005 and 2010, respectively,).
Table 1. Distribution of hypermucoviscosity (HV) phenotype between K. pneumoniae isolates with and without ESBLs
Moreover, 105 community-acquired blood KP isolates extracted from our previous study during 2003–2004,Citation1 which were almost non-ESBL isolates, showed similar prevalence of HV phenotype to 2004–2005 non-ESBL KP blood isolates (48.6% vs. 55%, p = 0.403), but were more HV-positive than ESBL-KP blood isolates obtained from 2007–2010 (48.6% vs. 22.3%, p < 0.0001,).
The prevalence of rmpA and rmpA2 genes among isolates
The rmpA was significantly lower in ESBL-KP isolates than that in non-ESBL KP isolates (21.1% vs. 58.6%. p < 0.0001,). Specifically, the rmpA-positive sputum isolates were more common in non-ESBL than ESBL group (p = 0.001). Similar to HV phenotype, the rmpA prevalence of ESBL-KP blood isolates (2007–2010) was significantly lower than the non-ESBL blood isolates (2004–2005 and 2010, respectively). Different to higher HV phenotype of blood isolates in ESBL group and rarer HV phenotype of urine isolates in non-ESBL group (), however, rmpA prevalence was not statistically different among various isolates within each group (). For further external validation, the rmpA prevalence of the community-acquired KP blood isolates (2003–2004)Citation1 was similar to non-ESBL blood isolates (2004–2005) but was higher than that of the ESBL-KP blood isolates (2007–2010,).
Table 2. Difference in distribution of rmpA and rmpA2 between K. pneumoniae isolates with and without ESBLs
The association of HV phenotype with rmpA and/or rmpA2
Overall, 96 of 226 KP isolates (2004–2005) were HV-positive. Of these, only 6 (KP331, KP 347, KP350, KP476, KP 495, KP511) did not harbor rmpA and/or rmpA2, as demonstrated by the inability to amplify the corresponding gene sequences by PCR. Meanwhile 100 of 130 HV-negative isolates did not harbor rmpA and/or rmpA2 (p < 0.0001). Similarly, 37 of 166 ESBK-KP blood isolates (2007–2010) were HV-positive. Of these, 10 did not harbor rmpA and/or rmpA2. Meanwhile 95 of 129 HV-negative isolates did not harbor rmpA and/or rmpA2 (p < 0.0001).
The prevalence of HV phenotype among rmpA-positive KP isolates
The HV prevalence of rmpA-positive KP isolates was significantly lower in the ESBL group than that in the non-ESBL group (33.3% vs. 91.9%. p < 0.0001,). Specifically, the rmpA- associated HV phenotype of sputum isolates was significantly rarer in the ESBL-KP group than non-ESBL group (p < 0.0001). No statistical difference of the HV prevalence was detected in the rmpA-positive ESBL-KP blood isolates between 2 periods (2004–2005 vs 2007–2010), but it was significantly higher in the non-ESBL groups (2004–2005 and 2010, respectively). Among the 99 rmpA-positive non-ESBL KP isolates, the HV prevalence of urine isolates was significantly lower than the sputum and blood isolates. Among the 12 rmpA-positive ESBL-KP isolates, the difference in the HV prevalence among various isolates was not statistically different. The blood isolates might have higher rate of HV phenotype, but did not reach statistical significance (p = 0.067).
Table 3. Proportion of hypermucoviscosity (HV) phenotype among rmpA-positive K. pneumoniae isolates with and without ESBLs from (implying expression rate or normal function of rmpA)
Genetic mutation in rmpA- and/or rmpA2-positive KP isolates
DNA sequencing of the rmpA and rmpA2 amplicons revealed the indel mutations (insertion or deletion of nucleotides) at poly(G) tract within rmpA gene as well as rmpA2 in all non-HV isolates but with rmpA and/or rmpA2 (; and ). Due to the triplet nature of gene expression by codons, a genetic mutation caused by indels of a number of nucleotides in a DNA sequence that is not divisible by 3 will result in a frameshift mutation and thereby a completely different translation from the original reading frame after the mutation to code for different amino acids. In similar, the truncated mutation occurring after a stop codon (TAA) in a reference rmpA2 sequence AB289643 will result in a truncated protein product ().
Table 4. The genotypes of rmpA and rmpA2 among K. pneumoniae without hypermucoviscosity phenotype
Figure 1. Alignment analysis of rmpA sequences. Multiple sequences are compared with the reference sequence of GenBank accession number AB289642, which defined plasmid pK2044 rmpA gene from the strain K. pneumoniae NTUH-K2044 (http://www.ncbi.nlm.nih.gov/nuccore/AB289642). The strains KP 309, 401, 420, and 425 represent isolates which were PCR-positive for rmpA and rmpA2 but negative for hypermucoviscosity phenotype. The strain KP291 represents isolates without hypermucoviscosity phenotype, which were PCR-positive for rmpA but PCR-negative for rmpA2. The strain KP449 represents isolates with hypermucoviscosity phenotype, which were PCR-positive for rmpA and rmpA2.

Figure 2. Alignment analysis of rmpA2 sequences. Multiple sequences are compared with reference sequences of GenBank accession numbers S64176 and AB289643. The S64176 defined plasmid rmpA2 gene from the strain K. pneumoniae, Chedid, O1:K2 serotype (http://www.ncbi.nlm.nih.gov/nuccore/S64176). The AB289643 revealed a truncated rmpA2 region and a nucleotide deletion in the poly-G tract of ORF KPP302 from the strain K. pneumoniae NTUH-K2044 (http://www.ncbi.nlm.nih.gov/nuccore/AB289643).

The frameshift mutations of rmpA and/or rmpA2 occurred in the HV-negative KP isolates that were PCR-positive for rmpA and/or rmpA2 genes (). In particular, among the 57 ESBL-KP isolates (2004–2005), K1 capsule serotype was found in 5 isolates, including 2 isolates with rmpA and rmpA2, 1 strain with rmpA but negative for rmpA2, and 2 isolates without rmpA and rmpA2. All 5 K1 isolates were HV-negative and the rmpA and rmpA2 were all mutated.
For HV phenotype-positive KP isolates, 10 isolates were randomly selected for sequencing analysis and the rmpA had not been mutated, but the rmpA2 had been mutated.
Genotypes and transference of ESBL genes
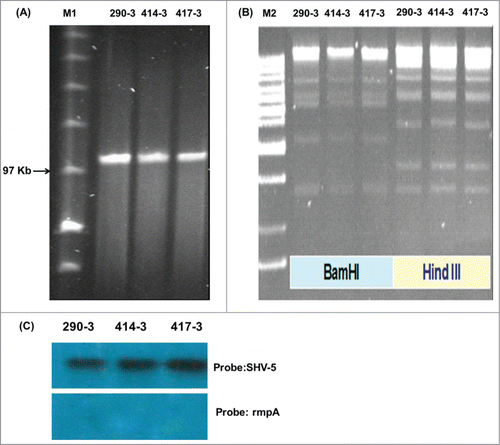
The 57 ESBL-KP isolates (2004 -2005) were further analyzed for the ESBL genes (). The identified ESBL types were predominately SHV-5, and then SHV-12 and CTX-M-3. Only 5 strains (4 producing SHV-5; 1 producing CTX-M-3 and SHV-11) exhibited HV phenotype. Conjugation experiments were performed for the 4 hypermucoviscous SHV-5-producers, of which 3 plasmids carrying blaSHV-5 (>97 kb) in 3 strains (KP290, KP414, KP417) were successfully conjugated into Escherichia coli J53–2 (). The plasmid DNA gel electrophoresis of the 3 transconjugational plasmids using either BamHI or Hind III restriction enzyme revealed that the 3 blaSHV-5-encoding plasmids were identical RFLP pattern (). Southern blotting hybridization confirmed location of the blaSHV-5 on the 3 transconjugational plasmids, but did not successfully hybrid rmpA gene in the plasmids of transconjugants (). Thus the blaSHV-5 and rmpA should be located in different plasmids. Moreover, PCR analysis for rmpA was positive in the above 3 ESBL-KP parent isolates, but was negative in their transconjugants.
Table 5. Distribution of ESBL types and virulence factors of the 57 ESBL-KP isolates from 2004 to 2005
Figure 3. DNA analysis for transconjugational plasmids of transconjugants 290–3, 414–3 and 417–3 by gel electrophoresis (A) without restriction enzyme (B) with restriction enzyme BamHI or Hind III. M1: MidRange Marker II (New England BioLabs, Ipswich, MA, USA); M2:1 kb DNA Ladder (Protech Technology Enterprise Co., Ltd, Taipei, Taiwan). Southern blotting for blaSHV-5 (C, upper) and rmpA (C, lower) on transconjugational plasmid of transconjugants was shown.
Virulence of KP isolates in the mouse lethality experiments
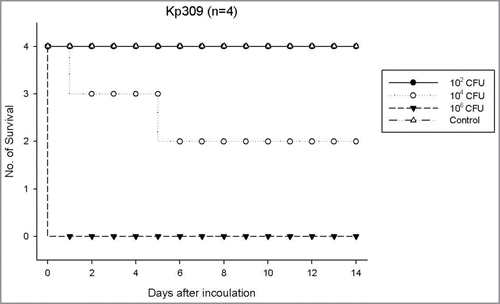
Twelve KP isolates (6 non-ESBL and 6 ESBL isolates) representative for capsule serotypes of K1, K2 and non-K1/K2 were selected for virulence tests (). Regardless of the capsule serotypes as well as ESBL or non-ESBL phenotype, in general, the isolates with HV phenotype exhibited the highest virulence for mouse lethality (LD50, ∼102- ∼ 103 CFU). The isolates without HV phenotype and negative for rmpA systems exhibited the lowest lethality (LD50, >5 × 107 CFU). The HV-negative isolates with mutated rmpA systems exhibited variable lethality (LD50, ∼103 - >5 × 107 CFU). For example, a smaller dose (102 CFU) of strain KP309 (non-HV rmpA mutant) could not cause mouse lethality, whereas a higher bacterial dose (105 -107 CFU) caused rapid mouse lethality on the next day of infection (). One exception of the above general rule was that a HV-positive ESBL strain (KP331) without harboring any rmpA system reduced its virulence to a LD50 of >5 × 107 CFU ().
Table 6. The influence of rmpA and/or rmpA2 mutation on the virulence (LD50) of KP isolates with various microbiologic characteristics
Discussion
In the present study, the prevalence of the HV phenotype was significantly lower in ESBL-KP isolates (8.8%) than that in non-ESBL KP isolates (53.8%). Specifically, the prevalence of HV phenotype in blood ESBL-KP isolates (20–30%) was lower than that in blood non-ESBL isolates (50–55%), but was higher than non-blood ESBL-KP isolates (4.2%). These findings might be applicable to the clinical scenario that ESBL-KP sputum and urine isolates are usually colonization. Whereas among non-ESBL KP isolates, the prevalence of HV phenotype of urine isolates (12.5%) was lower than that of the sputum (59%) and blood isolates. In clinical application, therefore, non-ESBL KP sputum isolates may be more pathogenic than ESBL-KP sputum isolates.
The prevalence of rmpA was significantly lower in ESBL-KP isolates (21.1%) than that in non-ESBL KP isolates (58.6%). This difference in rmpA prevalence was also validated in blood isolates between ESBL-KP and non-ESBL groups (or community-acquired isolates). However, the rmpA prevalence did not significantly differ among various clinical isolates either in ESBL-KP or in non-ESBL KP group, respectively. Yet, the HV phenotype was rarer in non-blood isolates among ESBL group and in urine isolates among non-ESBL group, respectively. These findings imply different rmpA expression rates leading to different HV prevalence among various clinical isolates.
In general for KP isolates, the absence of rmpA and rmpA2 (rmpA systems) correlated to negative HV phenotype. Nevertheless, it is out of context that some strains (6.3%) of HV-positive isolates were negative for rmpA systems, implying the presence of mucoid factors other than rmpA systems. This finding is similar to our previous report that 10% of HV-positive blood KP isolates did not harbor rmpA.Citation1 On the other hand, the association between HV phenotype and rmpA was significantly lower in the ESBL-KP isolates than non-ESBL KP isolates (implying rmpA expression rate, 33.3% vs. 91.9%. p < 0.0001). We proposed rmpA mutation responsive for the lower HV exhibition rate.
Overall, 64 (24.7%) of 259 HV-negative KP isolates were positive for rmpA system. Of these, all had frameshift mutation of rmpA systems. On the contrary, rmpA mutation was not found in the randomly selected HV-positive KP isolates. A frameshift mutation is not the same as a single-nucleotide polymorphism in which a nucleotide is replaced, rather than inserted or deleted. The earlier in the sequence the indel occurs, the more altered the protein. In general, the most common indels of rmpA mutants occurred at positions between 280–300 base areas (), about halfway on the rmpA complete codes (633 bases, GenBank accession number AB289642), indicating substantial alternations to the RmpA protein synthesis and leading to non-HV phenotype.
Therefore, the reasons of ESBL-KP isolates with low prevalence of HV phenotype were not only due to low prevalence of carrying rmpA systems, but also due to high prevalence of frameshift mutation in the rmpA systems. It could be speculated that ESBL-KP strains reduce genetic function of rmpA systems when they need to amplify ESBL genes under the antibiotic selective pressure. The mutation theory could also explain the discrepancy of HV phenotype exhibition in sputum isolates between ESBL-KP and non-ESBL KP (implying rmpA expression rate, 16.7% vs. 95.4%. p < 0.0001).
In fact, the spontaneous mutation of rmpA systems might not only be fortuitous. Lai et al. have found that some KP isolates produced frameshift mutation of rmpA2.Citation14 In addition, Cheng et al. also conceived an idea that DNA slip-strand synthesis in rmpA or rmpA2 may cause mutation and result in abnormal function of the 2 regulatory genes.Citation15 Together with our current results support our hypothesis that mutation of rmpA systems contributes to the loss of the HV phenotype in the KP isolates positive for rmpA systems.
In general view of the current study, ESBL-KP or non-ESBL KP isolates positive for both rmpA and HV phenotype corresponded to high virulence to mouse lethality. In contrast, non-HV isolates with negative rmpA systems exhibited low virulence to lethality. Unexpectedly, a rare instance of HV-positive isolate without rmpA systems exhibited low virulence (eg., KP331,), indicating that virulence is dependent on rmpA systems-related HV phenotype rather than other mucoid factors-mediated HV phenotype.
The HV phenotype with high serum resistance has been recognized as a virulence factor for KP.Citation1,2 Besides, the role of rmpA system in the HV-associated pathogenesis have previously been studied. However, different methods of the laboratory-constructed deletion of rmpA and/or A2 sometimes resulted in conflicting and confusing discrepancies, while interpreting the capability of the mutant strains in expressing the capsular polysaccharide production, hypermucoviscosity phenotype and virulence.Citation4,14–17 For examples, Cheng et al. demonstrated rmpA as a major mucoid factor because introducing rmpA-carrying plasmids in to the rmpA or rmpA2 mutants restored capsular polysaccharide production.Citation15 Hsu et al. demonstrated that plasmid-borne rmpA enhanced capsular polysaccharide synthesis, capsule production and mucoviscosity.Citation16 But the virulence of the rmpA mutant was not significantly reduced, with a LD50 of < 1 × 102 CFU.Citation16 Thus other factors like capsular types than rmpA per se might play some roles in virulence. However, in our current study, even for isolates with the most virulent K1 or K2 capsular type, concurrent mutation of rmpA and rmpA2 could lose the HV phenotype and then reduce virulence, with a LD50 of >5 × 107 CFU. We thought that the laboratory- constructed rmpA model might not fit to the true scenario of natural mutation in rmpA system.
In similar to the reports by Hsu et al.Citation16 and our previous study,Citation7 the LD50 of some HV-negative strains but with mutated rmpA systems ranged approximately between 103 to 104 CFU (eg., KP309,). It could be speculated that other virulence determinants on the same rmpA-encoded plasmid partly contributed to the virulence. The other virulence determinants than rmpA systems might be associated with the iron acquisition system, like aerobactin and kfu genes.Citation6,7,18 A high proportion of ESBL-KP or multidrug-resistant KP isolates were found with virulence factors like adhesins, siderophores, serum resistance, and hemolysin.Citation13,19 On the other hand, the LD50 of some HV-negative KP isolates with mutated rmpA systems were >5 × 107 (eg., KP291 and KP420,). It could be suggestive of absence of other co-existing virulence determinants on the same plasmid.
Only 3 HV-positive isolates harboring blaSHV-5 were selected for conjugation experiments and the blaSHV-5 and rmpA should be located on different plasmids, based on the different transference ability of their harboring plasmids, which was evidenced by Southern blotting and PCR analysis for rmpA (non-transferable) and blaSHV-5 (self-transferable) in the parent isolates and their transconjugants.
Conclusion
To our best knowledge, this is the first large-scale report that provided the in vivo evidence for the naturally-occurring rmpA and/or rmpA2 frameshift mutation in the HV-negative KP isolates that harbor rmpA and/or rmpA2 genes. Overall, ESBL genes and rmpA were located on different plasmids. Compared to non-ESBL KP isolates, the ESBL-KP isolates had generally lower prevalence of HV phenotype, lower carriage rate of rmpA, lower expression rate of rmpA, and higher mutation rate of rmpA. Frameshift mutation of rmpA corresponded to diminished exhibition of HV phenotype among KP isolates. Virulence of either ESBL-KP or non-ESBL KP to mouse lethality is dependent on rmpA-related HV phenotype. The limitation of the study included small numbers of urine isolates overall.
Materials and Methods
Clinical Isolates and Microbiological assays
The KP was identified by Phoenix system (Becton Dickinson Company, Baltimore, MD, USA) and API 20E system (bioMerieux, Marcy l’Etoile, France). The hypermucoviscosity phenotype was defined positive as a viscous string of > 5 mm of the colony on trypticase soy agar plate with 5% sheep blood (BD Diagnostics, MD, USA).Citation1,6 ESBL production was screened and confirmed by the double disc test using cefotaxime and ceftazidime along with an amoxicillin-clavulanate disc, in accordance with Clinical and Laboratory Standards Institute (CLSI) standards .20 Quality control was performed by testing Escherichia coli American Type Culture Collection (ATCC) 25922, K. pneumoniae ATCC 700603 and Pseudomonas aeruginosa ATCC 27853.
DNA manipulation, PCR amplification and sequencing
The genome of NTUH-K2044 (http://genome.nhri.org.tw/KP/) carries 3 different copies of rmpA: 2 (open reading frames [ORFs] KPP020 and KPP302) are on the 224-kb large plasmid pK2044, and the other (ORF KP3619) is on the chromosome.Citation14,21 In fact, the ORFs KPP020 and KPP302 were referred to rmpA and rmpA2, respectively, in previously published literature. Since the plasmid-borne rmpA copies (rmpA and rmpA2) have been well-known important virulent determinants,Citation1,4,6–8,14–16 we focused on the analysis of rmpA (ORF KPP020) and rmpA2 (ORF KPP302) in the current study.
Genomic DNA was extracted using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) for screening the genes for K capsule serotype and plasmid DNA was also extracted by QIAprep Spin Miniprep (Qiagen, Hilden, Germany) for detection of rmpA (ORF KPP020) and rmpA2 genes (ORF KPP302). GoTaq® DNA Polymerase (Promega Corporation, Madison, WI, USA) containing bacterially derived Tag DNA polymerase, dNTPs and MgCl2 in 2X Green GoTaq® Reaction Buffer (pH 8.5) was used for amplification of DNA templates by PCR.
The rmpA, rmpA2 and genes for K capsule genotypes were identified by PCR using specific primers as previous described and PCR products were confirmed by DNA sequencing. The primers for rmpA (forward, 5′- TAC ATA TGA AGG AGT AGT TAA T-3′; and reverse, 5′-GAG CCA TCT TTC ATC AAC-3') as well as forward, 5′-ACT GGG CTA CCT CTG CTT CA-3′; and reverse, 5′-CTT GCA TGA GCC ATC TTT CA-3′ were used to amplify rmpA (ORF KPP020) gene.Citation1 The rmpA2-specific primers were: forward, 5′-TGT GCA ATA AGG ATG TTA CAT TAG T-3′; and reverse, 5′-TTT GAT GTG CAC CAT TTT TCA-3′. The primers for magA (forward, 5′-GGT GCT CTT TAC ATC ATT GC-3′; and reverse, 5′-GCA ATG GCC ATT TGC GTT AG-3′) was used to amplify wzyKpK1, a capsule serotype K1-antigen–specific polymerase.Citation3,22 The primers for k2A (forward, 5′-CAA CCA TGG TGG TCG ATT AG-3′; and reverse, 5′-TGG TAG CCA TAT CCC TTT GG-3′) was used specifically to identify isolates with a capsule serotype K2.Citation23
The rmpA and rmpA2 amplicons were recovered for further sequencing. All amplicons were purified with PCR clean up kits (Roche Diagnostics, GmbH, Penzberg, Germany) and were ligated into pGEM-T-Easy vector (Promega Co., Madison, Wisconsin, USA) for further sequencing on an ABI PRISM 3730 sequencer analyzer (Applied Biosystems, Foster City, CA, USA). Sequence analyses were performed online at the Basic Local Alignment Search Tool (BLAST) website.Citation24 Multiple sequences alignment analysis was conducted by using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
PCR amplification for β-lactamase genes
Plasmid DNA was extracted as templates, PCR was used to amplify blaTEM, blaSHV, blaCTX-M and blaKPC using specific primers as previously published as follows: The blaTEM-specific primes were: TEM-forward (5′-ATAAAATTCTTGAAGACGAAA-3′) and TEM-reverse (5′-GACAGTTACCAATGCTTAATC-3′).Citation25 The blaSHV-specific primers were: SHV-forward (5′-GATCCACTATCGCCAGCAGG-3′) and SHV-reverse (5′-ACCACAATGCGCTCTGCTTTG-3′); SHV-12-forward (5′-ATG CGT TAT ATT CGC CTG TG-3') and SHV-12-reverse (5'-TTA GCG TTG CCA GTG CTC G-3').Citation25 The blaCTX-M-specific primers were: CTX-M-forward (5'-TGT TGT TAG GAA GTG TGC CGC-3'), CTX-M-reverse (5'-TCG TTG GTG GTG CCA TAG TC-3').26The blaKPC-specific primers were: KPC-2-forward (5'-ATG TCA CTG TAT CGC CGT CTA-3') and KPC-2-reverse (TTA CTG CCC GTT GAC GCC CA-3').Citation27
Conjugation experiments, plasmid DNA analysis and Southern hybridization
Conjugation of plasmid was performed by using the sodium azide-resistant E. coli J53–2 as the recipient. The transconjugants were selected on Luria-Bertani agar plates supplemented by sodium azide (100 μg/mL) and cefotaxime (2 μg/mL).Citation28 Plasmids in the transconjugants were extracted by QIAprep Spin Miniprep (Qiagen, Hilden, Germany) and then digested with restriction enzyme BamHI or Hind III (New England BioLabs, Ipswich, MA, USA), respectively. Restriction fragment length polymorphisms (RFLP) analysis was carried out by electrophoresis at 30 V for 2 h in 1% agarose gel. Moreover, Southern hybridization was carried out with a digoxigenin (DIG)-labeled probes for blaSHV and rmpA genes using a DIG system.
Mouse lethality assay
Determination of the virulence of KP in mouse lethality tests and the medium lethal dose (LD50, expressed as colony-forming units) was performed as previously described.Citation7 In brief, a graded dose of 102 to 107 CFU of each strain in 10-fold serial dilutions in 0.1 mL of normal saline was injected intraperitoneally into mice (4 mice for each dose of inoculum). The number of survivors was monitored and calculated every day during 14 d. Twelve KP strains (4 for capsule serotypes K1, 3 for K2 and 5 for non-K1/K2) were selected for the experiments, including 6 ESBL isolates (3 with HV and 3 without HV phenotype) and 6 non-ESBL isolates (3 with HV and 3 without HV phenotype) for comparison.
Ethical approval
All female BALB/c mice (6 to 7 weeks old; weighing 20–25g) were purchased from the animal breeding center, National Laboratory Animal Center (NLAC, Taiwan) and maintained under standard conditions of temperature, light and feeding according to NLAC guidelines. Mice were subjected to veterinary (Dr. Chih-Chan Lin) supervision for health and welfare during the whole period of the experiment and every effort was made to minimize suffering. Body weight and temperature were recorded daily throughout the experiment. Euthanasia was also performed at the end of the experiment at 14 d by using inhalant anesthetic (Isoflurane) according to NLAC guidance and the Institutional Animal Care and Use Committee of Chi Mei Medical Center approved protocol (Permit Number: 100120771).
Statistical analysis
The Chi-square or Fisher's exact test was used in the analysis of categorical variables. A two-tailed p value < 0.05 was considered statistically significant. All statistics were performed using Stata version 12.1 (Stata Press, College Station, TX, USA).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Miss. Li-Yin Liou, Department of Medical Research, Chi Mei Medical Center (Tainan, Taiwan), for technical assistance.
funding
This work was supported by the National Science Council of Taiwan (NSC-100–2314-B-384–002) and Chi Mei Medical Center Research Foundation [CMFHT10202, CMFHR10208].The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Yu WL, Ko WC, Cheng KC, Lee HC, Ke DS, Lee CC, Fung CP, Chuang YC. Association between rmpA and magA genes and clinical syndromes caused by klebsiella pneumoniae in taiwan. Clin Infect Dis 2006; 42:1351-8; PMID:16619144; http://dx.doi.org/10.1086/503420
- Ku YH, Chuang YC, Yu WL. Clinical spectrum and molecular characteristics of klebsiella pneumoniae causing community-acquired extrahepatic abscess. J Microbiol Immunol Infect 2008; 41:311-7; PMID:18787738
- Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 2004; 199:697-705; PMID:14993253; http://dx.doi.org/10.1084/jem.20030857
- Wacharotayankun R, Arakawa Y, Ohta M, Tanaka K, Akashi T, Mori M, Kato N. Enhancement of extracapsular polysaccharide synthesis in Klebsiella pneumoniae by RmpA2, which shows homology to NtrC and FixJ. Infect Immun 1993; 61:3164-74; PMID:8335346
- Yeh KM, Kurup A, Siu LK, Koh YL, Fung CP, Lin JC, Chen TL, Chang FY, Koh TH. Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for klebsiella pneumoniae liver abscess in singapore and taiwan. J Clin Microbiol 2007; 45:466-71; PMID:17151209; http://dx.doi.org/10.1128/JCM.01150-06
- Yu VL, Hansen DS, Ko WC, Sagnimeni A, Klugman KP, von Gottberg A, Goossens H, Wagener MM, Benedi VJ. Virulence characteristics of klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerg Infect Dis 2007; 13:986-93; PMID:18214169; http://dx.doi.org/10.3201/eid1307.070187
- Yu WL, Ko WC, Cheng KC, Lee CC, Lai CC, Chuang YC. Comparison of prevalence of virulence factors for klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis 2008; 62:1-6; PMID:18486404; http://dx.doi.org/10.1016/j.diagmicrobio.2008.04.007
- Lee CH, Liu JW, Su LH, Chien CC, Li CC, Yang KD. Hypermucoviscosity associated with klebsiella pneumoniae-mediated invasive syndrome: a prospective cross-sectional study in taiwan. Int J Infect Dis 2010; 14:e688-92; PMID:20547084; http://dx.doi.org/10.1016/j.ijid.2010.01.007
- Moon HW, Ko YJ, Park S, Hur M, Yun YM. Analysis of community- and hospital-acquired bacteraemia during a recent 5-year period. J Med Microbiol 2014; 63:421-6; PMID:24403599; http://dx.doi.org/10.1099/jmm.0.069054-0
- Lee JA, Kang CI, Joo EJ, Ha YE, Kang SJ, Park SY, Chung DR, Peck KR, Ko KS, Lee NY, et al. Epidemiology and clinical features of community-onset bacteremia caused by extended-spectrum β-lactamase-producing klebsiella pneumoniae. Microb Drug Resist 2011; 17:267-73; PMID:21388296; http://dx.doi.org/10.1089/mdr.2010.0134
- Lin TL, Tang SI, Fang CT, Hsueh PR, Chang SC, Wang JT. Extended-spectrum β-lactamase genes of Klebsiella pneumoniae strains in taiwan: recharacterization of shv-27, shv-41, and tem-116. Microb Drug Resist 2006; 12:12-5; PMID:16584302; http://dx.doi.org/10.1089/mdr.2006.12.12
- Li W, Sun G, Yu Y, Li N, Chen M, Jin R, Jiao Y, Wu H. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) klebsiella pneumoniae isolates in china. Clin Infect Dis 2014; 58:225-32; PMID:24099919; http://dx.doi.org/10.1093/cid/cit675
- El Fertas-Aissani R, Messai Y, Alouache S, Bakour R. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol Biol Paris 2013; 61:209-16; PMID:23218835; http://dx.doi.org/10.1016/j.patbio.2012.10.004
- Lai YC, Peng HL, Chang HY. RmpA2, an activator of capsule biosynthesis in klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J Bacteriol 2003; 185:788-800; PMID:12533454; http://dx.doi.org/10.1128/JB.185.3.788-800.2003
- Cheng HY, Chen YS, Wu CY, Chang HY, Lai YC, Peng HL. RmpA regulation of capsular polysaccharide biosynthesis in klebsiella pneumoniae CG43. J Bacteriol 2010; 192:3144-58; PMID:20382770; http://dx.doi.org/10.1128/JB.00031-10
- Hsu CR, Lin TL, Chen YC, Chou HC, Wang JT. The role of klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiol 2011; 157:3446-57; PMID:21964731; http://dx.doi.org/10.1099/mic.0.050336-0
- Cheng NC, Yu YC, Tai HC, Hsueh PR, Chang SC, Lai SY, Yi WC, Fang CT. Recent trend of necrotizing fasciitis in taiwan: focus on monomicrobial klebsiella pneumoniae necrotizing fasciitis. Clin Infect Dis 2012; 55:930-9; PMID:22715175; http://dx.doi.org/10.1093/cid/cis565
- Podschun R, Sievers D, Fischer A, Ullmann U. Serotypes, hemagglutinins, siderophore synthesis, and serum resistance of klebsiella isolates causing human urinary tract infections. J Infect Dis 1993; 168:1415-21; PMID:7902383; http://dx.doi.org/10.1093/infdis/168.6.1415
- Gundogan N, Citak S, Yalcin E. Virulence properties of extended spectrum β-lactamase-producing klebsiella species in meat samples. J Food Prot 2011; 74:559-64; PMID:21477469; http://dx.doi.org/10.4315/0362-028X.JFP-10-315
- Clinical and Laboratory Standards Institute. Performance Standards for antimicrobial susceptibility testing: twentieth informational supplement M100-S20. Wayne, PA: Clin Lab Stand Inst Press, 2010; vol. 30, p. 46.
- Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL, Chang SC. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis 2007; 45:284-93; PMID:17599305; http://dx.doi.org/10.1086/519262
- Yeh KM, Lin JC, Yin FY, Fung CP, Hung HC, Siu LK, Chang FY. Revisiting the importance of virulence determinant magA and its surrounding genes in klebsiella pneumoniae causing pyogenic liver abscesses: exact role in serotype K1 capsule formation. J Infect Dis 2010; 201:1259-67; PMID:19785524; http://dx.doi.org/10.1086/606010
- Yu WL, Fung CP, Ko WC, Cheng KC, Lee CC, Chuang YC. Polymerase chain reaction analysis for detecting capsule serotypes K1 and K2 of Klebsiella pneumoniae causing abscesses of the liver and other sites. J Infect Dis 2007; 195:1235-6; author reply 6; PMID:17357063; http://dx.doi.org/10.1086/512686
- Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, et al. BLAST: a more efficient report with usability improvements. Nucl Acid Res 2013; 41:W29-33; PMID:23609542; http://dx.doi.org/10.1093/nar/gkt282
- Su PA, Wu LT, Cheng KC, Ko WC, Chuang YC, Yu WL. Screening extended-spectrum β-lactamase production in enterobacter cloacae and serratia marcescens using antibiogram-based methods. J Microbiol Immunol Infect 2010; 43:26-34; PMID:20434120; http://dx.doi.org/10.1016/S1684-1182(10)60004-7
- Wu LT, Tsou MF, Wu HJ, Chen HE, Chuang YC, Yu WL. Survey of CTX-M-3 extended-spectrum β-lactamase (ESBL) among cefotaxime-resistant serratia marcescens at a medical center in middle taiwan. Diagn Microbiol Infect Dis 2004; 49:125-9; PMID:15183862; http://dx.doi.org/10.1016/j.diagmicrobio.2004.02.004
- Pillai DR, Melano R, Rawte P, Lo S, Tijet N, Fuksa M, Roda N, Farrell DJ, Krajden S. Klebsiella pneumoniae carbapenemase, canada. Emerg Infect Dis 2009; 15:827-9; PMID:19402984; http://dx.doi.org/10.3201/eid1505.081536
- Rice LB, Bonomo RA. Genetic and biochemical mechanisms of bacterial resistance to antimicrobial agents. In: Lorian V, ed. Antibiotics in laboratory medicine: Lippincott Williams & Wilkins Press, Maryland., 2005: p. 441-508.