Abstract
Th1-polarized host response, mediated by IFN-γ, has been associated with increased severity of periodontal disease as well as control of periodontal infection. The functional polymorphism TBX21-1993T/C (rs4794067) increases the transcriptional activity of the TBX21 gene (essential for Th1 polarization) resulting in a predisposition to a Th-1 biased immune response. Thus, we conducted a case-control study, including a population of healthy controls (H, n = 218), chronic periodontitis (CP, n = 197), and chronic gingivitis patients (CG, n = 193), to investigate if genetic variations in TBX21 could impact the development of Th1 responses, and consequently influence the pattern of bacterial infection and periodontitis outcome. We observed that the polymorphic allele T was significantly enriched in the CP patients compared to CG subjects, while the H controls demonstrated and intermediate genotype. Also, investigating the putative functionality TBX21-1993T/C in the modulation of local response, we observed that the transcripts levels of T-bet, but not of IFN-γ, were upregulated in homozygote and heterozygote polymorphic subjects. In addition, TBX21-1993T/C did not influence the pattern of bacterial infection or the clinical parameters of disease severity, being the presence/absence of red complex bacteria the main factor associated with the disease status and the subrogate variable probing depth (PD) in the logistic regression analysis.
Introduction
Periodontitis is a chronic infectious diseases characterized by the progressive and irreversible destruction of teeth-supporting structures. Periodontitis is initiated by the bacteria harbored in the teeth-attached biofilm that infiltrates the surrounding epithelial and connective periodontal tissues, triggering a host inflammatory immune response that leads to the subsequent lesion development.Citation1
While any bacteria can essentially trigger a host response, specific Gram negative anaerobic rods have a special capability to elicit and chronically sustain the host inflammatory immune response that mediates periodontal tissue destruction. Most prominent among periodontitis-associated bacteria are the so called red complex cluster (Porphyromonas gingivalis, Tanerella forsytia and Treponema denticola), which has been consistently associated with the occurrence, severity and negative response to treatment of the disease.Citation2 As previously mentioned, it is noteworthy that the sole bacterial infection is not sufficient to explain the complex pathological processes of periodontitis, being the nature and extent of host´s response ultimately responsible for the disease occurrence and outcome.Citation3
While microbial and environmental factors (lifestyle factors [as smoking and stress] or acquired diseases [as diabetes]) characteristically modulate host responses (and consequently periodontitis outcome), studies suggest that as much as 50% of the risk of disease can be determined by genetic factors,Citation4-6 and that numerous disease modifying genes may be involved in the pathogenesis of periodontal diseases by modulating the host's response and his susceptibility to infection.Citation3,7,8 Recently, the interaction between host genetic factors that can impact the ability of pathogens to invade and proliferate on host's tissues has been termed infectogenomics.Citation9 This concept highlights the close and intimate dependence of the host's immune mechanisms and the commensal and/or pathogenic microflora, which co-evolved in a mutually dependent fashion.Citation9
The Th1-polarized immune responses have long been implicated in the pathogenesis of periodontal diseases, since their characteristic cytokine products are overexpressed in the diseased tissues and in mononuclear cells isolated from periodontitis patients.Citation10 Also, from a mechanistic viewpoint, the prototypic Th1-cytokine IFN-γ amplify and exacerbate the pro-inflammatory activity of infiltrating phagocytes and resident cells, ultimately increasing matrix metalloproteinase (MMP) and receptor activator of NFκB ligand (RANKL) levels, leading to augmented tissue destruction.Citation11-13 While recent evidence points to the participation of other T helper subsets (such as Th17 and Thf) in the pathogenesis of osteolytic inflammatory lesions, numerous studies demonstrate that the Th1 subset can be independently responsible for the establishment and progression of periodontitis.Citation14
Th1 lineage commitment and differentiation involves the activation of the key transcription factor T-bet (a member of the T-box family of transcription factors)Citation15 and the secretion of the hallmark cytokine IFN-γ, involved in the activation of macrophage´s microbicide functions.Citation16 Indeed, in addition of modulating the periodontitis’ outcome from a tissue destruction perspective, it is also important to mention that Th1 responses have a fundamental role in the control of experimental periodontal infection.Citation13 Taking into consideration the essential role of T-bet in Th1 responses development, the TBX21 (17q21.32) -1993T/C polymorphism (rs4794067) of the promoter region of T-bet gene has been associated with an increased binding affinity to Sp1 transcription factor and increased transcriptional activity of the TBX21 gene, resulting in a predisposition to a Th-1 biased immune response characterized by increased IFN-γ secretion.Citation17,18 Indeed, numerous studies have associated the TBX21-1993T/C SNP with increased risk or early onset of different diseases, such as type I autoimmune hepatitis,Citation19 systemic lupus erythematosus,Citation18 childhood asthmaCitation20 and aspirin-induced asthma.Citation17 Despite the previous association of Th1-type responses with periodontitis development, no study has investigated the possible influence of this SNP in periodontitis, although it has a potential functional influence in Th1-polarization.
Therefore, it is possible to hypothesize that individual variations in TBX21 could impact the development of Th1 responses, and consequently could influence the pattern of bacterial infection and the outcome of chronic periodontitis. In this context, we conducted a case-control study to investigate whether the TBX21-1993T/C single nucleotide polymorphism (rs4794067) is associated with increased chronic periodontitis risk; and we also investigated the putative functionality of this SNP in the modulation of T-bet and IFN-γ levels in situ, as well as its potential impact in the susceptibility of developing infection caused by classic periodontal pathogens.
Results
TBX21-1993T/C SNP (rs4794067) frequency analysis
There were significant differences in the inflammatory and clinical periodontal parameters between all groups, as previously reported in detail.Citation21 A summary of the population´s demographics, clinical characteristics, genotype, and allele distribution is provided in . The CP and H subpopulations selected for microbiological analysis and quantification of T-bet and IFN-γ expression in gingival tissue were not significantly different from the total population sample (data not shown).
Genotype and allele frequencies of the studied polymorphism were in Hardy-Weinberg equilibrium in the H, CG and CP sample populations (P > 0.05) (). The allele distribution of the sample was T = 78.2% and C = 21.8%, resembling almost exactly the average allele distribution of the sample of the phase 1 ‘1000 Genomes Project’ (polymorphic T-allele = 78%; ancestral C-allele = 22%).Citation22 The frequency of the polymorphic allele homozygote genotype (TT) was significantly higher in the CP group, compared with the CG group (70.6% vs 59.1%, p = 0.0087). Similarly, the allele frequency of the polymorphic T-allele was significantly higher in the CP group compared with the CG group (18% vs 25.6%, p = 0.0051). Interestingly, the healthy control group (classically used as a control in periodontitis genetic studies) demonstrated an intermediary genotype, which was not significantly different from the CG or CP groups, as previously described for other SNPs.Citation21
Table 1. Demographic, clinical and microbiological characteristics in healthy controls (H), chronic periodontitis (CP) and chronic gingivitis (CG) patients
Table 2. Genotype and allele distribution of healthy controls (H), chronic periodontitis (CP) and chronic gingivitis (CG) patients. Fisher´s exact test and equality of proportions test were performed to evaluate the risk of suffering periodontitis associated with the genotype/allele
When the H and CP groups where compared, the recessive model rendered the lower Akaike information criteria (AIC), demonstrating the best fit with the data (), although failing to demonstrate any association between the genetic variants with the disease status, which was somewhat predictable taking into consideration the intermediate genotype of the H group. On the other hand, when the CG and CP groups where compared, theoretically representing the dichotomy between resistant and susceptible phenotypes,Citation21 the log-additive model demonstrated the best fit with the data and both the recessive and log-additive model provided the framework for a significant association between the genetic variants with the disease status, supporting the association of the T allele with the increased susceptibility to chronic periodontitis (). Power calculation demonstrated 97%, 57%, 86% and 16% power with tolerance for 0.001 false positives in the multiplicative, additive, recessive and dominant models, respectively.
Table 3. Inheritance models for TBX21 SNP (rs4794067) and association of genotype with disease. Lower AIC demonstrates best fit of the data with inheritance model. OR (95% CI) and associated p-value represent the risk of disease for subjects carrying the risk allele (polymorphic T allele) compared to ancestral allele (C) carriers according to each inheritance model
TBX21 genotype vs T-bet, IFN-γ levels and clinical parameters
We compared the profiles of T-bet and IFN-γ expression levels with the clinical parameters of periodontitis among the different genotypes of TBX21-1993T/C SNP. The expression of T-bet and IFN-γ was significantly higher in the CP in relation to the H group (≈15-fold, P < 0.001 and ≈2-fold, P < 0.001; respectively) []. In the CP and H groups the expression of T-bet was significantly higher in the polymorphic homozygotes (TT genotype) in comparison with both the heterozygotes (CT) and the ancient-allele homozygotes (CC) (p = 0.0128 and p = 0.021; respectively). Nevertheless, the IFN-γ expression levels only demonstrated a non-significant trend for increased expression in polymorphic homozygotes []. When T-bet and IFN-γ expression levels were compared irrespectively of the TBX21 genotypes, no correlation between T-bet and IFN-γ expression levels was observed in healthy control subjects, while a weak, but statistically significant correlation (r2 = 0.05, p = 0.01) was observed in chronic periodontitis patients []. Regarding the analysis of the possible impact of the TBX21-1993T/C SNP in chronic periodontitis development, no significant variations were observed in the clinical parameters of periodontitis (probing depth, mean probing depth, attachment loss and bleeding on probing) in the different TBX21 genotype carriers (). When T-bet and IFN-γ expression levels were correlated with the clinical parameters irrespectively of the TBX21 genotypes, no positive correlations were observed (data not shown).
Figure 1. T-bet and IFN-γ expression levels in gingival tissue from chronic periodontitis patients and healthy controls. Total RNA was extracted from gingival tissues of controls (H, n = 63) and chronic periodontitis (CP, N = 123) patients, and levels of T-bet and IFN-γ mRNA were determined by RealTimePCR (with normalization to β-actin using the Ct method). (A) T-bet expression in healthy controls and chronic periodontitis patients. * = P < 0.05. (B) IFN-γ expression in healthy controls and chronic periodontitis patients. * = P < 0.05. (C) T-bet expression in healthy controls and chronic periodontitis patients according to their genotype for TBX21 SNP (rs4794067). Different letters represent statistically significant difference between the groups. H and CP groups were tested independently. (D) IFN-γ expression in healthy controls and chronic periodontitis patients according to their genotype for TBX21 SNP (rs4794067). (E) Linear correlation of T-bet and IFN-γ expression levels in healthy subjects. (F) Linear correlation of T-bet and IFN-γ expression levels in chronic periodontitis patients (r2 = correlation coefficient).
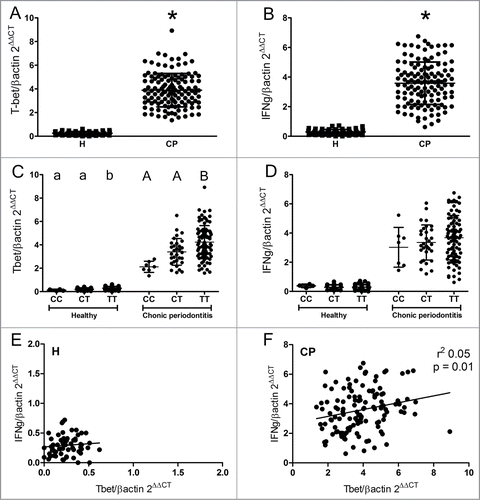
Figure 2. Clinical parameters of periodontal disease according to genotype for TBX21 SNP (rs4794067) in chronic periodontitis patients. Chronic periodontitis (CP) patients were subjected to periodontal examination, and the genotype of TBX21 SNP (rs4794067) was determined using Taqman® chemistry. (A) Probing depth of deepest site according to genotype for TBX21 SNP (rs4794067). (B) Probing depth mean according to genotype for TBX21 SNP (rs4794067). (C) Attachment loss mean according to genotype for TBX21 SNP (rs4794067). D) Bleeding on probing according to genotype for TBX21 SNP (rs4794067).
TBX21 genotype vs the pattern of bacterial infection, modulation of T-bet and IFN-γ levels, and clinical parameters
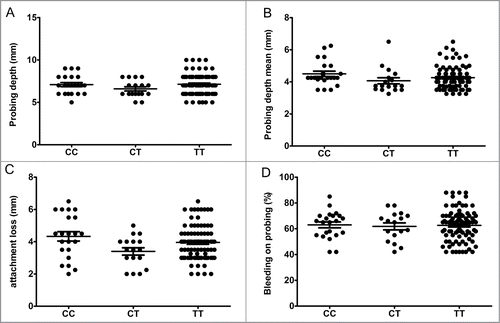
The detection frequency of the red complex's bacteria (), as well the bacterial load was significantly higher in the CP group in comparison to the H controls (P < 0.001). The dichotomic analysis of detection frequency (positive or negative) of P. gingivalis, T. forsytia, and T. denticola in H and CP groups demonstrated no influence of TBX21 genotype in the odds to suffer periodontal infection []. Since the groups/microbes stratification resulted in ‘zero’ subjects with CC genotype positive for T. forsytia and T. denticola (making the Fischer´s exact test analysis impossible) the comparative analysis was carried out based in the TBX21 C and T alleles instead of the genotypes []. Notwithstanding the lack of association of genotype with the red complex bacteria detection, we evaluated the possible variation in the bacterial load in the distinct TBX21 genotype carriers []. In this case, despite a trend toward higher bacterial levels in association with the T allele in CP patients, no statistically significant differences were observed []. Also, no significant association was observed in the load of red complex bacteria within the different TBX21 genotype carriers in the H group (data not shown). We then investigated if the presence and load of the pathogen bacteria could impact the gingival expression levels of T-bet and IFN-γ among the different genotypes [], dichotomously subdividing the samples in positive/negative for the detection of P. gingivalis, T. forsytia, and T. denticola. The analysis demonstrated no significant difference in T-bet and IFN-γ expression levels according to infection status, remaining the TBX21 genotype as the main factor associated with increased T-bet expression in the lesions []. Also, the bacterial load of P. gingivalis, T. forsytia, and T. denticola was not correlated with T-bet nor IFN-γ expression levels in the periodontal lesions (data not shown).
Table 4. Frequencies of detection of the studied pathogens in healthy control subjects and chronic periodontitis patients according to their TBX21 genotype. The values of Fisher´s exact test in the right column represent the increased risk of positive detection in chronic periodontitis subject compared to healthy controls (CC genotype vs TT genotype in the grey row and among alleles in the white rows). The values of the Fisher´s exact text in the healthy and chronic periodontitis columns represent the risk of positive detection of pathogens among T and C allele carriers; calculations were based on alleles and not in genotypes due to the existence of categories with zero subjects
Figure 3. Bacterial load of P. gingivalis, T. forsytia and T. denticola in total periodontal sites and shallow/deep sites (above and below the patient´s median) of chronic periodontitis patients (n = 123), according to TBX21 SNP (rs4794067) genotype. The presence and load of the periodontopathogens were determined by RealTimePCR. (A) P. gingivalis. (B) T. forsytia. (C) T. denticola.
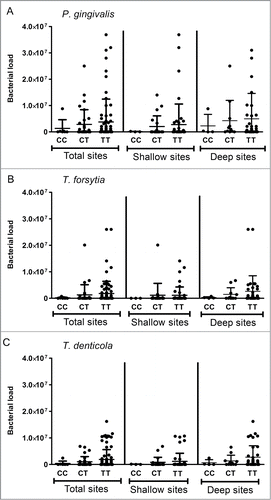
Figure 4. Quantitative assessment of T-bet and IFN-γ mRNA expression in the presence or absence of periodonto pathogens associated with the genotypes of TBX21 SNP (rs4794067) in CP patients. Total RNA was extracted from gingival tissues, and levels of T-bet and IFN-γ mRNA were measured quantitatively by RealTimePCR, and the results are presented as expression of the individual mRNAs, with normalization to β-actin. The presence of the periodontopathogens P. gingivalis, T. forsythia and T. denticola was investigated by RealTimePCR. The graphs depict the expression of T-bet and IFN-γ in CP patients regarding their TBX21 SNP (rs4794067) genotype concomitantly with their positiveness or not to the detection of each periodonto pathogen. (A) T-bet mRNA expression in positive/negative subjects for P. gingivalis. (B) T-bet mRNA expression in positive/negative subjects for T. forsythia. (C) T-bet mRNA expression in positive/negative subjects for T. denticola. (D) IFNγ mRNA expression in positive/negative subjects for P. gingivalis. (E) IFN-γ mRNA expression in positive/negative subjects for T. forsythia. (F) IFN-γ mRNA expression in positive/negative subjects for T. denticola.
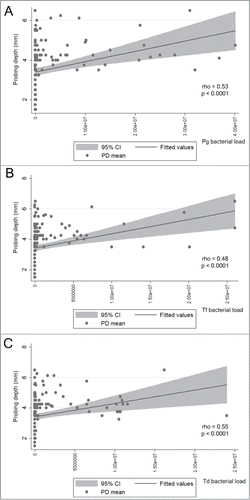
Finally, since T-bet genetic variants were not associated with CP clinical parameters [], we tested the possible association of the red complex bacteria with CP clinical parameters []. Our results demonstrated a strong monotonic correlation between the absolute load of each individual red complex bacteria with the disease subrogate PD mean []. The correlation coefficients (rho) were 0.53 for P. gingivalis (P < 0.0001); 0.48 for T. forsytia (P < 0.0001); and 0.55 for T. denticola (P < 0.0001). In the logistic regression model, when the genetic data was analyzed in interaction with the presence/absence of red complex bacteria as environmental co-variable, the combined effect on disease status was significant for each bacteria species individually: P. gingivalis showed and odds ratio of 9.44 (CI 3.58-24.89) and 10.32 (CI 2.03-52.52) in association with TT and CT genotypes, respectively; T. forsytia showed and odds ratio of 12.41 (CI 4.06-37.88) and 24.94 (CI 2.94-211.06) in association with TT and CT genotypes, respectively; while T. denticola showed and odds ratio of 12.41 (CI 4.06-37.88) and 8.44 (CI 2.02-35.26) in association with TT and CT genotypes, respectively. Nevertheless, the Cochrane-Armitage test for interaction in the trend for the genetic/microbiologic association failed to detect a significant interaction (0.84; 0.29 and 0.86 for P. gingivalis, T. forsytia and T. denticola, respectively).
Figure 5. Correlation between the load of each pathogen and the disease subrogate clinical measure probing depth in chronic periodontitis patients. Quantitative assessment of bacterial load was performed by RealTimePCR. The graphs depict the lineal regression and the 95% confidence interval (in gray) for the bacterial load and the PD. Regression coefficients (rho) and corresponding p-values were calculated. (A) P. gingivalis bacterial load correlation to PD (rho = 0.53, P-value < 0.0001). (B) T. forsythia bacterial load correlation to PD (rho = 0.48, P-value < 0.0001). (C) T. denticola bacterial load correlation to PD (rho = 0.53, P-value < 0.0001).
The ordinary least squared (OLS) regression model including simultaneously the detection of red complex bacteria and genotype (with PD as response variable) failed to detect a significant contribution of the genetic variants in the changes of the dependent variable, while the microbiological data proved significant to explain the variations of PD (P < 0.0001; R-squared 0.3531). When the genotype was fitted in the OLS regression and the logistic regression as the sole explanatory variable it did not demonstrate a significant effect in the outcome (data not shown).
Discussion
Periodontitis is a complex disease in which microbial, environmental and genetic factors interact increasing or decreasing the predisposition to periodontal tissue inflammatory destruction. The principal causative factor recognized of the disease is the periodontal infection by pathogen bacteria, specifically the so called red complex bacteria (P. gingivalis, T. forsytia and T. denticola), which are characterized by their ability to elicit and chronically sustain the host's inflammatory immune response that mediates tissue destruction. Th1-type immune responses have been mechanistically associated with tissue destruction and with the control of experimental periodontal infection.Citation11-13 Since the functional TBX21-1993T/C SNP directly impacts Th1-type response development, we conducted parallel case-control and functional studies to address the possible influence of this genetic variant in the susceptibility of developing chronic periodontitis.
We employed a previously validated strategy to increase the power of the case-control study, selecting a “resistant” control population opposed to the “susceptible” subjects presenting chronic periodontitis.Citation21 The “resistant” control population, represented by chronic gingivitis patients (exposed to microbial challenge without developing the clinical signs of periodontitis) proved significantly different from the “susceptible” chronic periodontitis sample, both in allele frequencies and allele distribution, while the “classical” healthy controls (individuals presetting periodontal health but with a low exposure to chronic microbial challenge due to proper oral hygiene methods) did not prove genetically different from the CP or the CG subjects. These results further support the notion that the “classical” healthy control population usually enrolled in case-control studies in periodontitis is unsuitable for direct comparison with the susceptible patients, since it is arguably composed of a heterogeneous mixture of genetically “susceptible” and “resistant” subjects, whose real phenotype remains unrevealed by the relatively low microbial challenge to which they are exposed.Citation21 An analogous alternative strategy that has been recently proposed is the enrolment of the “highly susceptible” phenotype as the cases, clinically characterized as aggressive periodontitis patients, increasing the power of the study, but without addressing the issue of the relatively low exposure of the healthy control group.Citation23
Our results demonstrated that TBX21-1993 polymorphic T allele carriers were more prevalent in the disease group, proving that the TBX21 SNP polymorphism exerts some influence in disease susceptibility. Previous studies demonstrated that the increased expression of T-bet in polymorphic subjects predispose to a Th1-biased adaptive immune response, which is characterized by a robust and stronger production of pro-inflammatory mediators.Citation24 Accordingly, the Th1-polarized immune responses have long been implicated in the pathogenesis of periodontal diseases, linked with increased disease severity owing to the pro-inflammatory and catabolic activities mediated by the prototypic Th1 cytokine IFN-γ.Citation11-13 Indeed, the expression of both T-bet and IFN-γ was significantly higher in diseased than in healthy tissues, reinforcing the assumed association of Th1 response with periodontitis development.
While the case-control data from TBX21 SNP genotypes/alleles frequency in “susceptible” and “resistant” subjects suggests its involvement in periodontitis susceptibility/resistance, additional experimental approaches were conducted to support such potential association from the functional and mechanistic viewpoints. In this context, our data demonstrate that polymorphic homozygote subjects (TT) exhibited significantly increased expression of T-bet (2-fold change compared to ancestral homozygotes), providing supporting evidence of the functionality of the investigated SNP. In this aspect our result are in accordance with published evidence.Citation25,26 However, it is important to notice that despite the strong influence of T allele/genotypes over T-bet expression (≈2 fold), the subsequent impact over IFN-γ expression was significantly lower (1.21-fold), and proved statistically non-significant.
The weak influence of TBX21 genetic variance in the Th1-response expression (represented by IFN-γ mRNA levels) reinforces the concept of the complex nature of the leukocyte polarization. Indeed, the local Th1 response (as measured in this study in the gingival biopsies) is the result of leukocyte polarization, probably in the lymph nodes, followed by their migration to periodontal tissues, local stimulation by antigen presenting cells under the influence of a complex local cytokine milieu, followed by the continuous regulation of Th1 cells by the local cytokine networks.Citation27,28 As a matter of fact, while Th1 cells exhibit epigenetic stability at signature transcription factor loci level, a number of other factors such as chromatin modifications and intergenic noncoding RNAs (such as Tmevpg1) can decisively impact IFN-γ production.Citation29,30
Nevertheless, it is worth noting that regardless of the TBX21 genotype, CP subjects exhibited an overall ≈10-fold increase in IFN-γ expression compared with the H group, and T-bet and IFN-γ levels presented a significant statistical correlation (even though such correlation was relatively weak) in periodontal lesions, reinforcing the association of T-bet with Th1 responses in situ. However, no clear association was observed between the TBX21 genotype with the clinical parameters of chronic periodontitis, or between T-bet and IFN-γ expression levels and the clinical parameters of periodontitis. Such lack of a significant association between clinical phenotype and genotype (and T-bet and IFN-γ levels) could be explained by the multifarious and overlapping layers of finely tuned processes which intervene in the pathogenesis of the disease. A single SNP (even one accounting for a major change in an important pathway of immune response, such as TBX21-1993T/C) is unlikely to produce such a large phenotype change to be evident in a routine clinical periodontal examination.Citation4 Accordingly, despite the proven association between pro-inflammatory cytokines (such as IL-1β and TNF-α) with periodontitis onset and progression,Citation31 previous studies have failed to provide a strong correlation between the levels of such mediators and the clinical parameters of periodontitis.Citation32-34
Interestingly, in spite of the demonstrated association between IFN-γ and the control of experimental periodontal infection,Citation13 our results indicated a lack of significant interaction in the trend between microbiological data and genotype. One possible explanation derives from the diluted impact of TBX21 variants on the Th1 response outcome, as measured by the local IFN-γ levels. Additionally, we must consider that other cytokines, such as TNF-α and IL-6, are supposed to contribute to the control of periodontal infection, as suggested by experimental models and human studies data.Citation33-35 It is also important to consider that the experimental evidence linking IFN-γ with the control of periodontal infection derives from IFN-KO mice, which obviously represent an extreme situation that does not reflect the variation in IFN-γ levels observed in humans.Citation13 Correspondingly, published evidence from our group demonstrated that after the host´s immune response reach a minimal threshold that confers protection, the increase of host responsiveness in degree/intensity does not provide additional protection and, conversely result in heightened tissue damage.Citation36
It is also important to mention that, contrary to previously described for TNF-α and IL-1β, the local levels of T-bet (and in a lower extent, the IFN-γ levels) are not influenced by the presence/absence or load of the red complex pathogens.Citation33,34 While TNF-α and IL-1β production is supposed to be derived by immediate stimulation of leukocytes and resident cells by microbial components in the lesions, our results suggests that Th1 polarization (represented by T-bet expression) is probably a process centered in the lymph nodes, with a minor influence from the gingival environment per se. Accordingly, modification in the lymph nodes environment by co-morbidities, such as arthritis, can trigger or exacerbate periodontitis.Citation37-39 On the other hand, the local expression of Th1 markers in the lesions (represented by IFN-γ levels), as previously discussed, is influenced by numerous other factors, including the complex local cytokine milieu.Citation27,28
Indeed, the complexity of local cytokine milieu in infectious chronic inflammatory osteolytic lesions, such as periodontitis and periapical lesions, is far from being completely understood. In a recent study that investigated various Th expression markers, we demonstrated that multiple cytokine clusters are accountable for the activity of osteolytic lesions, including Th1-, Th17- and Thf-biased clusters.Citation14
While the immune response pattern associated with a clear disease outcome seems to vary significantly, the common point among progressive lesions (in both periodontitis and periapical lesions) seems to be the presence of recognized pathogens.Citation40-43 Therefore, the data presented herein demonstrates a disproportionate large effect of the presence/absence of red complex bacteria with the disease status and the periodontitis subrogate variable PD. Even though the T-allele carriers were enriched in the diseased group, and the TT and CT genotypes were overrepresented in the CP patients group (indicating an association of the SNP with the disease status); the environmental microbiological covariate obscures this association in the logistic regression model. This is also true for the OLS regression model, where the microbiological data accounted for a 35% of the variation of the response variable PD, and the genetic variance proved insignificant when fitted into the model. It is also worth noting that the genetic data failed to demonstrate a significant effect in the OLS regression (PD as outcome) and in the logistic regression (disease as outcome) when tested as the sole explanatory variable.
In summary, the data presented supports the previous indicatives that TBX21-1993T/C (rs4794067) polymorphism is in fact functional, and that genotypes carrying the T allele are associated with a noticeable increase in T-bet transcription as well as a significant (while modest) increase in the risk to develop chronic periodontitis. Interestingly, the marked impact of TBX21-1993 T allele is not reflected in a similar magnitude in the IFN-γ transcription, and does not influence the pattern of pathogen bacterial infection or the clinical parameters of disease severity, being the presence/absence of red complex bacteria the main factor associated with the disease status and subrogate variable PD in the logistic regression analysis. However, it is important to consider that the cross-sectional nature of the study design poses limitations on the conclusions that can be inferred from the data. Further, it is possible that uncontrolled variables in the recruitment phase of the study could have influenced the results. Particularly, although the voluntaries were age and gender matched, no socio-economical information was recorded. Since socio-economic factors can impact the development of periodontitis, it is possible that this uncontrolled confounder exerted some impact in the reported results. Nevertheless, it is worth considering that the population that uses the services from the dental clinic of the Ribeirão Preto Dentistry School is fairly homogeneous in socio-economic terms, so the effect of this confounder is arguably limited.
Also, it is possible that the single time point sampling caused the lack of association between genotype and microbiological data and that a time-series sampling strategy could overcome this issue. Indeed, the very nature of the pathogenesis of periodontal disease impedes a simple straightforward association between a particular genotype and disease phenotype, requiring for its development the cumulative effect of multiple genes and environmental causal factors, each of which are neither necessary nor sufficient to individually cause the disease.Citation44,45 The simultaneous analysis of numerous functional SNPs and multiple microbiological, metabolic and immune markers through hypothesis-free tools (such as cluster analysis) becomes necessary in order to unveil the complicated and intermingled paths that tip the balance from health to disease. The way to future discovery and insight in the paths that leads to disease must contemplate the simultaneous association of known causative factors (such as red complex infection), with multiple putative disease markers (genetic, environmental, metabolic, inflammatory, etc.) in order to construct valid explanatory and prognosis models. The knowledge gained from these kinds of approaches will provide the tools for the development of more specific diagnostic protocols and therapeutic interventions.
Materials and Methods
Study population and sample collection
Patients and controls from the São Paulo state, south-eastern region of Brazil, scheduled for treatment at the Dentistry School of University of Ribeirão Preto (UNAERP), were submitted to anamnesis, radiographic study and clinical examination by an experienced periodontist (scored for bleeding on probing, probing depth and clinical attachment loss). All enrolled subjects signed an informed consent form that was approved by the Institutional Review Board, and received supra gingival prophylaxis. Exclusion criteria was applied as follows: not providing informed consent; medical history indicating evidence of known systemic modifiers of periodontal disease, and having received periodontal therapy in the previous 2 years. Current and former smokers were specifically excluded. No strategy was used to identify subpopulations (population stratification) or population relatedness among the recruited subjects. After the diagnostic phase, patients were subsequently categorized into healthy (H; n = 218) (the classic control), chronic gingivitis (CG; n = 193)(the ‘resistant’ phenotype control) or chronic periodontitis (CP; n = 197)(the susceptible subjects, the case-study group) groups, as previously described.Citation21,33
Epithelial buccal cells were sampled scrapping the inner cheek buccal mucosa after a mouthwash with 3% glucose in all groups; while biopsies of gingival tissue and microbiological samples were collected from fractions of CP and H groups as previously described in detail.Citation21,33 Patients in the chronic periodontitis group (CP; n = 197) presenting moderate to advanced periodontitis (at least one teeth per sextant with probing depth > 6mm and clinical attachment loss >3 mm plus radiographic evidence of extensive bone loss [>30% alveolar bone height in at least 50% of teeth]), received basic periodontal therapy. Biopsies of gingival tissue (one sample from each patient) were obtained during surgical therapy of the sites that exhibited no improvement in clinical condition (i.e. persistent bleeding on probing and increased probing depth) 3–4 weeks after the basic periodontal therapy (n = 123). Gingival biopsies were taken during surgical therapy as previously describedCitation34 and comprised junctional epithelium, pocket epithelium and gingival connective tissue. The healthy control group (H; n = 218) included subjects with clinically healthy gingival tissues (<10% of bleeding on probing; no sites with probing depth >3 mm, no clinical attachment loss and no radiographic evidence of alveolar bone loss) scheduled to undergo restorative dentistry procedures. A representative fraction of the control group (n = 63) was also scheduled to surgical procedures for restorative/prosthetic reasons, when biopsies of healthy gingival tissue were taken. Gingival biopsies were taken from sites showing no bleeding on probing and probing depth <3 mm during surgical procedures due to esthetics, orthodontic and/or prosthetic reasons, and also comprised junctional epithelium, gingival crevicular epithelium and gingival connective tissue, as previously described.Citation34 The chronic gingivitis group (CG; n = 193), representing a ‘resistance phenotype’ to case-control analysis,Citation21 was composed of subjects with clinical history of poor oral hygiene, bleeding on probing >70% of periodontal sites and no clinical attachment loss (CAL) or radiographic evidence of alveolar bone loss. Due to ethical restrictions (lack of indication of surgical therapy) no gingival samples were collected from CG group. The clinical features of the groups are summarized in .
In order detect and quantify P. gingivalis, T. forsythia and T. denticola, periodontal crevice/pocket biofilm samples were collected with a sterile paper point ISO #40 from the same site biopsied immediately before the periodontal surgical procedure (H n = 63; CP n = 123), as described elsewhere.Citation46 Since no surgical procedures were conducted in CG group due to ethical restrictions as previously mentioned, no samples for microbiological analysis were collected from CG group.
Analysis of TBX21-1993T/C SNP (rs4794067)
DNA was extracted from epithelial buccal cells with sequential phenol/chloroform solution and precipitated with salt/ethanol solution, as described elsewhere.Citation47 Extracted DNA was immediately used for genotyping. DNA integrity was checked and the allelic discrimination of TBX21-1993T/C SNP (rs4794067) variants was performed in 3 μL reactions using Taqman (Applied Biosystems, Warrington, UK) chemistry as previously described.Citation48,49 Genotyping was performed blinded to group status. For reaction quality control, a sample of known genotype was included in the plate and a no DNA template sample was included as negative control. Only genotypes with an automatic call rate >95% were considered, error rate was <3%. Samples that failed to provide a genotype were repeated in additional reactions. Genotyping procedures, from isolation to allelic discrimination and data analysis was performed in the laboratory of Osteoinmunology of the Bauru Faculty of Dentistry of the University of Sao Paulo.
Real-Time PCR reactions – IFN-γ and T-bet mRNA quantification
The extraction of total RNA from periodontal tissues samples was performed with Trizol reagent (Invitrogen), and the cDNA synthesis were accomplished as previously described.Citation34 RealTime-PCR mRNA or DNA analyses were performed in an MiniOpticon system (BioRad, Hercules, CA, USA), using SybrGreen MasterMix (Invitrogen), specific primers (IFN-γ: sense ATGAAATATACAAGTTATATCATGC, antisense TGTTTCGAGGTCGAAGAGCATCCCAGTAA; T-bet: sense CCTCTTCTATCCAACCAGTAT, antisense CTCCGCTTCATAACTGTG; Beta-actin: sense ATGTTTGAGACCTTCAACAC, antisense CACGTCADACTTCATGATGG) and 2.5ng of cDNA in each reaction, as previously described.Citation32,33 The standard PCR conditions were 95°C (10 minutes), and then 40 cycles of 94°C (1 minute), 56°C (1 minute), and 72°C (2 minutes), followed by a standard denaturation curve. Negative controls without cDNA and without the primer/probe sets were also performed. Calculations for determining the relative levels of gene expression were made from triplicate measurements of the target gene, with normalization to β-actin in the sample, using the cycle threshold (Ct) method and the 2ΔΔct equation, as previously described.Citation33
Real-Time PCR reactions – bacterial DNA quantification
In order to detect P. gingivalis, T. forsythia and T. denticola, periodontal crevice/pocket biofilm samples were collected with a sterile paper point ISO #40 from the same site biopsied immediately before the periodontal surgical procedure (H n = 63 and CP n = 123), as described elsewhere.Citation46 Bacterial DNAs were extracted from plaque samples by DNA Purification System (Promega).Citation34 RealTime-PCR mRNA or DNA analyses were performed in a MiniOpticon system (BioRad), using SybrGreen MasterMix (Invitrogen), specific primers (P. gingivalis: sense TGCAACTTGCCTTACAGAGGG, antisense ACTCGTATCGCCCGTTATTC; T. forsythia: sense GGGTGAGTAACGCGTATGTAACCT, antisense ACCCATCCGCAACCAATAAA; T. denticola: sense AGAGCAAGCTCTCCCTTACCGT, antisense TAAGGGCGGCTTGAAATAATGA), and 5ng of DNA in each reaction.Citation33 The standard PCR conditions were 95°C (10 minutes), and then 40 cycles of 94°C (1 minute), 56°C (1 minute), and 72°C (2 minutes), followed by a standard denaturation curve. The positivity to bacteria detection and the bacterial counts in each sample were determined based on the comparison with a standard curve comprised by specific bacterial DNA (109 to 102 bacteria) and negative controls, similar to previously described,Citation21,33 and then adjusted for sample dilution in the assay to obtain the copy numbers in each site (i.e., absolute load). The sensibility range of bacteria detection and quantification of our RealTime-PCR technique was of 103 to 108 bacteria to each of the 3 putative periodontal pathogens tested.
Statistical analysis
The Shapiro-Wilk test was performed to test the distribution of all test groups prior to comparative analysis; P-value >0.05 was considered indicative of normal distribution. The differences in the demographic and clinical data for the study populations were tested with the Fisher´s exact test, rank sum Wilcoxon test (Mann-Whitney U test) and one-way ANOVA or Kruskal-Wallis test, depending of the data distribution. The intra examiner agreement was tested by Cohen's kappa 4 times a year during the recruiting phase of the project using the repeated measures strategy, a kappa value >0.81 was considered the critical value (regarded as almost perfect agreement). Genotype and allele distribution among groups was tested by Fisher´s exact test and the equality of proportions test. The bacterial load between CP and H groups was compared by the rank sum Wilcoxon test (Mann-Whitney U test); the microbiological data was compared between genotype groups inside the CP and H groups by the Kruskal-Wallis test. The T-bet and IFN-γ expression levels were compared by the unpaired t test and rank sum Wilcoxon test (Mann-Whitney U test). All tests were performed in Stata11.1 (College Station, TX USA) or GraphPad Prism 5.01 (San Diego, CA USA). Power calculation of the study was performed using CaTS software (Department of Biostatistics and Center for Statistical Genetics, University of Michigan) as elsewhere describedCitation50 using Multiplicative, Additive, Dominant and Recessive models. Chi-squared test with 1 degree of freedom was performed to test the Hardy-Weinberg equilibrium in the allele frequencies of the study populations, using the Hardy-Weinberg equilibrium calculator including analysis for ascertainment bias online-based tool.Citation51 The association between genetic and microbiological data was tested by logistic regression and Cochrane-Armitage interaction in the trend analysis using the SNPStats web-based tool.Citation52 A P-value < 0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This research was funded by FAPESP grant 2014/03276-0.
References
- Cekici A, Kantarci A, Hasturk H, Van Dyke TE. Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontol 2000 2014; 64:57-80; PMID:24320956; http://dx.doi.org/10.1111/prd.12002
- Offenbacher S. Periodontal diseases: pathogenesis. Ann Periodontol 1996; 1:821-78; PMID:9118282; http://dx.doi.org/10.1902/annals.1996.1.1.821
- Garlet GP. Destructive and protective roles of cytokines in periodontitis: a re-appraisal from host defense and tissue destruction viewpoints. J Dent Res 2010; 89:1349-63; PMID:20739705; http://dx.doi.org/10.1177/0022034510376402
- Laine ML, Crielaard W, Loos BG. Genetic susceptibility to periodontitis. Periodontol 2000 2012; 58:37-68; PMID:22133366; http://dx.doi.org/10.1111/j.1600-0757.2011.00415.x
- Michalowicz BS, Diehl SR, Gunsolley JC, Sparks BS, Brooks CN, Koertge TE, Califano JV, Burmeister JA, Schenkein HA. Evidence of a substantial genetic basis for risk of adult periodontitis. J Periodontol 2000; 71:1699-707; PMID:11128917; http://dx.doi.org/10.1902/jop.2000.71.11.1699
- Michalowicz BS, Aeppli D, Virag JG, Klump DG, Hinrichs JE, Segal NL, Bouchard TJ, Jr., Pihlstrom BL. Periodontal findings in adult twins. J Periodontol 1991; 62:293-9; PMID:2072240; http://dx.doi.org/10.1902/jop.1991.62.5.293
- Nibali L, Donos N, Farrell S, Ready D, Pratten J, Tu YK, D'Aiuto F. Association between interleukin-6 -174 polymorphism and Aggregatibacter actinomycetemcomitans in chronic periodontitis. J Periodontol 2010; 81:1814-9; PMID:20681812; http://dx.doi.org/10.1902/jop.2010.100084
- Loos BG, John RP, Laine ML. Identification of genetic risk factors for periodontitis and possible mechanisms of action. J Clin Periodontol 2005; 32(Suppl 6):159-79; PMID:16128836; http://dx.doi.org/10.1111/j.1600-051X.2005.00806.x
- Nibali L, Donos N, Henderson B. Periodontal infectogenomics. J Med Microbiol 2009; 58:1269-74; PMID:19528161; http://dx.doi.org/10.1099/jmm.0.012021-0
- Takeichi O, Haber J, Kawai T, Smith DJ, Moro I, Taubman MA. Cytokine profiles of T-lymphocytes from gingival tissues with pathological pocketing. J Dent Res 2000; 79:1548-55; PMID:11023273; http://dx.doi.org/10.1177/00220345000790080401
- Ford PJ, Gamonal J, Seymour GJ. Immunological differences and similarities between chronic periodontitis and aggressive periodontitis. Periodontol 2000 2010; 53:111-23; PMID:20403108; http://dx.doi.org/10.1111/j.1600-0757.2010.00349.x
- Ferreira SB, Jr., Repeke CE, Raimundo FM, Nunes IS, Avila-Campos MJ, Ferreira BR, Santana da Silva J, Campanelli AP, Garlet GP. CCR5 mediates pro-osteoclastic and osteoclastogenic leukocyte chemoattraction. J Dent Res 2011; 90:632-7; PMID:21245464; http://dx.doi.org/10.1177/0022034510395021
- Garlet GP, Cardoso CR, Campanelli AP, Garlet TP, Avila-Campos MJ, Cunha FQ, Silva JS. The essential role of IFN-gamma in the control of lethal Aggregatibacter actinomycetemcomitans infection in mice. Microbes Infect 2008; 10:489-96; PMID:18403243; http://dx.doi.org/10.1016/j.micinf.2008.01.010
- Araujo-Pires AC, Francisconi CF, Biguetti CC, Cavalla F, Aranha AM, Letra A, Trombone AP, Faveri M, Silva RM, Garlet GP. Simultaneous analysis of T helper subsets (Th1, Th2, Th9, Th17, Th22, Tfh, Tr1 and Tregs) markers expression in periapical lesions reveals multiple cytokine clusters accountable for lesions activity and inactivity status. J Appl Oral Sci 2014; 22:336-46; PMID:25141207; http://dx.doi.org/10.1590/1678-775720140140
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 2000; 100:655-69; PMID:10761931; http://dx.doi.org/10.1016/S0092-8674(00)80702-3
- Suzuki Y, Orellana MA, Schreiber RD, Remington JS. Interferon-gamma: the major mediator of resistance against Toxoplasma gondii. Science 1988; 240:516-8; PMID:3128869; http://dx.doi.org/10.1126/science.3128869
- Akahoshi M, Obara K, Hirota T, Matsuda A, Hasegawa K, Takahashi N, Shimizu M, Nakashima K, Cheng L, Doi S, et al. Functional promoter polymorphism in the TBX21 gene associated with aspirin-induced asthma. Hum Genet 2005; 117:16-26; PMID:15806396; http://dx.doi.org/10.1007/s00439-005-1285-0
- You Y, Zhao W, Chen S, Tan W, Dan Y, Hao F, Deng G. Association of TBX21 gene haplotypes in a Chinese population with systemic lupus erythematosus. Scand J Rheumatol 2010; 39:254-8; PMID:20429676; http://dx.doi.org/10.3109/03009740903347983
- Chen S, Zhao W, Tan W, Luo X, Dan Y, You Z, Kuang X, Wang Y, Deng G. Association of TBX21 promoter polymorphisms with type 1 autoimmune hepatitis in a Chinese population. Hum Immunol 2011; 72:69-73; PMID:20977921; http://dx.doi.org/10.1016/j.humimm.2010.10.019
- Suttner K, Rosenstiel P, Depner M, Schedel M, Pinto LA, Ruether A, Adamski J, Klopp N, Illig T, Vogelberg C, et al. TBX21 gene variants increase childhood asthma risk in combination with HLX1 variants. J Allergy Clin Immunol 2009; 123:1062-8; 1068.e1-8; PMID:19362357; http://dx.doi.org/10.1016/j.jaci.2009.02.025
- Garlet GP, Trombone AP, Menezes R, Letra A, Repeke CE, Vieira AE, Martins W, Jr., Neves LT, Campanelli AP, Santos CF, et al. The use of chronic gingivitis as reference status increases the power and odds of periodontitis genetic studies: a proposal based in the exposure concept and clearer resistance and susceptibility phenotypes definition. J Clin Periodontol 2012; 39:323-32; PMID:22324464; http://dx.doi.org/10.1111/j.1600-051X.2012.01859.x
- Genomes Project C, Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature 2010; 467:1061-73; PMID:20981092; http://dx.doi.org/10.1038/nature09534
- Vaithilingam RD, Safii SH, Baharuddin NA, Ng CC, Cheong SC, Bartold PM, Schaefer AS, Loos BG. Moving into a new era of periodontal genetic studies: relevance of large case-control samples using severe phenotypes for genome-wide association studies. J Periodontal Res 2014; 49:683-95; PMID:24528298; http://dx.doi.org/10.1111/jre.12167
- Repeke CE, Ferreira SB, Jr., Vieira AE, Silveira EM, Avila-Campos MJ, da Silva JS, Santos CF, Campanelli AP, Trombone AP, Garlet GP. Dose-response met-RANTES treatment of experimental periodontitis: a narrow edge between the disease severity attenuation and infection control. PLoS One 2011; 6:e22526; PMID:21799885; http://dx.doi.org/10.1371/journal.pone.0022526
- Li JR, Li JG, Deng GH, Zhao WL, Dan YJ, Wang YM, Chen S. A common promoter variant of TBX21 is associated with allele specific binding to Yin-Yang 1 and reduced gene expression. Scand J Immunol 2011; 73:449-58; PMID:21272048; http://dx.doi.org/10.1111/j.1365-3083.2011.02520.x
- Morita M, Watanabe M, Inoue N, Inaoka C, Akamizu T, Tatsumi KI, Hidaka Y, Iwatani Y. Functional polymorphisms in TBX21 and HLX are associated with development and prognosis of Graves' disease. Autoimmunity 2012; 45:129-36; PMID:22014209; http://dx.doi.org/10.3109/08916934.2011.622013
- Cavalla F, Araujo-Pires AC, Biguetti CC, Garlet GP. Cytokine networks regulating inflammation and immune defense in the oral Cavity. Curr Oral Health Rep 2014; 1:104-13; http://dx.doi.org/10.1007/s40496-014-0016-9
- Harris NL, Watt V, Ronchese F, Le Gros G. Differential T cell function and fate in lymph node and nonlymphoid tissues. J Exp Med 2002; 195:317-26; PMID:11828006; http://dx.doi.org/10.1084/jem.20011558
- Cohen CJ, Crome SQ, MacDonald KG, Dai EL, Mager DL, Levings MK. Human Th1 and Th17 cells exhibit epigenetic stability at signature cytokine and transcription factor loci. J Immunol 2011; 187:5615-26; PMID:22048764; http://dx.doi.org/10.4049/jimmunol.1101058
- Aune TM, Collins PL, Collier SP, Henderson MA, Chang S. Epigenetic activation and silencing of the gene that encodes IFN-gamma. Front Immunol 2013; 4:112; PMID:23720660; http://dx.doi.org/10.3389/fimmu.2013.00112
- Graves DT, Cochran D. The contribution of interleukin-1 and tumor necrosis factor to periodontal tissue destruction. J Periodontol 2003; 74:391-401; PMID:12710761; http://dx.doi.org/10.1902/jop.2003.74.3.391
- Repeke CE, Trombone AP, Ferreira SB, Jr., Cardoso CR, Silveira EM, Martins W, Jr., Trevilatto PC, Silva JS, Campanelli AP, Garlet GP. Strong and persistent microbial and inflammatory stimuli overcome the genetic predisposition to higher matrix metalloproteinase-1 (MMP-1) expression: a mechanistic explanation for the lack of association of MMP1-1607 single-nucleotide polymorphism genotypes with MMP-1 expression in chronic periodontitis lesions. J Clin Periodontol 2009; 36:726-38; PMID:19659894; http://dx.doi.org/10.1111/j.1600-051X.2009.01447.x
- Trombone AP, Cardoso CR, Repeke CE, Ferreira Jr SB, Martins Jr W, Campanelli AP, Avila-Campos MJ, Trevilatto PC, Silva JS, Garlet GP. Tumor necrosis factor-alpha -308G/A single nucleotide polymorphism and red-complex periodontopathogens are independently associated with increased levels of tumor necrosis factor-alpha in diseased periodontal tissues. J Periodontal Res 2009; 44(5):598-608; http://dx.doi.org/10.1111/j.1600-0765.2008.01166.x
- Ferreira SB, Jr., Trombone AP, Repeke CE, Cardoso CR, Martins W, Jr., Santos CF, Trevilatto PC, Avila-Campos MJ, Campanelli AP, Silva JS, et al. An interleukin-1beta (IL-1beta) single-nucleotide polymorphism at position 3954 and red complex periodontopathogens independently and additively modulate the levels of IL-1beta in diseased periodontal tissues. Infect Immun 2008; 76:3725-34; PMID:18541658; http://dx.doi.org/10.1128/IAI.00546-08
- Garlet GP, Cardoso CR, Campanelli AP, Ferreira BR, Avila-Campos MJ, Cunha FQ, Silva JS. The dual role of p55 tumour necrosis factor-alpha receptor in Actinobacillus actinomycetemcomitans-induced experimental periodontitis: host protection and tissue destruction. Clin Exp Immunol 2007; 147:128-38; PMID:17177972
- Trombone AP, Ferreira SB, Jr., Raimundo FM, de Moura KC, Avila-Campos MJ, Silva JS, Campanelli AP, De Franco M, Garlet GP. Experimental periodontitis in mice selected for maximal or minimal inflammatory reactions: increased inflammatory immune responsiveness drives increased alveolar bone loss without enhancing the control of periodontal infection. J Periodontal Res 2009; 44:443-51; PMID:18973535; http://dx.doi.org/10.1111/j.1600-0765.2008.01133.x
- Queiroz-Junior CM, Madeira MF, Coelho FM, de Oliveira CR, Candido LC, Garlet GP, Teixeira MM, de Souza Dda G, Silva TA. Experimental arthritis exacerbates Aggregatibacter actinomycetemcomitans-induced periodontitis in mice. J Clin Periodontol 2012; 39:608-16; PMID:22582749; http://dx.doi.org/10.1111/j.1600-051X.2012.01886.x
- Queiroz-Junior CM, Madeira MF, Coelho FM, Costa VV, Bessoni RL, Sousa LF, Garlet GP, Souza Dda G, Teixeira MM, Silva TA. Experimental arthritis triggers periodontal disease in mice: involvement of TNF-alpha and the oral Microbiota. J Immunol 2011; 187:3821-30; PMID:21890656; http://dx.doi.org/10.4049/jimmunol.1101195
- Trombone AP, Claudino M, Colavite P, de Assis GF, Avila-Campos MJ, Silva JS, Campanelli AP, Ibanez OM, De Franco M, Garlet GP. Periodontitis and arthritis interaction in mice involves a shared hyper-inflammatory genotype and functional immunological interferences. Genes Immun 2010; 11:479-89; PMID:20428191; http://dx.doi.org/10.1038/gene.2010.13
- Hernandez M, Dutzan N, Garcia-Sesnich J, Abusleme L, Dezerega A, Silva N, Gonzalez FE, Vernal R, Sorsa T, Gamonal J. Host-pathogen interactions in progressive chronic periodontitis. J Dent Res 2011; 90:1164-70; PMID:21471325; http://dx.doi.org/10.1177/0022034511401405
- Silva N, Dutzan N, Hernandez M, Dezerega A, Rivera O, Aguillon JC, Aravena O, Lastres P, Pozo P, Vernal R, et al. Characterization of progressive periodontal lesions in chronic periodontitis patients: levels of chemokines, cytokines, matrix metalloproteinase-13, periodontal pathogens and inflammatory cells. J Clin Periodontol 2008; 35:206-14; PMID:18269660; http://dx.doi.org/10.1111/j.1600-051X.2007.01190.x
- Wang J, Jiang Y, Chen W, Zhu C, Liang J. Bacterial flora and extraradicular biofilm associated with the apical segment of teeth with post-treatment apical periodontitis. J Endod 2012; 38:954-9; PMID:22703660; http://dx.doi.org/10.1016/j.joen.2012.03.004
- Ricucci D, Siqueira JF, Jr. Biofilms and apical periodontitis: study of prevalence and association with clinical and histopathologic findings. J Endod 2010; 36:1277-88; PMID:20647081; http://dx.doi.org/10.1016/j.joen.2010.04.007
- Kinane DF, Hart TC. Genes and gene polymorphisms associated with periodontal disease. Crit Rev Oral Biol Med 2003; 14:430-49; PMID:14656898; http://dx.doi.org/10.1177/154411130301400605
- Hajishengallis G. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol 2014; 35:3-11; PMID:24269668; http://dx.doi.org/10.1016/j.it.2013.09.001
- Nonnenmacher C, Dalpke A, Mutters R, Heeg K. Quantitative detection of periodontopathogens by real-time PCR. J Microbiol Methods 2004; 59:117-25; PMID:15325758; http://dx.doi.org/10.1016/j.mimet.2004.06.006
- Trevilatto PC, Line SR. Use of buccal epithelial cells for PCR amplification of large DNA fragments. J Forensic Odontostomatol 2000; 18:6-9; PMID:11324090
- Carvalho FM, Tinoco EM, Deeley K, Duarte PM, Faveri M, Marques MR, Mendonca AC, Wang X, Cuenco K, Menezes R, et al. FAM5C contributes to aggressive periodontitis. PLoS One 2010; 5:e10053; PMID:20383335; http://dx.doi.org/10.1371/journal.pone.0010053
- Claudino M, Trombone AP, Cardoso CR, Ferreira SB, Jr., Martins W, Jr., Assis GF, Santos CF, Trevilatto PC, Campanelli AP, Silva JS, et al. The broad effects of the functional IL-10 promoter-592 polymorphism: modulation of IL-10, TIMP-3, and OPG expression and their association with periodontal disease outcome. J Leukoc Biol 2008; 84:1565-73; PMID:18725394; http://dx.doi.org/10.1189/jlb.0308184
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 2006; 38:209-13; PMID:16415888; http://dx.doi.org/10.1038/ng1706
- Rodriguez S, Gaunt TR, Day IN. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. AmJ Epidemiol 2009; 169:505-14; PMID:19126586; http://dx.doi.org/10.1093/aje/kwn359
- Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics 2006; 22:1928-9; PMID:16720584; http://dx.doi.org/10.1093/bioinformatics/btl268
