P. aeruginosa is the fifth most frequent pathogen worldwide, the second in nosocomial pneumonia, the third in urinary infections, the forth in post-surgical infections and the seventh pathogen responsible for sepsis.Citation1 It is also the major cause of mortality in cystic fibrosis patients and the most prevalent Gram-negative multidrug-resistant bacteria in the airway of mechanically ventilated patientsCitation2 and in pediatric patients hospitalized in intensive care units (ICU).Citation1 Infections by P. aeruginosa are diverse and difficult to treat due to its intrinsic ability to develop resistance under antibiotic pressure and to produce a variety of virulence factors (VF), like adhesins, proteases, phenazines and exotoxins.Citation3 Several studies report that the presence or expression of virulence traits is related to resistance to antibiotics.Citation2,4,5,6 Some suggest these relations are antagonisticCitation5,6 while others report enhanced resistance in isolates with high levels of VF.Citation2,4 Also, literature indicates that inhibitors of specific VF may be good therapeutic alternatives to treat P. aeruginosa infections.Citation7,8 With present work, we intend to address possible associations between VF and antibiotic resistance in P. aeruginosa, aiming to provide information that may be useful for the development of alternative therapies using VF inhibitors.
Seventy-six P. aeruginosa clinical isolates were randomly selected from sputum (43%), respiratory tract aspirates (21%), urine (20%), exudates (12%) and blood (4%) of inpatients and outpatients of a Portuguese central hospital, with consent from the patients. Antibiotic susceptibility tests were performed by disk diffusion method, using imipenem (IP), meropenem (MP), ceftazidime (CAZ), cefepime (FEP), aztreonam (AZT), piperacillin (PIP), amikacin (AMK) and ciprofloxacin (CIP). Results were interpreted according to CLSI recommendations,Citation9 with intermediate results considered resistant. MDR classification was performed according to Magiorakos et al. (2011).Citation10 Twelve virulence phenotypic characteristics were screened: swimming, swarming, twitching motility and production of rhamnolipids,Citation11 elastase,Citation12 protease,Citation13 lipase, lecithinaseCitation14 and pyocyanin.Citation15 Production of phospholipase C, pyoverdine and biofilms were semi-quantified and classified as described elsewhere,Citation16,17,18 considering 4 classes of production: absent, reduced, moderate and high. Screening for the genes flaG, orfF, pilA, pilB, associated to motility; lecA, lecB, involved in lectin production; apr, lasA, lasB, encoding protease and elastases; phzH, phzM, phzS, phzI, phzII, from the biosynthetic pathway of phenazines; exoA, encoding exotoxinA and exoS, exoT, exoU and exoY, encoding type III secretory system effector proteins, was carried out as previously described.Citation3 exoA primers annealing temperature was 68°C with expected amplified fragments of 368 bp.Citation19 Data were analyzed using SPSS® version 21.0 (IBM) software. Logistic regression (LR) modeling of antibiotic resistance as function of VF presence was performed retaining the predictors statistically significant in Wald test (p < 0.05). Models were selected according to the usual fit criteria: overall significance of the model (p < 0.05), Cox and Snell coefficient, Nagelkerke coefficient, Hosmer and Lemeshow test (p > 0.05) and the percentage of correctly classified cases.Citation20 Although using the entire sample for LR analysis, we previously checked the isolates' clonality by PFGE, as previously describedCitation3 and performed a preliminary LR analysis on the selected clones. With low robustness results provided by the used fit criteria, we tested the VF association to ATB by qui-square test (p < 0.05) of the clones and compared it with the entire sample. Since no differences were observed in the VF-ATB association results of the clones and the entire sample, we performed a final LR analysis with the entire sample, that returned more robust results, presented in .
Table 1. Odds ratios for antibiotic resistance predicted by the presence of virulence factors (phenotypes or genes) in clinical Pseudomonas aeruginosa isolates
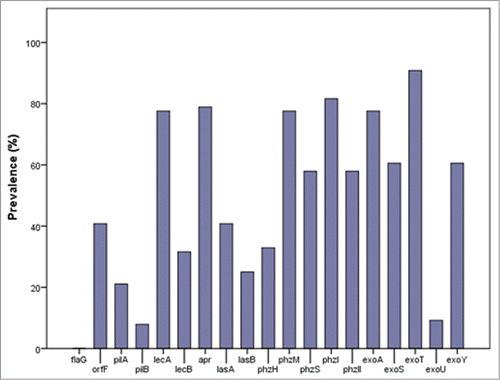
The studied P. aeruginosa isolates showed high resistance levels, with 52.6% (n = 40) classified as multidrug-resistant.Citation10 More than half of the population was resistant to IP, AZT, PIP and CIP (62.3%, 61%, 61% and 59.7%, respectively); 49.4% resisted to MP; 41.6% to CAZ and 40.3% to FEP. AMK was the antibiotic with better activity, with a resistance rate of 9.1%. and present the prevalence of the tested virulence phenotypes and genes, respectively. All clinical isolates were able to swim and other binary phenotypes ranged from 84.4% (twitching) to 44.2% (pyocyanin) (). 90.9% of all isolates presented high production of biofilms, while phospholipase C and pyoverdine reduced production were more common (). All virulence genes were observed, except flaG (). exoT (89.6%) and phzI (80.5%) were the most prevalent genes and exoU (9.1%) and pilB (7.8%) the least. However, it is important to highlight that PCR-failure in protein variants is an acknowledged constraint of the technique. Multiple sets of primers or DNA hybridization could have been used to confirm the negative results, but technical constraints impeded its application in this prospective study. Although PCR duplicate reactions of negative results, analysis of the DNA quality and quantity prior to the reactions and use of positive controls in every PCR-reaction were routinely performed during the study, some false negative results could still have occurred and are worth to explore in future similar studies.
Figure 1. Prevalence (%) of the tested virulence phenotypes in the clinical P. aeruginosa isolates: a – nominal phenotypes (presence or absence); b – ordinal phenotypes (absent, reduced, moderate or high production).
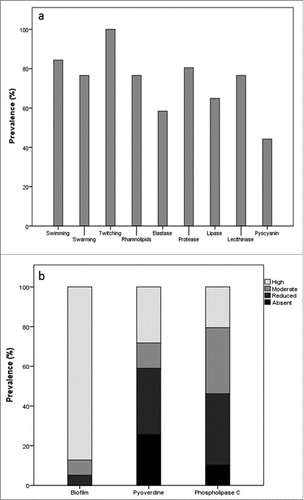
Figure 2. Prevalence (%) of the tested virulence encoding genes in the clinical P. aeruginosa isolates.
Results from LR are presented in . AMK did not provide good regression models, due to its low resistance rate, being excluded from the study. Pyoverdine was the only VF present in all LR models, with its moderate or high production predicting the resistance to all antibiotics except PIP. Pyoverdine is a siderophore involved in iron acquisition, participate in biofilm formationCitation21 and its chelating activity may be important in antibiotics resistance. A positive association between pyoverdine production and antibiotic resistance in P. aeruginosa was previously demonstrated,Citation4 which corroborates our findings. lecA gene was also a good predictor of resistance to the majority of antibiotics and lecB gene had an opposite influence (). Balasubramanian et al.Citation22 evidenced the co-regulation of biofilm formation, where lectins participate, with resistance to β-lactams. Although not finding in the literature any evidence that explains the opposite behavior of lecA and lecB genes, the different biochemical conformations of LecA and LecB may have some influence in this difference, as they present distinct specific affinity to galactose and fucose, respectively, but further studies are required. Being both present in the cytoplasm and in the outer membrane of P. aeruginosa, apart from participating in biofilm formation, lectins also have different roles in bacterial virulence, with LecA more associated to cytotoxic effects and higher absorption of exotoxin A and LecB to pilus biogenesis and protease IV activity.Citation8 These distinct virulence mechanisms presumably associated with distinct predictive drug resistances, as found in our study, deserve future attention as they may contain new pathways for drug targeting.
Rhamnolipids production, also involved in biofilm formation,Citation22 was considered a good predictor for cephalosporins susceptibility and CIP resistance. The diminished expression of rhamnolipids in isolates with penicillinases and cephalosporinases has been reported,Citation6 which concurs with our results. Regarding CIP resistance, one of the resistance mechanisms is the over-production of efflux pumps. Jeannot et al.Citation5 refer rhamnolipids diminished expression in mutants with over-production of the efflux pump MexCD-OprJ, responsible for CIP efflux. In current study, presence of rhamnolipids was considered a good CIP resistance predictor, contradicting these authors. Other VF that intervened as antibiotic resistance predictors were only significant in few regression models (), for which their overall relevance is lower. Also, we must highlight that the majority of the tested virulence phenotypes and genes were not considered statistical significant predictors of resistance.
Overall, the observed predictive relation between VF genotypes or phenotypes and ATB resistance may suggest possible mechanistic relations between VF and ATB resistance that deserve being explored. It is relevant to emphasize that some VF associated to biofilm formation was considered good predictors of ATB resistance. Considering the fact that biofilm forms of pathogens are generally known to better resist antibiotherapy,Citation3 the mechanisms involved in biofilm formation may have unexplored ATB resistance mechanisms that deserve higher attention in the future.
Some authors refer that the use of compounds that inhibit or attenuate the action of specific VF is a good therapeutic alternative for the treatment of P. aeruginosa infections.Citation7,8 However, this strategy can only be effective if the targeted VF is actually present in the bacterial cell, particularly in drug resistance strains. Otherwise the currently available antibiotics still remain as a good therapeutic option, as bacteria can be destroyed by them, leaving the new drugs to be used only when needed. This is a strategy of paramount importance to prevent development of resistance to new pharmaceutical chemicals. Regarding present results, suggesting a significant role of pyoverdine in P. aeruginosa resistance to several antibiotics, we think it is relevant to replicate similar studies in bigger samples of clinical populations of diverse origins to confirm this observation and provide further information for clinical practice and for the development of VF inhibitors as new therapeutic strategies. These studies should focus mainly in multidrug resistant isolates, particularly extensively and pan-drug resistant strains, as those are the more concerning,Citation10 due to the eminent lack of therapeutic options to treat the infections they cause. Similar approach for other pathogens should also be performed, as it may also provide relevant information to direct research efforts on new virulence inhibition drugs.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This study was supported by a doctoral fellowship (SFRH/BD/46668/2008) of the Portuguese Foundation for Science and Technology to Sónia Gonçalves Pereira.
References
- Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromossomally encoded resistance mechanisms. Clin Microbiol Rev 2009; 22: 582-610; PMID:19822890; http://dx.doi.org/10.1128/CMR.00040-09
- Fricks-Lima J, Hendrickson CM, Allgaier A, Zhuo H, Wiener-Kronish JP, Lynch SV, Yang K. Differences in biofilm formation and antimicrobial resistance of Pseudomonas aeruginosa isolated from airways of mechanically ventilated patients and cystic fibrosis patients. Int J Antimicrob Agents 2011; 37: 309-15; PMID:21382698; http://dx.doi.org/10.1016/j.ijantimicag.2010.12.017
- Pereira SG, Rosa AC, Ferreira AS, Moreira LM, Proença DN, Morais PV, Cardoso O. Virulence factors and infection ability of Pseudomonas aeruginosa isolates from a hydropathic facility and respiratory infections. J Appl Microbiol 2014; 116: 1359-68; PMID:24484457; http://dx.doi.org/10.1111/jam.12463
- Finlayson EA, Brown PD. Comparison of antibiotic resistance and virulence factors in pigmented and non-pigmented Pseudomonas aeruginosa. West Indian Med J 2011; 60: 24-32; PMID:21809707
- Jeannot K, Elsen S, Köhler T, Attree I, Van Delden C, Plésiat P. Resistance and virulence of Pseudomonas aeruginosa clinical strains overproducing the MexCD-OprJ efflux pump. Antimicrob Agents Chemother 2008; 52: 2455-62; PMID:18474583; http://dx.doi.org/10.1128/AAC.01107-07
- Ramisse F, Van Delden C, Gidenne S, Cavallo J, Hernandez E. Decreased virulence of a strain of Pseudomonas aeruginosa O12 overexpressing a chromosomal type 1 β-lactamase could be due to reduced expression of cell-to-cell signaling dependent virulence factors. FEMS Immunol Med Microbiol 2000; 28: 241-5; PMID:10865177
- Cathcart GRA, Quinn D, Greer B, Harriot P, Lynas JF, Gilmore BF, Walker B. Novel inhibitors of the Pseudomonas aeruginosa virulence factor LasB: a potential therapeutic approach for the attenuation of virulence mechanisms in pseudomonal infection. Antimicrob Agents Chemother 2011; 55: 2670-8; PMID:21444693; http://dx.doi.org/10.1128/AAC.00776-10
- Chemani C, Imberty A, de Bentzmann S, Pierre M, Wimmerová M, Guery BP, Faure K. Role of LecA and LecB lectins in Pseudomonas aeruginosa-induced lung injury and effect of carbohydrate ligands. Infect Immun 2009; 77: 2065-75; PMID:19237519; http://dx.doi.org/10.1128/IAI.01204-08
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Nineteenth Informational Supplement. M100-S19. Wayne, PA, 2009
- Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2011; 18: 268-81; PMID:21793988; http://dx.doi.org/10.1111/j.1469-0691.2011.03570.x
- Inoue T, Shingaki R, Fukui K. Inhibition of swarming motility of Pseudomonas aeruginosa by branched-chain fatty acids. FEMS Microbiol Lett 2008; 281: 81-6; PMID:18318842; http://dx.doi.org/10.1111/j.1574-6968.2008.01089.x
- Smibert RM, Krieg NR. General characterization. Washington DC: American Society for Microbiology, c1981, p. 409-433 Gerhardt P, Murray RGE, Costilow RNE, Nester W, Wood WA, Krieg NR, Phillips GH, editors. Manual methods for general microbiology
- Finnan S, Morrissey JP, O'Gara F, Boyd EF. Genome diversity of Pseudomonas aeruginosa isolates from cystic fibrosis patients and the hospital environment. J Clin Microbiol 2004; 42: 5783-92; PMID:15583313; http://dx.doi.org/10.1128/JCM.42.12.5783-5792.2004
- Janda JM, Bottone EJ. Pseudomonas aeruginosa enzyme profiling: predictor of potential invasiveness and use as an epidemiological tool. J Clin Microbiol 1981; 14: 55-60; PMID:6790569
- Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol 2001; 183: 6454-65; PMID:11591691; http://dx.doi.org/10.1128/JB.183.21.6454-6465.2001
- Berka RM, Gray GL, Vasil ML. Studies of phospholipase C (heat-label hemolysin) in Pseudomonas aeruginosa. Infect Immun 1981; 34: 1071-4; PMID:6800952
- Choy MH, Stapleton F, Willcox MDP, Zhu H. Comparison of virulence factors in Pseudomonas aeruginosa strains isolated from contact lens- and non-contact lens-related keratitis. J Med Microbiol 2008; 57: 1539-46; PMID:19018027; http://dx.doi.org/10.1099/jmm.0.2008/003723-0
- McMorran BJ, Kumara HMCS, Sullivan K, Lamont IL. Involvement of a transformylase enzyme in siderophore synthesis in Pseudomonas aeruginosa. Microbiology 2001; 147: 1517-24; PMID:11390682
- Kaszab E, Kriszt B, Atzél B, Szabó G, Szabó I, Harkai P, Szoboszlay S. The occurrence of multidrug-resistant Pseudomonas aeruginosa on hydrocarbon-contaminated sites. Microb Ecol 2010; 59: 37-45; PMID:19597862; http://dx.doi.org/10.1007/s00248-009-9551-7
- Field A, Logistic regression. London: Sage Publications Ltd, c2005, p. 760-813 Field A, editor. Discovering Statistics Using SPSS Statistics, 2th edition
- Lamont IL, Konings AF, Reid DW. Iron acquisition by Pseudomonas aeruginosa in the lungs of patients with cystic fibrosis. Biometals 2009; 22: 53-60; PMID:19130260; http://dx.doi.org/10.1007/s10534-008-9197-9
- Balasubramanian D, Schneper L, Kumari H, Mathee K. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res 2012; 41: 1-20; PMID:23143271; http://dx.doi.org/10.1093/nar/gks1039
