Intracellular pathogens manipulate host cells in myriad ways. Coxiella burnetii, a slow-growing (generation time ˜11 h), obligately intracellular bacterial pathogen inhibits host cell apoptosis in order to generate a replicative vacuole. This subversion is critical to its pathogenicity; however, very little mechanistic details are known about how the pathogen usurps the host cell's regulatory machinery. It is imperative that we gain better insights into Coxiella's pathogenicity because the select agent causes human Q fever and chronic endocarditis.Citation1 C. burnetii is found worldwide, and a recent outbreak in Netherlands involved thousands of human cases and resulted in the culling of a large number of goats (a) primary reservoir.Citation2 Additionally, untreated chronic Q fever, usually presented as endocarditis, is associated with high mortality rates.Citation1
Apoptosis occurs through a complex cascade of events that involves both extrinsic and intrinsic pathways. The extrinsic pathway acts through the cell surface death receptor signaling, whereas the intrinsic pathway is activated through the mitochondrial membrane proteins.Citation3 Coxiella is known to manipulate multiple factors along both the extrinsic and intrinsic apoptosis pathways.Citation4 For instance, Coxiella activates anti-apoptosis kinases AKT, ERK1/2 and PKA that prevent the onset of apoptosis, and induces the expression of several apoptosis-related microRNAs.Citation5-8 Additionally, 3 effector proteins (AnkG, CaeA and CaeB) secreted through Coxiella's Type IV Secretion System (T4SS) inhibit apoptosis, although their mechanisms of action are not fully understood.Citation9,10 AnkG was shown to interact with the mitochondrial protein p32, indicating that it inhibits intrinsic apoptosis by decreasing cytochrome c release. Intriguingly, this protein (along with p32) is translocated to the host cell nucleus for as yet unknown functions.Citation9,10 CaeA and CaeB inhibit apoptosis at the mitochondrial level but in a p32-independent fashion.Citation11 In this issue, Bisle et al. further clarifies the mechanism of action of CaeA. They show that the effector protein inhibits both extrinsic and intrinsic apoptotic pathways, and reveal that its anti-apoptotic activity is dependent on an EK (glutamic acid, lysine) repeat motif.Citation12
Throughout its prolonged infectious cycle, C. burnetii continuously manipulates host processes, including host cell death, thus creating a stable intracellular niche.Citation3 This is an important virulence property that allows this obligate intracellular pathogen to maintain host viability despite inducing stress that would normally activate apoptosis.Citation13 Coxiella encodes a T4SS that is essential for intracellular replication,Citation14,15 and over a hundred effector proteins secreted through this apparatus have been identified (for e.g.,Citation16-21). However, the functions of most of these substrates are not understood. Legionella pneumophila, a close relative, also encodes a similar secretion system, but only a few T4SS substrates are shared between the 2 pathogens, making it difficult to use a comparative approach to understand the functions of effector proteins secreted by Coxiella into the host cell.Citation9,22 Similarly, different strains of C. burnetii encode diverse repertoires of effector proteins,Citation23 indicating that T4SS substrates evolve rapidly in a species- or even strain-specific manner. In accord with this view, Bisle et al. show that caeA from 25 strains of C. burnetii contain variable number of EK repetition motif, and that the number of EK repeats embedded within the coiled-coil domain of CaeA appears to have an effect on apoptosis inhibition.Citation12
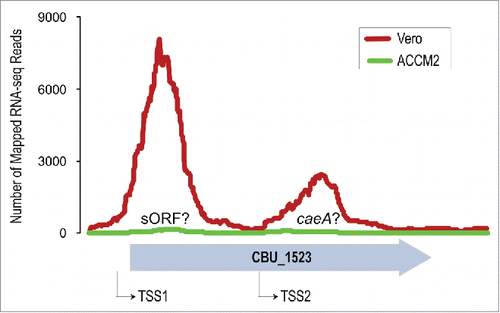
Because C. burnetii is a highly infective pathogen that requires a Biosafety Level-3 facility for conducting experiments, most studies have been performed using the Nine Mile Phase II (NMII) strain, which can be handled under Biosafety Level-2 containment. But the genome of the NMII strain has not yet been sequenced, and hence most researchers utilize the published genome of Nine Mile Phase I strain (NMI), which sometimes lead to inconclusive results. For instance, caeA (CBU_1524) is not annotated as a protein-coding gene in the current version of the NMI genome (NC_002971.3), instead, it is now part of the pseudogene CBU_1523. To check whether the putative pseudogene is transcribed, I analyzed a transcriptome dataset generated from Vero cells infected with C. burnetii NMII.Citation24 As shown in , CBU_1523 is indeed expressed during intracellular growth, and appears to contain 2 separate transcription start sites that correspond, respectively, to a small open reading frame of unknown function, and to caeA. Additionally, both proteins seem to have important infection-specific roles because they are expressed 5–10 fold higher intracellularly than in ACCM2 medium (). The RNA-seq data also suggests that several other presumed pseudogenes in NMII could be functional, thereby explaining —at least partially—why transposon insertions in several presumed pseudogenes resulted in reduced intracellular growth.Citation25 Based on these observations, it is clear that a fully annotated genome of the laboratory workhorse NMII strain is urgently required. Having access to the correct genome sequence will further the tremendous progress the research community has already made in comprehending Coxiella's biology and virulence using genome-scale methodologies (for e.g.,Citation26-30).
Figure 1. Expression patterns of the annotated pseudogene CBU_1523 (gray wide arrow) in C. burnetii grown in the cell-free medium ACCM2 (green line), and in Vero cells (red line) for 72 hours. Two apparent transcription start sites (TSSs) that possibly correspond to a small open reading frame (sORF) and to caeA are indicated by black thin arrows.
Although it is clear that CaeA traverses to host cell nucleus and blocks apoptosis, the complete molecular details of this process are yet to be worked out. Further studies are required to identify the proteins that possibly interact with CaeA, and to determine whether the interacting proteins are required for nuclear localization and/or for inhibiting apoptosis. Furthermore, the recent development of a genetic system,Citation31 should enable the generation of caeA-deletion strains of C. burnetii, and to directly analyze the importance of this protein to Coxiella-mediated inhibition of apoptosis. This is especially important because, as noted by the authors,Citation12 the orthologs of caeA in a majority of Coxiella strains seem to contain frame-shift mutations that would render CaeA non-functional. Collectively, the approaches used by Luhrmann and colleaguesCitation9-13 should serve as a model for future studies aimed at uncovering the functions of all effector proteins secreted by C. burnetii. Because of the lack of conservation of Coxiella effector proteins both within and between species, this will be a slow and painstaking process, which could be accelerated using genome- and transcriptome-enabled approaches. This is an important endeavor that will enlighten us about the distinct biology of an enigmatic pathogen that has uniquely figured out how to make a living in an inhospitable lysosome-derived intracellular compartment.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Maurin M, Raoult D. Q fever. Clin Microbiol Rev 1999; 12:518-53; PMID:10515901.
- Dijkstra F, van der Hoek W, Wijers N, Schimmer B, Rietveld A, Wijkmans CJ, Vellema P, Schneeberger PM. The 2007–2010 Q fever epidemic in The Netherlands: characteristics of notified acute Q fever patients and the association with dairy goat farming. FEMS Immunol Med Microbiol 2012; 64:3-12; PMID:22066649; http://dx.doi.org/10.1111/j.1574-695X.2011.00876.x.
- Voth DE, Howe D, Heinzen RA. Coxiella burnetii inhibits apoptosis in human THP-1 cells and monkey primary alveolar macrophages. Infect Immun 2007; 75:4263-71; PMID:17606599; http://dx.doi.org/10.1128/IAI.00594-07.
- Moffatt JH, Newton P, Newton HJ. Coxiella burnetii: turning hostility into a home. Cell Microbiol 2015; 17:621-31; PMID:25728389; http://dx.doi.org/10.1111/cmi.12432.
- Macdonald LJ, Graham JG, Kurten RC, Voth DE. Coxiella burnetii exploits host cAMP-dependent protein kinase signaling to promote macrophage survival. Cell Microbiol 2014; 6:146-59; http://dx.doi.org/10.1111/cmi.12213.
- Voth DE, Heinzen RA. Sustained activation of Akt and Erk1/2 is required for Coxiella burnetii antiapoptotic activity. Infect Immun 2009; 77:205-13; PMID:18981248; http://dx.doi.org/10.1128/IAI.01124-08.
- Hussain SK, Broederdorf LJ, Sharma UM, Voth DE. Host kinase activity is required for Coxiella burnetii parasitophorous vacuole formation. Front Microbiol 2010; 1:137; PMID:21772829; http://dx.doi.org/10.3389/fmicb.2010.00137.
- Millar JA, Valdes R, Kacharia FR, Landfear SM, Cambronne ED, Raghavan R. Coxiella burnetii and Leishmania mexicana residing within similar parasitophorous vacuoles elicit disparate host responses. Front Microbiol 2015; 6:794; PMID:26300862; http://dx.doi.org/10.3389/fmicb.2015.00794.
- Luhrmann A, Nogueira CV, Carey KL, Roy CR. Inhibition of pathogen-induced apoptosis by a Coxiella burnetii type IV effector protein. Proc Natl Acad Sci U S A 2010; 107:8997-9001; http://dx.doi.org/10.1073/pnas.1004380107.
- Eckart RA, Bisle S, Schulze-Luehrmann J, Wittmann I, Jantsch J, Schmid B, Berens C, Lührmann A. Antiapoptotic activity of Coxiella burnetii effector protein AnkG is controlled by p32-dependent trafficking. Infect Immun 2014; 82:2763-71; PMID:24733095; http://dx.doi.org/10.1128/IAI.01204-13.
- Klingenbeck L, Eckart RA, Berens C, Luhrmann A. The Coxiella burnetii type IV secretion system substrate CaeB inhibits intrinsic apoptosis at the mitochondrial level. Cell Microbiol 2012; 15:675-87; PMID:23126667; http://dx.doi.org/10.1111/cmi.12066.
- Bisle S, Klingenbeck L, Borges V, Sobotta K, Schulze-Luehrmann J, Menge C, Heydel C, Gomes JP, Luhrmann A. The inhibition of the apoptosis pathway by the Coxiella burnetii effector protein CaeA requires the EK repetition motif, but is independent of survivin. Virulence 2016; 7(4): 400-412; PMID:26760129.
- Luhrmann A, Roy CR. Coxiella burnetii inhibits activation of host cell apoptosis through a mechanism that involves preventing cytochrome c release from mitochondria. Infect Immun 2007; 75:5282-9; PMID:17709406; http://dx.doi.org/10.1128/IAI.00863-07.
- Carey KL, Newton HJ, Luhrmann A, Roy CR. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog 2011; 7:e1002056; PMID:21637816; http://dx.doi.org/10.1371/journal.ppat.1002056.
- Beare PA, Gilk SD, Larson CL, Hill J, Stead CM, Omsland A, Cockrell DC, Howe D, Voth DE, Heinzen RA. Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. MBio 2011; 2:1-10; http://dx.doi.org/10.1128/mBio.00175-11.
- Pan X, Luhrmann A, Satoh A, Laskowski-Arce MA, Roy CR. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 2008; 320:1651-4; PMID:18566289; http://dx.doi.org/10.1126/science.1158160.
- Chen C, Banga S, Mertens K, Weber MM, Gorbaslieva I, Tan Y, Luo ZQ, Samuel JE. Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc Natl Acad Sci U S A 2010; 107:21755-60; PMID:21098666; http://dx.doi.org/10.1073/pnas.1010485107.
- Maturana P, Graham JG, Sharma UM, Voth DE. Refining the plasmid- encoded type IV secretion system substrate repertoire of Coxiella burnetii. J Bacteriol 2013; 195:3269-76; PMID:23687269; http://dx.doi.org/10.1128/JB.00180-13.
- Weber MM, Chen C, Rowin K, Mertens K, Galvan G, Zhi H, Dealing CM, Roman VA, Banga S, Tan Y, Luo ZQ, Samuel JE. H. Identification of Coxiella burnetii type IV secretion substrates required for intracellular replication and Coxiella-containing vacuole formation. J Bacteriol 2013; 195:3914-24; PMID:23813730; http://dx.doi.org/10.1128/JB.00071-13.
- Larson CL, Beare PA, Voth DE, Howe D, Cockrell DC, Bastidas RJ, Valdivia RH, Heinzen RA. Coxiella burnetii effector proteins that localize to the parasitophorous vacuole membrane promote intracellular replication. Infect Immun 2015; 83:661-70; PMID:25422265; http://dx.doi.org/10.1128/IAI.02763-14.
- Weber MM, Faris R, McLachlan J, Tellez A, Wright WU, Galvan G, Luo ZQ, Samuel JE. Modulation of the host transcriptome by Coxiella burnetii nuclear effector Cbu1314. Microbes Infect 2016; in press; PMID:26827929.
- Broederdorf LJ, Voth DE. Cheating death: a Coxiella effector prevents apoptosis. Front Microbiol 2011; 2:43; PMID:21747783; http://dx.doi.org/10.3389/fmicb.2011.00043.
- Beare, PA, Unsworth N, Andoh M, Voth DE, Omsland A, Gilk SD, Williams KP, Sobral BW, Kupko JJ 3rd, Porcella SF, et al. Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella. Infect Immun 2009; 77:642-56; PMID:19047403; http://dx.doi.org/10.1128/IAI.01141-08.
- Warrier I, Hicks LD, Battisti JM, Raghavan R, Minnick MF. Identification of novel small RNAs and characterization of the 6S RNA of Coxiella burnetii. PLoS ONE 2014; 9:e100147; PMID:24949863; http://dx.doi.org/10.1371/journal.pone.0100147.
- Martinez E, Cantet F, Fava L, Norville I, Bonazzi M. Identification of OmpA, a Coxiella burnetii protein involved in host cell invasion, by multi-phenotypic high-content screening. PLoS Pathog 2014; 10:e1004013; PMID:24651569; http://dx.doi.org/10.1371/journal.ppat.1004013.
- Seshadri R, Paulsen IT, Eisen JA, Read TD, Nelson KE, Nelson WC, Ward NL, Tettelin H, Davidsen TM, Beanan MJ et al. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc Natl Acad Sci U S A 2003; 100:5455-60; PMID:12704232; http://dx.doi.org/10.1073/pnas.0931379100.
- Omsland A, Beare PA, Hill J, Cockrell DC, Howe D, Hansen B, Samuel JE, Heinzen RA. Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl Environ Microbiol 2011; 77:3720-5; PMID:21478315; http://dx.doi.org/10.1128/AEM.02826-10.
- Smith TA, Driscoll T, Gillespie JJ, Raghavan R. A Coxiella-like Endosymbiont is a potential vitamin source for the Lone Star Tick. Genome Biol Evol 2015; 7:831-8; PMID:25618142; http://dx.doi.org/10.1093/gbe/evv016.
- McDonough JA, Newton HJ, Klum S, Swiss R, Agaisse H, Roy CR. Host pathways important for Coxiella burnetii infection revealed by genome-wide RNA interference screening. MBio 2013; 4:e00606-12; PMID:23362322; http://dx.doi.org/10.1128/mBio.00606-12.
- Mahapatra S, Ayoubi P, Shaw EI. Coxiella burnetii Nine Mile II proteins modulate gene expression of monocytic host cells during infection. BMC Microbiol 2010; 10:244e57; http://dx.doi.org/10.1186/1471-2180-10-244.
- Beare, PA, Larson CL, Gilk SD, Heinzen RA. Two systems for targeted gene deletion in Coxiella burnetii. Appl Environ Microbiol 2012; 78:4580-9; PMID:22522687; http://dx.doi.org/10.1128/AEM.00881-12.
