ABSTRACT
Legionella pneumophila is the primary respiratory pathogen and mostly transmitted to human through water cooling systems and cause mild to severe pneumonia with high mortality rate especially in elderly both in hospitals and community. However, current Legionella risk assessments may be compromised by uncertainties in Legionella detection methods. Here, we investigated the presence of L. pneumophila mip gene in water samples collected from different hospitals cooling towers, nursing homes and building/hotels water coolants from two geographical locations of Iran (Kerman and Bam cities) during summer season of 2015 by both nested and real-time PCR methods. Analysis of the 128 water samples for presence of the mip gene by nested-PCR revealed, 18 (23%) positive cases in Kerman and 7(14%) in Bam. However, when samples were tested by real-time PCR, we identified 4 more new cases of L. pneumophila in the hospitals as well as nursing homes water systems that were missed by nested-PCR. The highest rate of contamination was detected in water obtained from hospitals cooling towers in both the cities (p≤0.05). Dendrogram analysis and clonal relationship by PCR-base sequence typing (SBT) of the L. pneumophila genomic DNAs in Kerman water samples showed close clonal similarities among the isolates, in contrast, isolates identified from Bam city demonstrated two fingerprint patterns. The clones from hospital water samples were more related to the L. pneumophila serogroup- 1.
Legionella pneumophila is the primary human pathogen and is the causative agent of Legionnaires' disease, also known as legionellosis.Citation1 Potable water and water coolant containers are important source of both nosocomial and community acquired Legionella infections.Citation2 Upon transmission to human, L. pneumophila infect and replicate within alveolar macrophages and spread to blood stream causing mortality rates approaching 30–40%.Citation3 Outbreaks have been linked to a range of sources, including natural environments such as ground water as well as in technical water carrying systems like cooling towers, household coolers, spas, showerheads and drinking water.Citation4-6 For these reasons some countries specifically regulate the surveillance and control of L. pneumophila in water regularly and assess its presence. In a survey conducted in Spain, the prevalence of L. pneumophila was found to be 66.6% of total water samples collected (449 confirmed cases of legionellosis).Citation7 Furthermore, 42% of Italian hotels water cooling systems of different sizes were contaminated by L. pneumophila.Citation8 Prevalence of L. pneumophila in water distribution systems in hospitals and public buildings of the Lublin region of eastern Poland was found to be 166 (74.77%) of hot water samples.Citation9 In other study, more than 1100 cases of legionellosis in Japan, caused by contaminated artificial whirlpool spas or natural hot springs were presented in Infectious Agents Surveillance Report 2014.Citation10
Only a few factors have been detected and characterized that contribute to survival of the L. pneumophila in eukaryotic cells. The macrophage infectivity potentiator (mip) gene is described as a virulence factor necessary for optimal intracellular survival of this bacterium.Citation11 The mip gene was first L pneumophila virulence-associated gene that required for efficient host cell infection. This gene encode a protein belongs to the class of FK 506-binding proteins catalyzing the slow cis/trans interconversion of polypeptide bonds in oligopeptides and well conserved in L. pneumophila.Citation4
Isolation of Legionella by culture method is considered the gold standard, but have several limitations namely long incubation period, the presence of viable but non-culturable (VBNC) cells and co-contamination with other microorganisms. Furthermore, the sensitivity of Legionella detection based on culture methods depends largely on the physiological state of the cells.Citation12 Preliminary evidence indicates that PCR sensitivity and specificity are comparable to those of cell culture.Citation13 Therefore, PCR- based techniques may best substitute for detection of this bacterium in water systems. Molecular methods described thus far targeted a number of sequences such as the 16S rRNA, the 23S–5S spacer regions and the mip gene of L. pneumophila.Citation14-16 However, current Legionella risk assessments may be compromised by uncertainties in Legionella detection methods.Citation16,17
There is paucity of information regarding contamination rate of L. pneumophila in water system of different sources in Iran.Citation17,18 Here, we investigated presence of L. pneumophila mip gene in a total of 128 water samples collected from different hospitals towers, nursing homes and building/hotels water coolants of 2 Iranian cities (Kerman and Bam) during summer season of 2015 (May to August) by both nested and real-time PCR. We also studied genetic relationship among the L. pneumophila genomic DNAs from these water samples by PCR-base sequencing method. Selection of the mip gene for screening purposes was based on its discriminatory power and frequent usage in other studies. Geographical locations and sampling sources are illustrated in . A total volume of one liter of each water sample was aseptically collected from the bottom or side of the vessels or reservoirs in 1.5 liter capacity polypropylene containers and placed in the sealed plastic bag in a temperature controlled box (the water systems were not treated with biocides). The samples were then transferred to the microbiology laboratory in less than 4 hour and kept in the refrigerator (4°C) for further analysis. Physico-chemical parameters of each water sample such as temperature, pH, turbidity, biological oxidation demand (BOD), chemical oxidation demand (COD) and total chloride content were examined according to standard method for the examination of water and wastewater treatment.Citation19 250 ml each water sample was then passed through 0.4 µm pore diameter membrane filter (Millipore, Bedford, USA), the filter coat was scraped by pipet tips and suspended in 5 ml of sterile TE-buffer (10 mM Tris-HCl, 1 mM EDTA) pH-8.0. One ml of each suspension was then transferred to sterile Eppendorf tubes (Eppendorf, Germany) and centrifuged at 7000 × g for 10 min at 4°C, supernatant discarded and pellet was kept at refrigeration condition (4°C) for further analysis. DNA extraction was carried out with a commercially available kit (Thermo Scientific, Vilnius, Lithuania) according to the manufacturer instructions. The quality of isolated DNA was measured by determination of absorbancy at the wave lengths A260 nm and A280 nm (ratio of these values between 1.7 and 1.9 indicates a high quality of the product). The conventional PCR reaction was carried out with 5 µl of a 340-bp extracted DNA fragment of mip gene (sequence was obtained from GenBank database; http://www.ncbi.nlm.nih.gov/GeneBank), 20 pmol forward (5′-AAAGGCATGCAAGACGCTAT-3′) and reverse (5′-ACGTTGCTGGCTTAGCAGTT-3′) primers (GeneRay, Shanghai, China), 2U Taq DNA polymerase (Ampliqon, Denmark) with 3 µl deoxynucleoside triphosphates (dNTPs) in 10X reaction buffer containing 1.5 mmol l−1 MgCl2 in a total volume of 25 μl. Sterile distilled water was used as the negative control. DNA ladder was a ready to use plasmid double digest sized range 100- 3000 bp obtained from SMOBIO Technology (Hsinchu, Taiwan). Specifity of the primers were checked by Primer Quest software tool (http://www.ncbi.nlm.nih.gov/GeneBank). Amplification was conducted in temperature gradient thermal cycler (Biometra-T300, Gottingen, Germany) with initial denaturation temperature at 95°C for 2 min, followed by 40 cycles of 94°C for 30 s, annealing 53°C for 30 s, extension 72°C for 30 s and a final extension at 72°C for 5 min. Nested-PCR was employed to amplify 124-bp mip DNA fragment by using 2 specific primers (forward 5′-TTTGATGGCAAAGCGTACTG-3′ and reverse 5′-TTGCAAACCACTTGGCAATA-3′) as described previously.Citation20 Here, the PCR condition was consisted of an initial denaturation at 95°C for 2 min, followed by 40 cycles at 94°C for 30 s, 55°C for 30 s, 72°C for 30 s and a final extension at 72°C for 5 min, respectively. Presence of the mip gene in all PCR experiments was checked with positive control consist of L. pneumophila serogroup-1 (GenBank accession number CR628336) kindly obtained from Department of Bacteriology, Tarbiat Modarres University, Tehran, Iran.
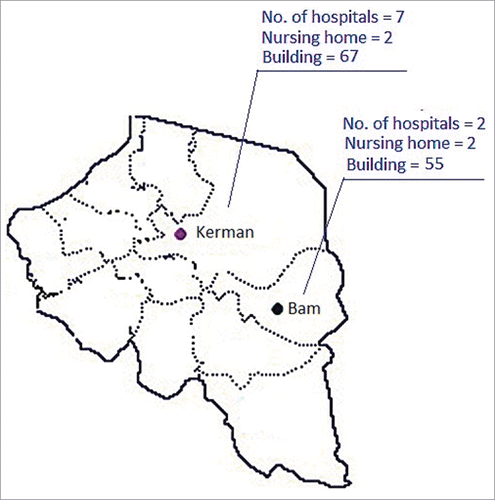
Figure 1. The map and sources of water samples analyzed for detection of L. pneumophila in this study. Both the Kerman and Bam cities were located in south east of Iran. Number of hospitals, building (homes / hotels) and nursing houses that water samples were taken are included in this figure.
The specificity of the nested-PCR was further confirmed by real-time PCR. Here, a specific set of primers were used for detection of L. pneumophila mip gene.Citation16 The reaction mixtures were consisted of 2 X real-time PCR Master Mix Green with no Rox dye (Ampliqon, Denmark) along with 5 µl DNA template in 20 µl reaction mixture composed of 2.5 μl (0.5 µM) forward (5′-ACCGAACAGCAAATGAAAGA-3′) and reverse (5′-AACGCCTGGCTTGTTTTTGT-3′) primers and 3.0 mM MgCl2. Five µl of nuclease-free DD/W was taken as a negative control for each run. The experimental LightCycler protocol consisted of an initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 10 s, annealing for 15 s at 55°C, extension for 30 s at 72°C and a cooling step (40°C for 2 min) with acquisition of data following extension step using Rotor-Gene 6000 (Corbett Research, Australia). PCR performance was confirmed to be reproducible at the threshold cycles (Ct) <35. For determination sensitivity and specifity of primers, a serial dilution in the range from 100 ng to 1 fg of purified L. pneumophila serogroup -1 DNA was used. All samples were tested by 3-fold repetitions. Results were analyzed with the use of standard slope, provided by the producer (slope points were: 100,000, 10,000, 1,000, 100, 10, 1). Following amplification, melting curve was performed on the SYBER channel (at gain 70) using a ramping rate 0.5° C/10 s for 65–95°C. Melt (65–95°C) hold secs on the first step, hold 5 secs on next steps. One cycle melting curve was carried out and optimized using Rotor Gene software (QIAGEN, Hilden, Germany) for analysis of PCR amplification as described by manufacturer guidelines. L. pneumophila serogroup -1 mip gene was used to generate standard curve. The experimental points aligned in a straight line and correlation coefficients (R) was ascertained at 0.91658 (R2=0.84) by following calibration equation was obtained Y = 0.34908x + 29.66623.Citation8,16 The slope of 0.34908 corresponded to an amplification efficiency of 99.8%. Tm (temperature melting) value of the products was 87°C.
In order to confirm the in-silico findings, we performed sequencing of 340-bp PCR product of the mip gene amplified from all positive cases. Sequencing was carried out by the Bioneer Company (Seoul, Korea) with Sanger dideoxy chain termination method using Applied Biosystems 3730/3730Xl DNA Analyzers (Applied Biosystems, Foster City, CA, USA). Both strands of the amplicons were sequenced. For sequencing we used PFU polymerase instead of Taq DNA polymerase. Similarity searches for the Legionella mip gene sequences verified using database provided by the European Working Group for Legionella Infections (http://www.ewgli.org/). Genetic relationship of L. pneumophila genomic DNAs among Kerman and Bam isolates were performed directly by PCR- sequence based typing (SBT) method as described previously.Citation22 Bands were arbitrarily chosen to range from 200 to 4000 bp. Strains that had fingerprinting patterns more than one band difference in terms of size or intensity were considered distinct types. Banding patterns were analyzed by UPGMA (unweighted pair-group method with arithmetic averages) clustering method using Gel Compare II software version 4.0 (Applied Maths, Sint-Matens-latem, Belgium).Citation21 Degree of homology was determined by Dice coefficient. Isolates that clustered >95% were considered related. All statistical analysis was performed using SPSS 17.0 (SPSS, Chicago, IL, USA). p-value greater than 0.05 was considered as statistically significant for 2-tailed test.
The average water temperature in Bam city (38 ± 0.3°C) was higher as compared to Kerman (33 ± 0.3°C). The pH of both water samples were alkaline (pH- 8.7). Turbidity was higher in Bam (NTU = 14.1), however, total hardness was higher in water samples collected from Kerman (THD = 699.0 mg l−1) as compared to Bam city (THD = 563.1 mg l−1). Average chloride ion concentration of water samples obtained from Kerman (826 mg l−1) was higher than Bam (787 mg l−1). Water conductivity was more or less same in both the cities (4750 µScm−1). The average BOD and COD of both waters were approximately similar (7 ± 0.2 mg l−1). Analysis of the water samples for presence of the mip gene by nested-PCR revealed, 18 (23%) positive cases in Kerman and 7 (14%) in Bam (, ). However, when samples were tested by real-time PCR, we identified 4 more new cases L. pneumophila in the hospitals as well as nursing homes water samples that were missed by nested-PCR. The highest rate of contamination was detected in water obtained from hospitals cooling towers in both the cities. For building (homes/hotels), the number was much lower (p ≤ 0 .05). The data obtained by real-time PCR are shown in and . This results were further supported by sequencing of the mip DNA extracted from each L. pneumophila isolates. A blast search of the GenBank database demonstrates a high specificity, with the only cross-reacting bacteria being L. worsleiensis (GenBank accession number LWU60164) with 89% homology. Pairwise alignments with mip sequences in NCBI database searched by neighbor joining method showed 98% of homology with L. pneumophila accession number NC_002942.5. The dendrogram analysis and clonal relationship of L. pneumophila genomic DNA showed 2 patterns of fingerprints among isolates obtained from Bam water samples (). Indeed, DNAs from water samples 6 and 7 demonstrated different clonal patterns while, clone 5 was a singleton. In contrast, most members of the DNA isolated from Kerman hospital and nursing home water coolants especially isolates 9-13 were closely related (clones 1 and 18 were identical) as the predominant type, and distributed commonly throughout the majority of environmental facilities.
Figure 2. Agarose gel electrophoresis amplification of L. pneumophila by A) conventional-PCR (340bp) and B) nested –PCR (124bp) of the mip gene detected in cooling water samples in this study. Lanes 7-9 and 12 are water samples from Bam. Lane 11 is positive control (340bp). Lanes 1 and 14 are ladder consist of 100 base pairs DNA fragments. NC = negative control.
Figure 3. The real-time PCR detection of L. pneumophila mip gene isolated from water cooling systems investigated in this study. Panel: A) Real-time PCR of DNA extracted from water samples of Kerman and Bam. Panel: B) The standard curve with the CT plotted against the concentration of the starting quantity of template for each dilution
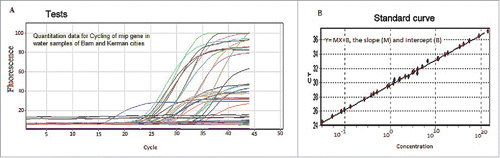
Figure 4. Dendrogram analysis of whole L. pneumophila genomic DNAs obtained from water samples of Bam and Kerman cities. Banding patterns were analyzed by UPGMA (unweighted pair-group method with arithmetic averages) clustering method using Gel Compare II software version 4.0 (Applied Maths, Sint-Matens-latem, Belgium). Degrees of homology were determined by Dice coefficient. Isolates that clustered >95% were considered related.
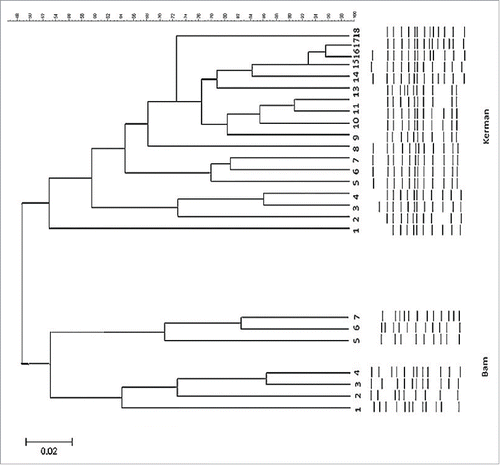
Table 1. Distribution of L. pneumophila mip gene detected in water samples collected from different cooling water systems in Kerman and Bam cities by nested and real-time PCR.
Water cooling systems are frequently used during summer season in 2 cities of Kerman and Bam situated near the central desert of Iran. No information exists on rate of contamination of different water systems in this region. For this reason we attempted to analyzed water systems of 2 main cities of this region for the presence of L. pneumophila by molecular techniques. Evaluation of physico-chemical parameters of water samples revealed average change in COD, BOD, total dissolved solids, alkaline pH and chlorine had not influenced on the presence of L. pneumophila DNA and survival of organism in different cooling water systems (P > 0.05; the χ2 test). Legionella are presently identified by comparing their 16S rRNA or mip gene sequences, with known sequences deposited in GenBank.Citation17 To validate the nested –PCR, we performed real-time PCR analysis targeted the mip gene, we found 4 more cases of L. pneumophila corresponds to approximately one-two genome more equivalent per reaction as compared to nested-PCR. Furthermore, real-time PCR offers a rapid amplification of several samples simultaneously and diminishes the likelihood of laboratory contamination.Citation22,23 Indeed, real-time PCR can supplement gold standard culture based detection of Legionella in environmental samples.Citation13 The yearly incidence of Legionnaires' disease seem to be associated with climate changes, only 4% of cases were associated with a known outbreak or possible cluster.Citation24 The dendrogram analysis of the L. pneumophila genomic DNAs investigated in this study revealed close proximity among the lineages in environmental water samples of Kerman city, while, 2 patterns of fingerprints were observed in dendrogram obtained from Bam city indicating clonal divergence in these strains. Our results showed the clones 5, 6 and 7 were isolated from hotels water samples showed close banding pattern and suggest the isolates were transferred from visiting individuals. L. pneumophila serogroup -1 had entirely different banding pattern. This may suggest that our environmental isolates especially from building/hotel water coolants are not closely related to serogroup- 1. However, 3 isolates obtained from hospital towers water samples showed almost similar fingerprint with L. pneumophila Serogroup -1. Similar cases were observed from Kerman isolates. Georghiou et al.,Citation24 studied molecular fingerprinting of Legionella species from different water systems and revealed substantial variation among the fingerprints of different Legionella species and serogroups. More limited, but distinct, polymorphisms of the fingerprint were observed among epidemiologically unrelated isolates of L. pneumophila serogroup 1. It has also been found in several studies (8, 18, 23) that endemic clones of L. pneumophila causing apparently unrelated cases of legionellosis have the same molecular genotype. In a study carried out by sequence-based typing using 6 loci, flaA, pilE, asd, mip, mompS, and proA in Japan, indicated that all 10 isolates from cooling towers clustered into a unique type, which was distinct from strains of other environmental sources.Citation25
In conclusion, L. pneumophila represents a potential pathogen, especially for some risk groups such as elderly, ICU patients. This study highlights the need of continuous monitoring, and risk assessment of water supplies of large buildings and hospitals. One advantage of the molecular detection is that it enables rapid interventions to limit infections caused by L. pneumophila. Molecular typing showed closed lineage similarities between environmental isolates but hospital isolates yielded distinct clusters and similarity with serogroup-1. Further research must be carried out on stereo-structure of MIP protein.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The physico-chemical parameters analyses of waters obtained in this study were performed by environmental quality center, Kerman, Iran. We also thank hospital stuffs and owners of buildings / hotels for their cooperation for collection of water samples.
Funding
This study was funded by Bam University of Medical Sciences, Iran (grant number 11/94).
Reference
- Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol 1980; 33(12):1179-83; PMID:7451664
- Newton HJ, Ang DK, van Drie IR, Hartland EL. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev 2010; 23:274-298; PMID:20375353; http://dx.doi/10.1128/CMR.00052-09
- Dominguez A, Alvarez J, Sabria M, Carmona G, Torner N, Oviedo M, Cayla J, Minguell S, Barrabeig I, Sala M, et al. Factors influencing the case-fatality rate of Legionnaires' disease. Int J Tuberc Lung Dis 2009; 13(3):407-12; PMID:19275805
- Walser SM, Gerstnera DG, Brennera B, Höllerb C, Liebla B, Her CE. Assessing the environmental health relevance of cooling towers – A systematic review of legionellosis outbreaks. Int J Hyg Environ Health 2014; 217(2-3):145-54; PMID:24100053; http://dx.doi.org/10.1016/j.ijheh.2013.08.002
- Greig JE, Carnie JA, Tallis GF, Ryan NJ, Tan AG, Gordon IR, Zwolak B, Leydon JA, Guest CS, Hart WG. An outbreak of Legionnaires' disease at the Melbourne Aquarium, April 2000: investigation and case–control studies. Med J Aust 2004; 180(11):566-72; PMID:15174987
- Atlas RM. Legionella: from environmental habitats to disease pathology, detection and control. J Environ Microbiol 1999; 1:283-93; PMID: 11207747; http://dx.doi.org/10.1046/j.1462-2920.1999.00046.x
- Grúas C, Lambi S, Arruga MV. Detection of Legionella spp. and Legionella pneumophila in water samples of Spain by specific realtime PCR. Arch Microbiol 2014; 196(1):63-71; PMID:24264468; http://dx.doi.org/10.1007/s00203-013-0934-2
- Bonetta S, Ferretti E, Balocco F, Carraro E. Evaluation of Legionella pneumophila contamination in Italian hotel water systems by quantitative real-time PCR and culture methods. Appl Microbiol 2010; 108:1576-83; PMID:19796090; http://dx.doi.org/10.1111/j.1365-2672.2009.04553.x
- Sikora A, Wojtowicz-Bobin M, Koziol-Montewka M, Magrys A, Gladysz I. Prevalence of Legionella pneumophila in water distribution systems in hospitals and public buildings of the Lublin region of eastern Poland. Ann Agri Environ Med 2015; 22:195-201; PMID:26094507; http://dx.doi.org/10.5604/12321966.1152064
- Edagawa A, Kimura A, Kawabuchi-Kurata T, Adachi S, Furuhata K, Miyamoto H. Investigation of Legionella contamination in bath water samples by culture, Amoebic co-culture, and real-time quantitative PCR methods. Int J Environ Res Public Health 2015; 12(10):13118-30; PMID:26492259; http://dx.doi.org/10.3390/ijerph121013118
- Wintermeyer E, Ludwig B, Steinert M, Schmidt B, Fischer G, Hacker J. Influence of site specifically altered Mip proteins on intracellular survival of Legionella pneumophila in eukaryotic cells. Infect Immun 1995; 63:4576-83; PMID:7591108
- Mérault N, Rusniok C, Jarraud S, Gomez-Valero L, Cazalet C, Marin M, Brachet E, Aegerter P, Gaillard JL, Etienne J, et al. Specific real-time PCR for simultaneous detection and identification of Legionella pneumophila serogroup 1 in water and clinical samples. Appl Environ Microbiol 2011; 77:1708-17; PMID:21193672; http://dx.doi.org/10.1128/AEM.02261-10
- Collins S, Jorgensen F, Willis C, Walker J. Real-time PCR to supplement gold-standard culture-based detection of Legionella in environmental samples. J Appl Microbiol 2015; 119(4):1158-69; PMID:26218315; http://dx.doi.org/10.1111/jam.12911
- Herpers BL, de Jongh BM, vander -Zwaluw K, van Hannen EJ. Real-time PCR assay targets the 23S–5S spacer for direct detection and differentiation of Legionella spp. and Legionella pneumophila. J Clin Microbiol 2003; 41:4815-6; PMID:14532229; http://dx.doi.org/10.1128/JCM.41.10.4815-4816.2003.
- Yong SFY, Tan SH, Wee J, Tee JJ, Sansom FM, Newton HJ, Hartland EL. Molecular Detection of Legionella: Moving on From mip. Front Microbiol 2010; 1:123; PMID:3109421; http://dx.doi.org/10.3389/fmicb.2010.00123.
- Wilson DA, Yen-Lieberman B, Reisch U, Gordon SM, Procop GW. Detection of Legionella pneumophila by real-time PCR for the mip gene. J Clin Microbiol 2003; 41:3327-30; PMID:12843084; http://dx.doi.org/10.1128/JCM.41.7.3327-3330.2003.
- Yasliani S, Mobarez AM, Fatolahzadeh B, Feizabadi MM. Colonization of hospital water systems by Legionella pneumophila, Pseudomonas aeruginosa, and Acinetobacter in ICU wards of Tehran hospitals. Indian J Pathol Microbiol 2012; 1:65-7; PMID:23032830; http://dx.doi.org/10.4103/0377-4929.101743.
- JalaliMoghadam A, Honarmand HR, Meshginshahr AS, SoltaniTehrani B, Nojavan M. Frequency of Legionella pneumophila in tap water and water of infant incubators in Guilan hospitals, Iran. J Mazandaran Univ Med Sci 2013; 23(98):312-21.
- APHA. Standard method for the examination of water and wastewater 1995; 15th ed. Pp,27-32 APHAWEF and AWWA, Washington DC
- Fiume L, BucciSabattini MA, Poda G. Detection of Legionella pneumophila in water samples by species-specific real-time and nested- PCR assays. Lett Appl Microbial 2005; 41:470-5; PMID:16305672; http://dx.doi.org/10.1111/j.1472-765X.2005.01779.x
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013; 30:2725-9; PMID:24132122; http://dx.doi.org/10.1093/molbev/mst197
- Hayden RT, Uhl JR, Qian X, Hopkins MK, Aubry MC, Limper AH, Lloyd RV, Cockerill FR. Direct detection of Legionella species from bronchoalveolar lavage and open lung biopsy specimens: comparison of LightCycler PCR, in situ hybridization, direct fluorescence antigen detection, and culture. J Clin Microbiol 2001; 39:2618-26; PMID:11427579; http://dx.doi.org/10.1128/JCM.39.7.2618-2626.2001
- Cunha BA, Burillo A, Bouza E. Legionnaires' disease. The Lancet 2016; 387:376-85; PMID:26231463; http://dx.doi.org/10.1016/S0140-6736(15)60078-2
- Georghiou PR, Doggett AM, Kielhofner MA, Stout JE, Watson DA, Lupski JR, Hamill RJ. Molecular fingerprinting of Legionella species by repetitive element PCR. J Clin Microbiol 1994; 32(12):2989-94; PMID:7883887
- Amemura-Maekawa J, Kura F, Chang B, Watanabe H. Legionella pneumophila serogroup 1 isolates from cooling towers in Japan form a distinct genetic cluster. Microbiol Immunol 2005; 49: 1027-33; PMID:16365527; http://dx.doi.org/10.1111/j.1348-0421.2005.tb03699.x
