ABSTRACT
Klebsiella pneumoniae is an opportunistic pathogen that causes several kinds of infections, including pneumonia, bacteremia, urinary tract infection and community-acquired pyogenic liver abscess (PLA). Adhesion is the critical first step in the infection process. Our previous work demonstrated that the transcellular translocation is exploited by K. pneumoniae strains to migrate from the gut flora into other tissues, resulting in systemic infections. However, the initial stages of K. pneumoniae infection remain unclear. In this study, we demonstrated that a K. pneumoniae strain deleted for yfgL (bamB) exhibited reduced adherence to and invasion of host cells; changed biogenesis of major β-barrel outer membrane proteins; decreased transcriptional expression of type-1 fimbriae; and increased susceptibility to vancomycin and erythromycin. The yfgL deletion mutant also had reduced ability to against neutrophil phagocytosis; exhibited decreased induction of host IL-6 production; and was profoundly attenuated for virulence in a K. pneumoniae model of bacteremia. Thus, the K. pneumoniae YfgL lipoprotein mediates in outer membrane proteins biogenesis and is crucial for anti-phagocytosis and survival in vivo. These data provide a new insight for K. pneumoniae attachment and such knowledge could facilitate preventive therapies or alternative therapies against K. pneumoniae.
Introduction
Klebsiella pneumoniae is an opportunistic Gram-negative bacterium that causes several hospital-acquired and community-acquired infections.Citation1-4 Recently, community-acquired pyogenic liver abscess (PLA) caused by K. pneumoniae has emerged, with 10–13% of PLA patients also exhibiting metastatic meningitis and/or endophthalmitis.Citation4-10 Mortality rates are 10% for those with K. pneumoniae PLA alone, and 30–40% among those with metastatic meningitis.Citation5-8, Citation11 Cases suffering from K. pneumoniae endophthalmitis usually became blind in the affected eyes.
K. pneumoniae belongs to human microbiota, and gut-resident strains have been shown to possess genotypes similar to those detected in liver abscess and bacteremia strains.Citation12,13 Our recent study suggested that transcellular translocation is exploited by K. pneumoniae to migrate from the gut flora into other tissues, resulting in systemic infections.Citation14 However, the initial stages of K. pneumoniae infection remain unclear.
Bacterial adherence to epithelial cell surfaces is believed to be an important first step in the initiation of infection. Several early studies have demonstrated that type-1 fimbriae, type-3 fimbriae, KPF-28 fimbriae, and nonfimbrial protein CF29K of K. pneumoniae all mediate in vitro adherence to human lung and bladder cells.Citation15-19 In in vivo competition assays, type-1 fimbriae did not influence the ability of K. pneumoniae to colonize the intestine or infect the lung, but type-1 fimbriae were a significant virulence factor in K. pneumoniae urinary tract infection.Citation20 However, the influence of the other K. pneumoniae fimbriae on virulence has not been evaluated in vivo.
Gram-negative bacteria have an outer membrane (OM) that functions as a barrier to protect the cell from toxic compounds such as antibiotics and detergents. The folding and insertion of β-barrel proteins in the OM is mediated by the β-barrel assembly machinery (BAM) complex, which is composed of an integral membrane protein BamA (YaeT) and 4 accessory lipoproteins, BamB (YfgL), BamC (NlpB), BamD (YfiO) and BamE (SmpA).Citation21-23 Of those proteins solely BamA and BamD were found to be essential for viability in E. coli.Citation22,23 Although discrepancies exist in the literature, BAM complex proteins have recently been renamed.Citation22,24, Citation25 YfgL (BamB) is an OM lipoprotein, which is anchored to the periplasmic face of the OM Citation22 and displays a role in membrane permeability and antibiotic resistance of E. coli and S. enterica serovar Enteritidis.Citation26,27 A previous study demonstrated that the Salmonella YfgL lipoprotein is essential for expression of the type-III secretion system.Citation27 Several putative secretion systems, but not a type-III secretion system, have already been identified in the K. pneumoniae NTUH-K2044 genome.Citation28 Additionally, some OM proteins contribute significantly to the virulence of K. pneumoniae, by conferring protection against host's innate immunity.Citation29,30
In this study, we demonstrate that YfgL is the major facilitator of K. pneumoniae adherence. Moreover, the YfgL lipoprotein altered integrity of the OM that affects transcriptional expression of the fim operon and production of type-1 fimbriae in K. pneumoniae. The yfgL mutation increased susceptibility to vancomycin and erythromycin and is crucial for bacterial anti-phagocytosis and survival in vivo.
Results
Identification of the E. coli recombinant strains elevated in adhesion
In an epidemiologic study of 42 PLA-associated K. pneumoniae strains, we found that K1 was the dominant capsular type of 9 isolates from patients with K. pneumoniae PLA complicated with endophthalmitis (9/9, 100%). We initially analyzed these 9 PLA-endophthalmitis-associated K. pneumoniae strains for adhesion to ARPE-19 cells. The NTUH-K2044 showed a higher adhesion rate than other tested K. pneumoniae strains (). In a previous study, we generated a NTUH-K2044 genomic library (with 3- to 5-kb inserts).Citation31 To identify the possible adhesion molecules of the PLA-endophthalmitis-associated K. pneumoniae strains, adherence of DH5α E. coli harboring the NTUH-K2044 genomic library was analyzed. Eleven clones showed a hyperadherent phenotype; the NTUH-K2044 DNA inserts in these clones were sequenced ().
Figure 1. Adhesion of K. pneumoniae to human retinal epithelial cells. (A) Adhesion to ARPE-19 cells by PLA-endophthalmitis-associated K. pneumoniae clinical isolates (as indicated). NTUH-A4528 was used as a K2 type control strain. The adhesion rate was expressed as the proportion of the inoculum that adhered (∼1 × 10−4 of the inoculum). Data are presented as the mean ± SEM from 3 independent trials. (B) ARPE-19 adhesion of the K. pneumoniae NTUH-K2044 wild-type and its isogenic mutants (as indicated). The adhesion rate was expressed as the proportion of the NTUH-K2044 wild-type strain that adhered. Data are presented as the mean ± SEM from 3 independent trials. *, P < 0.05; **, P < 0.01. (C) Mapping of the transcription start site for the yfgL gene. The promoter region and the proposed −10 and −35 regions are indicated by gray shading. The transcription start site is indicated by +1 with arrowhead. The primers shown here indicate the relative positions in construction of the yfgL complementation strain. (D) Growth experiments with the wild type, yfgL, clpS and fim mutant strains in LB broth or MM. Overnight cultures of the NTUH-K2044 wild-type, yfgL, clpS, and fim mutant strains were inoculated (separately) into fresh LB medium or MM and grown at 37°C, respectively. The growth of bacteria was monitored hourly by plating of serial dilutions on LB agar and counting of CFUs following overnight growth of the plates. Data are presented as the mean ± SEM from 3 independent trials.
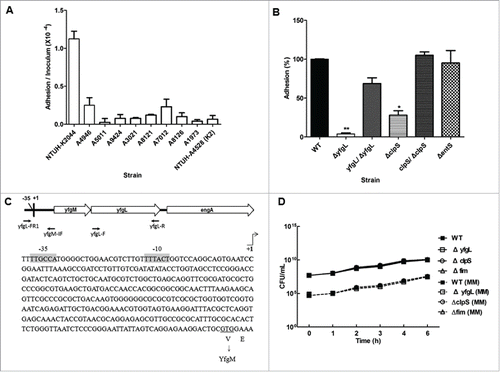
Table 1. The K. pneumoniae NTUH-K2044 hyperadherent clones.
The role of the YfgL lipoprotein in K. pneumoniae adhesion
Among the resulting protein candidates, we focused on a subgroup that included an ATP-dependent Clp protease adaptor protein (ClpS), an enterobactin exporter (EntS), and a subunit (YfgL) of an outer membrane protein assembly complex. All three of these proteins are supposed to display at the cell surface and their functions in adhesion have not been characterized in K. pneumoniae. To assess the putative contribution of these protein candidates during the initial host cell interaction, we constructed in-frame deletion mutants of clpS, entS, and yfgL, and examined the adherence to ARPE-19 cells by each of these 3 single mutants (). The yfgL mutant exhibited a 15-fold and highly significant decreased in ability to adhere to ARPE-19 cells. The clpS mutant strain also was significantly attenuated (3-fold) in adherence to ARPE-19 cells. In contrast, the entS mutant strain displayed adherence similar to that of the wild-type strain.
The sequence features of yfgL suggested that yfgL and its upstream gene yfgM are transcribed as an operon; the two ORFs are separated only by a very short intergenic region consisting of 10 base-pairs between the postulated stop codon of yfgM and the postulated ATG of yfgL. To avoid the failure of yfgL complementation resulting from the loss of the endogenous promoter, we determined the transcription initiation site of the yfgL gene by the 5′-RACE method. As expected, the transcription start site and promoter region of yfgL were located upstream of the yfgM ORF (). The adherence defects of the yfgL and clpS mutants were significantly restored by complementation with the corresponding gene ().
To determine whether the impaired adherence of the yfgL mutant resulted from a growth defect, bacterial growth assays were performed. As assayed in either LB broth or minimal medium culture, the growth rates of the yfgL mutant showed no significant difference compared to these of the wild-type strain (). Moreover, we further examined whether the YfgL protein was surface exposed on K. pneumoniae. Firstly, we extracted the OM proteins of K. pneumoniae and verified the specificity of anti-YfgL polyclonal antibodies by western blots (). Then we utilized these specific anti-YfgL antibodies for the YfgL detection by immunofluorescence microscopy. As expected, the YfgL of the NTUH-K2044 was not accessible to the anti-YfgL antibodies (). Thus, these data indicate that the lipoprotein YfgL is not surface exposed on K. pneumoniae and not likely act as an adhesin in cell adherence.
Figure 2. The effects of YfgL on K. pneumoniae adherence. (A) Specificity of rabbit polyclonal antibodies raised against YfgL. Total outer membrane protein extracts from normalized bacterial suspensions (4 × 109 CFU) of the NTUH-K2044 wild-type, ompA, yfgL mutant, and yfgL complementation strains grown in LB were analyzed by 12% SDS-PAGE, and stained with Coomassie brilliant blue (left), respectively. ompA deletion strain was used as a control strain. For western blots, proteins were probed with polyclonal antibodies raised against YfgL (1:10,000). The positions of OmpA and YfgL are indicated. (B) The NTUH-K2044 was incubated with rabbit polyclonal anti-K1 antibodies (as a positive control) or with rabbit polyclonal anti-YfgL antibodies, respectively. After incubation, Alexa Fluor 488 anti-rabbit IgG were added for staining, and then the fluorescence was observed. Scale bar, 10 μm. (C) Adhesion of the K. pneumoniae NTUH-K2044 wild-type, yfgL, and fim mutant strains (as indicated) to ARPE-19, Caco-2, or A549 cells. The adhesion rate was expressed as the proportion of the NTUH-K2044 wild-type strain that adhered. Data are presented as the mean ± SEM from 3 independent trials. *, P < 0.05; **, P < 0.01. (D) Immunofluorescence image analysis showing the adhesion of ARPE-19 cells by the NTUH-K2044 and the yfgL mutant. Cultured cells were infected by a high MOI of the NTUH-K2044 or the yfgL mutant for 15 min. Adherence K. pneumoniae (green) were incubated with rabbit anti-K1 polyclonal antibodies (1:1000) and then stained with Alexa Fluor 488 anti-rabbit IgG (Invitrogen). The actin cytoskeleton and DNA were stained with rhodamine-phalloidin (red) and DAPI (blue). Scale bar, 8 μm.
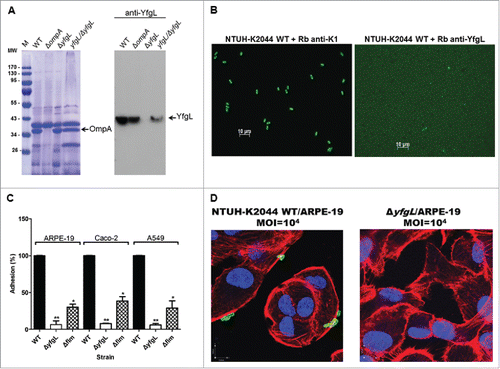
To further confirm the role of YfgL involved in K. pneumoniae host cell attachment, we performed similar cell adhesion assays using different cell lines. The yfgL mutant also exhibited significantly reduced adherence to 2 other human tissue cell lines Caco-2 and A549 (). Additionally, we obtained similar results with another invasive K. pneumoniae NTUH-A4528 yfgL-mutant strain (Figure S1A). The intimate attachment of the NTUH-K2044 onto the ARPE-19 cells was clearly observed at a higher MOI using immunofluorescence microscopy. In contrast, the adherence of the yfgL mutant onto the ARPE-19 cells was hardly observed ().
Transcriptome analysis of the K. pneumoniae yfgL deletion mutant
To clarify the impacts of the yfgL-gene deletion in K. pneumoniae, the transcriptomes of the yfgL mutant and the wild-type strain were compared. Among a total 2,848 cloned sequences covering 99.9% of the entire genome sequence of NTUH-K2044, the expression of 9 cloned sequences was down-expressed and 3 cloned sequences were up-expressed at a cut-off value of 2. Genes that were highly affected by YfgL are listed in . Levels of the fim transcript (encoding type-1 fimbriae) were reduced in the yfgL mutant compared to the wild-type strain, implying that the transcriptional regulation of the fim gene cluster might depend on YfgL. In addition to the fim gene cluster, expression of genes encoding amino acid metabolism (e.g., the tyrosine decarboxylase (tdc) gene cluster), peptidoglycan synthesis (e.g., yraM), and a putative phosphoenopyruvate:carbohydrate phosphotransferase system (PTS; the kp0760-kp0763 (pts) gene cluster) also appeared to depend on YfgL.
Table 2. Effect of yfgL deletion on transcription of genes in the K. pneumoniae NTUH-K2044 by microarray.
Transcriptional regulation of the K. pneumoniae fim gene cluster by YfgL
Using sequence analysis of the DNA upstream of the NTUH-K2044 fimA gene, we identified a putative invertible DNA element containing the fimA promoter in this strain. In K. pneumoniae C3091 and E. coli, inversion of such element regulates expression of the type-1 fimbriae-encoding genes Citation20,32; we inferred that fim expression is regulated by a similar mechanism in K. pneumoniae NTUH-K2044. Indeed, we showed that the NTUH-K2044 did not express type-1 fimbriae-encoding genes when the strain was cultivated in statically LB broth. In contrast, expression of type-1 fimbriae-encoding genes was observed when the strain was cultivated in LB broth with shaking by fimbrial switch orientation assays (). In this context, we noted that one early study indicated that a PTS system is involved in type-1 fimbriae expression in extraintestinal pathogenic E. coli (ExPEC).Citation33 We therefore sought to determine whether the NTUH-K2044 kp0760-kp0763 gene cluster (which, as noted above, encodes a PTS system and is positively affected by YfgL) was involved in type-1 fimbriae expression. We compared the fim phase switching of the yfgL mutant with that of the wild-type strain; the yfgL complementation strain; and a pts mutant (kp0760-kp0763) strain. Only DNA fragments corresponding to the “off” orientation were detected in the yfgL mutant, indicating that type-1 fimbriae expression was down-regulated in the yfgL mutant. When deleting pts, it looks that there is as much as “on” than “off” orientation (). We obtained similar results by real-time qPCR. Transcripts for fimC and fimD were detected at lower levels (less than 7% of the wild-type) under culturing in LB broth with shaking in the yfgL mutant than that in the wild-type strain. In contrast, the transcript for yfgL was present in the fim mutant at similar level to that in the wild-type strain. In the yfgL complementation strain, transcriptions of the yfgL gene and fim operon were approximately 20% and 40% restored, respectively (). In addition, the expression of type-1 fimbriae can be detected by mannose-sensitive yeast agglutination (MSYA), which attests the ability of type-1 fimbriated bacteria to bind to mannosides-containing receptors on the surface of yeast cells. In this assay, the fim mutant served as a negative control, which did not synthesized type-1 fimbriae and no MSYA was observed (). Consistent with the results of the fim transcription, MSYA assays demonstrated that the yfgL mutant synthesized 32-fold lower titers of type-1 fimbriae than the parent strain. In contrast, the levels of type-1 fimbriae expressed in the yfgL complementation strain were restored to within 4-fold of the titer seen in the wild-type strain.
Figure 3. The roles of YfgL in type-1 fimbriae production and cell invasion of K. pneumoniae. (A) Orientation of the fim phase-switch element in the K. pneumoniae NTUH-K2044 strains following overnight culture in either static or shaking LB broth. Depending on the orientation of the fim invertible element, this method generates paired fragments of different sizes (613 and 404 bp when in the “on” orientation, 840 and 177 bp when in the “off” orientation). Lane M contains DNA molecular size markers. (B) Effect of mutations in the yfgL or fim genes on yfgL and fim transcription in the wild-type, yfgL, fim mutants and yfgL complementation strains. Quantitative real-time expression of the yfgL and fim genes in response to LB broth shaking cultures in the wild-type strain, yfgL, fim mutants and yfgL complementation strains. The data represent averages from 3 independent trials, and the error bars represent the SEM. (C) Three-dimensional image analysis showing the invasion of ARPE-19 cells by K. pneumoniae NTUH-K2044. Cultured cells were infected by a high MOI of the NTUH-K2044 carrying TA-GFP (encoding greenfluorescent protein; green) for 2 h. The actin cytoskeleton and DNA were stained with rhodamine-phalloidin (red) and DAPI (blue). Images from confocal microscopy with z-stacking were analyzed by XY, XZ, and YZ sections. Intracellular K. pneumoniae are indicated by white arrows. Scale bar, 8 μm. (D) Invasion of ARPE-19 cells by NTUH-K2044 wild-type, yfgL mutant, and yfgL complemented strains. The invasion rate was expressed as the proportion of the NTUH-K2044 wild-type strain that invaded. Data are presented as the mean ± SEM from 3 independent trials. **, P < 0.01. (E) Effects of yfgL deletion on outer membrane protein level of K. pneumoniae. Total outer membrane protein extracts from normalized bacterial suspensions (4 × 109 CFU) of K. pneumoniae NTUH-K2044 wild-type, ompA, yfgL mutant, and yfgL complementation strains grown in LB or NB broth were analyzed by 12% SDS-PAGE, and stained with Coomassie brilliant blue, respectively. ompA deletion strain was used as a control strain. The positions of OmpA and OmpK35/OmpK36 are indicated.
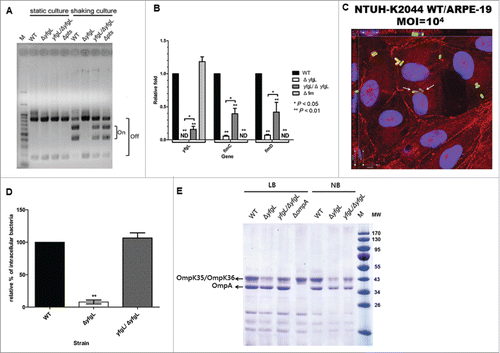
Table 3. Characteristics of the NTUH-K2044 wild-type and its derivatives.
To check whether type-1 fimbriae of the NTUH-K2044 were contributing to adherence, we compared the adherence of the fim mutant with that of the wild-type strain. The fim mutant had significantly (approximately 3-fold) decreased in adherence when compared with the wild-type strain in various human epithelial cells (). To analyze the impacts on adherence of the other yfgL down-expressed genes, we examined the adherence to ARPE-19 cells in tdc, yraM, and pts deletion mutants. These three mutant strains displayed adherence that was statistically indistinguishable from that of the wild-type strain (Figure S1B). Therefore, the attenuated adherence of the yfgL mutant was due in part to reduction in production of type-1 fimbriae.
The role of the YfgL lipoprotein in K. pneumoniae invasion
Our previous study demonstrated that the NTUH-K2044 strain has in vivo invasion ability, permitting penetration of the intestinal epithelium.Citation14 Cell invasion assays confirmed that NTUH-K2044 also invaded ARPE-19 cells. The presence of intracellular K. pneumoniae within ARPE-19 cells was clearly observed for the NTUH-K2044 (). The capacity of the yfgL mutant to invade ARPE-19 cells was significantly reduced compared to that of the wild-type strain. Complementation of the yfgL gene, allowed the yfgL mutant to recover its ability to invade cells (). These findings suggest that the YfgL lipoprotein is required for cell adhesion and invasion of K. pneumoniae.
Antibiotic sensitivity of the yfgL deletion mutant
To analyze and compare the levels of OM proteins in the yfgL mutant and the wild-type strains, we observed a slightly decreased of the OmpA level and marked reduction of the OmpK35/OmpK36 levels in the yfgL mutant compared to those in the wild-type strain when bacteria were cultured in either LB or NB broth. The OM protein reduction of the yfgL mutant was significantly restored by complementation with the yfgL gene (). To determine whether the yfgL mutant of K. pneumoniae exhibited altered OM properties, the sensitivity of this mutant was tested against selected antimicrobial agents (including colistin, polymyxin B, and 3 other antibiotics). The yfgL mutant exhibited 32-fold and 16-fold decreased in resistance to vancomycin and erythromycin, respectively (). These two compounds need to cross the bacterial membrane to reach their targets. This altered antibiotic resistance phenotype was partially complemented by introducing of the yfgL gene. In contrast, no change in sensitivity to colistin, polymyxin B or imipenem was seen in the yfgL mutant compared to the wild-type strain. In the case of other tested mutants, including clpS, fim, tdc, yraM, and pts, no difference in antibiotic resistance to these antimicrobial agents was observed. Thus, the increased susceptibility of the yfgL mutant to vancomycin and erythromycin reflects an increase in the membrane permeability of K. pneumoniae. We conclude that, as in E. coli and S. enterica serovar Enteritidis, YfgL plays a role in outer membrane biogenesis in K. pneumoniae.
The role of the YfgL lipoprotein in virulence
To test the sensitivity to the serum's bactericidal effect of the yfgL mutant, serum killing assay was performed. Killing of the yfgL mutant by non-immunized healthy human serum showed no significant difference compared to that of the wild-type strain (). To test the opsonophagocytic activity of the yfgL mutant, healthy human neutrophils were used. The bacterial number of the yfgL mutant was increased in the human neutrophils. However, the yfgL complemented and the wild-type strains were rarely present inside the human neutrophils (). Neutrophil-mediated killing of these strains was also examined. Compared to the number of wild-type bacteria, the number of yfgL mutant was decreased after incubation with human neutrophils, and the complemented strain restored the resistance to neutrophil killing (). Thus, the YfgL in K. pneumoniae confer protection against neutrophil-mediated phagocytosis and killing.
Figure 4. The roles of YfgL in virulence during K. pneumoniae infection. (A) Serum sensitivity assays of resistance to killing by nonimmune healthy human serum of the NTUH-K2044 wild-type and the yfgL-gene deletion mutant strains. The data represent the means of 3 independent trials; the error bars represent the standard deviations. A mean survival ratio ≥1 corresponds to serum resistance. ** P <0.01 by Student's t test (compared to the wild-type strain). The magA-gene deletion mutant is a CPS-deficient strain of NTUH-K2044 and used as a control. ((B)and C) Determination of phagocytosis resistance of the wild type, the yfgL-gene mutant and the yfgL complementation strains by human neutrophil. The pal-gene deletion strain is sensitive to human neutrophil phagocytosis and killing and used as a control. Bacteria carrying the GFP plasmid were incubated with human neutrophils for 45 min and observed under a confocal microscope. Bacteria phagocytosed by human neutrophils were counted under a confocal microscope. Data are presented as mean ± SEM of 3 independent trials. ** P < 0.01 or * P < 0.05 by Student's t test (compared to the wild-type strain). A representative confocal section through the middle of the cell was shown for observation of intracellular bacteria. Arrows denote intracellular bacteria. Scale bar, 10 μm. (D) Bacterial susceptibilities to killing by human neutrophils of the wild type, the yfgL-gene mutant and the yfgL complementation strains after 45 min of incubation are presented. Survival rate indicates percent survival of wild-type or mutant strains calculated on the basis of viable counts relative to those for the no-neutrophil controls. Data are presented as mean ± SEM of 3 independent trials. ** P < 0.01 or * P < 0.05 by Student's t test (compared to the wild-type strain). (E) BALB/c mice (4 mice per group) were infected with the NTUH-K2044 wild-type and isogenic mutant strains at an IP dose of 1 × 103 CFU/animal. Survival of mice was monitored for 4 weeks. ▪, NTUH-K2044; □, yfgL mutant (yfgL vs. parent, **, P = 0.008; log-rank test); ○, clpS mutant; △, fim mutant; ▴, yfgL complementation strain. ((F)and G) BALB/c mice (4 mice per group) were inoculated by IP injection with equivalent doses (1 × 103 CFU) of the NTUH-K2044 wild-type, yfgL mutant, or fim mutant strains. Surviving animals were sacrificed at 20 hours after challenge. Bacterial loads were measured in the liver and spleen (F); IL-6 levels were measured in the serum (G). Tissue counts (log10 CFU) were standardized per 0.1 g wet organ weight. IL-6 levels were measured by ELISA. Data are presented as mean ± SEM. **, P < 0.01 by Student's t-test (compared to the wild-type strain); other comparisons were not statistically significant (P ≥ 0.05).
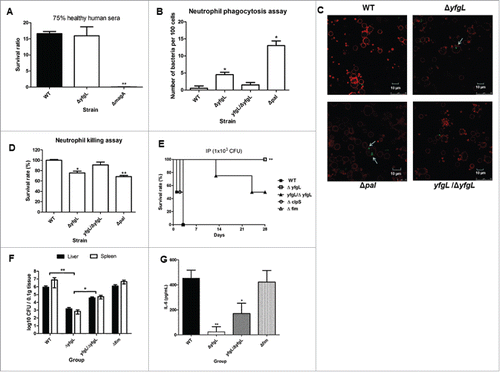
To evaluate the contribution of the YfgL lipoprotein to K. pneumoniae bacteremia, we challenged mice IP with the wild-type, yfgL mutant, the yfgL complementation and fim mutant strains, respectively. In this murine infection model, the yfgL mutant exhibited attenuated virulence compared to the wild-type, with respective LD50 values of 9.8 × 104 CFU and < 1 × 102 CFU, respectively ( and ). Moreover, complementation of the yfgL mutation significantly restored virulence in mice (LD50 = 1 × 103 CFU). In contrast, the fim mutant and other tested mutant strains yielded LD50 values resembling those of the wild-type (). To explore the contribution of the YfgL protein in bacterial dissemination and proinflammatory response, we challenged mice with a given dose (1 × 103 CFU) of the wild-type, yfgL mutant, the yfgL complementation or fim mutant strain. BALB/c mice infected with the yfgL mutant strain yielded significantly fewer colony counts in both the liver and spleen and lower cytokine IL-6 levels compared with those of the wild-type or the fim mutant strain (P≤0.001; ). Complementation of the yfgL mutant with the yfgL gene partially restored the colony counts in both in the liver and spleen. These data indicate that the yfgL mutant strain is impaired in the ability to spread in vivo and to induce IL-6 responses. We infer that YfgL contributes to virulence, not mediated by type-1 fimbriae, during K. pneumoniae infection.
Discussion
The YfgL lipoprotein has been demonstrated to play pleiotropic roles in bacterial OM biogenesis, antibiotic resistance, invasivity and virulence.Citation26,27, Citation34 In S. enterica serovar Enteritidis, the yfgL mutant had reduced invasivity for enterocytes compared to the wild-type strain.Citation34 Similarly, deletion of the yfgL gene in an adherent-invasive E. coli strain LF82 resulted in a decreased invasivity for intestinal epithelial cells.Citation26 In the present study, we demonstrated that YfgL serves as a mediator and is essential for K. pneumoniae adhesion and invasion of host cells. These findings indicate that the YfgL homologs of S. enterica serovar Enteritidis, E. coli, and K. pneumoniae have similar functions in adhesion to and invasion of host cells.
An earlier finding indicated that E.coli BL21 cells overproducing YfgL resulted in induction of at least 6 other unidentified host proteins.Citation35 In this study, we performed an expression library screen of hyperadherent E.coli DH5α harboring a PLA-causing K. pneumoniae genomic library. Multicopy effect of the K. pneumoniae's YfgL presumably changed E.coli's protein biogenesis and then increased its adherence to retinal pigment epithelial cells. A recent study indicated that that mutation of an ATP-dependent protease ClpS affected antibiotic resistance, swarming motility and biofilm formation in P. aeruginosa. The P. aeruginosa clpS mutant formed slightly less (approximately 70% of the wild-type) biofilm formation and this difference was due to a defect during the initial stages of biofilm formation.Citation36 In the present study, deletion of the clpS gene in K. pneumoniae resulted in a significantly attenuated (about 30% of the wild-type) in adherence to ARPE-19 cells. However, the patterns of cell adhesion, antibiotic resistance and virulence varied when the K. pneumoniae yfgL or clpS gene was individual deleted, suggesting that these 2 gene-encoded proteins were independently determined.
Previous works also have suggested that the yfgL gene product plays a role in the regulation of gene expression. The E. coli LF82 yfgL deletion mutant synthesized fewer type-1 fimbriae than did the wild-type strain; however, this mutant engineered to overexpress type-1 fimbriae did not exhibit increased invasivity.Citation26 That report indicated that the yfgL gene is necessary for the full ability of strain LF82 to invade intestinal epithelial cells, regardless of the level of production of type-1 fimbriae. Using microarray analysis, we showed in this study that the deletion of yfgL in K. pneumoniae also led to decreased expression of the fim transcript. In three distinct host cell models, the K. pneumoniae fim mutant exhibited a 3-fold reduction in adherence compared to the wild-type strain, whereas the yfgL mutant strain exhibited a more severe defect in adherence. Hence, the significant reduction of adherence exhibited by the K. pneumoniae yfgL mutant was not attributable solely to decreased expression of type-1 fimbriae.
In general, the lipoprotein YfgL is not a transcriptional factor and does not directly affect transcriptional regulation of the fim operon. Indeed, deletion of the yfgL gene altered integrity of the outer membrane that affects biogenesis and stability of major β-barrel OM proteins and type-1 fimbriae on the K. pneumoniae's cell wall. These results are consistent with an earlier finding that the E. coli FimD levels were strongly reduced (∼5-fold) in a mutant lacking the accessory lipoprotein BamB. That finding suggested that a functional BAM complex is needed for folding of FimD, the fimbrial usher of type-1 fimbriae.Citation37 Thus, our findings reflect that deletion of yfgL in K. pneumoniae led to compensate for post-transcriptional expression of the fim operon. It could be that the fim proteins are made but not inserted and accumulated in the yfgL mutant, which leading to a down-regulation of gene expression. Another related study also indicated that most of their isolated K. pneumoniae strains produced the E. coli common pilus (ECP), which is an adhesive structure produced by all E. coli pathogroups and encoded by the ecpABCDE operon, during adhesion to cultured epithelial cells.Citation38 That study highlights the heterogeneity of K. pneumoniae strains and their potential to produce multiple pili types during host colonization. Thus, not only type-1 fimbriae are involved in adherence of K. pneumoniae strains but the reduced adherence of the yfgL mutant could also be attributed to deficient assembly of type-1 fimbriae.
In Gram-negative bacteria, OM serves as the outermost barrier that an antimicrobial agent must overcome when interacting with its target. In this study, the expression levels of major β-barrel OM proteins, such as OmpA and OmpK35/OmpK36, were significantly reduced in the yfgL mutant. Additionally, deletion of K. pneumoniae yfgL perturbed the OM permeability barrier, rendering the mutant more susceptible to vancomycin and erythromycin, but not more susceptible to imipenem. Consistent with these findings, a previous study indicated that an ompK36 deletion mutant harboring a CMY-2 β-lactamase-expressing plasmid could significantly decrease susceptibility to imipenem compared to the corresponding strain without an additional plasmid bearing a CMY-2 β-lactamase gene in K. pneumoniae.Citation39 Therefore, the K. pneumoniae YfgL protein is important for the maintenance of cell envelope integrity and outer-membrane protein assembly. Some previous studies have reported that a significantly increased in susceptibility to neutrophil phagocytosis was observed in an ompK36 deletion K. pneumoniae mutant.Citation39,40 Our previous data also demonstrated that capsular polysaccharide (CPS) and OM proteins Pal and LppA, but not OmpA, all play important roles in a PLA K. pneumoniae strain resistance to phagocytosis.Citation30,41 We showed here that the K. pneumoniae YfgL confers protection against neutrophil phagocytosis, implying that reduced biogenesis of OmpK35/OmpK36 might account for loss of in vivo virulence in the yfgL mutant.
Because YfgL is anchored to the periplasmic face of the OM, it is not likely to function as an adhesin. We have confirmed in this study that YfgL is not surface exposed on K. pneumoniae. However, these results do not exclude the importance that YfgL plays crucial roles during K. pneumoniae infection. Thus, the pleomorphic effects of the K. pneumoniae YfgL lipoprotein, such as cell adhesion, outer-membrane protein assembly, type-1 fimbriae expression, anti-phagocytosis and in vivo virulence; should not jump to conclusion as consequences of aberrant OM protein assembly. More detailed studies will be needed to clarify that whether the functions of YfgL are inter-related or whether all of them require the interaction of YfgL with the BAM complex in K. pneumoniae.
The most common complications in PLA patients are metastatic meningitis and/or endophthalmitis. Our previous work demonstrated that PLA strains harbor a pathogenesis-specific genotype and transcriptional profile.Citation31 We therefore hypothesized that a novel adhesin may be produced in PLA-causing K. pneumoniae. However, our in vitro observations suggested that K. pneumoniae has a broad tissue tropism and can infect various types of human epithelial cells. The non-specificity of K. pneumoniae for adherence to various host tissues is not surprising, given that this pathogen causes several kinds of infections, including pneumonia, bacteremia, urinary tract infection, and PLA.Citation1-3 Strains with the K1 and K2 capsular types have been identified as the predominant virulent types and are prevalent in K. pneumoniae PLA.Citation4,10 Moreover, brain and eyes are immune privileged sites; once these tissues are invaded by pathogens, brain and eyes would be highly susceptible to infection. The present study suggests that the PLA-associated K. pneumoniae might spread through the bloodstream, thereby invading the liver, brain, and eyes.
As K. pneumoniae belongs to the endogenous microbiota in humans, the invasion rate would not be as high as those of invasive pathogens. In agreement with this, invasion assays showed that the K. pneumoniae NTUH-K2044 invaded ARPE-19 cells, as well as Caco-2 cells, at a rate (˜1 × 10−6 of the inoculum) lower than that of the invasive pathogen S. typhimurium (˜1 × 10−2 of the inoculum) across Caco-2 monolayers.Citation42 Thus, our results indicate that K. pneumoniae invades host cells at a low frequency, with the YfgL lipoprotein playing a critical role of K. pneumoniae invade to host cells.
Since YfgL homologs are present in the genomes of many Gram-negative bacteria, it is tempting to suggest that YfgL plays a preponderant role in the virulence of other pathogenic bacteria. Additionally, further point-mutation studies are warranted to realize whether interaction with the other BAM complex proteins was critical for YfgL function. Such knowledge could be useful for the development of preventive therapies or alternative therapies against these potential pathogens.
Materials and methods
Ethics statement. All animal procedures were approved under application number 20140062 of the Institutional Animal Care and Use Committee (IACUC) of the National Taiwan University College of Medicine (NTUCM). Procedures were consistent with the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and of Taiwan's Animal Protection Act. These studies used BALB/cByl mice that were bred and housed in specific pathogen–free rooms within the animal care facilities of the NTUCM and the Laboratory Animal Center at the National Laboratory Animal Center (NLAC).
Bacterial strains and culture conditions. K. pneumoniae and Escherichia coli strains were cultured in Luria-Bertani (LB) medium or LB medium supplemented with 50 µg/mL kanamycin. Bacterial strains, plasmids, and primers used in this study are listed in Table S1.Citation30, 31, 41, 43, 51
Gene deletion and complementation. K. pneumoniae mutated in yfgL, clpS, entS, fim, yraM, or pts were constructed using the previously described unmarked deletion method.Citation31 For cis-complementation, the intact yfgL, or clpS gene was amplified by polymerase chain reaction (PCR). To generate the yfgL complementation strain, a PCR fragment amplified by primers yfgL-FR1 and yfgL-R was cloned into a pGEM-T Easy vector (Promega). yfgM-IF and yfgL-F primers were used for inverse PCR with KOD xtreme polymerase (Takara) (). PCR-amplified fragments were phosphorylated by use of a polynucleotide kinase (New England Biolabs) for self-ligation. Then the inverse PCR-amplified plasmid was cut with NotI (New England Biolabs) to generate fragments and cloned into the intergenic region of the 2 open reading frames (ORFs), pgpA and yajO, in a pKO3-Km-pgpA-yajO recombinant vector used previously Citation43 (Table S1). Resulting plasmids were transformed into their corresponding isogenic mutant strains. The primer pairs for the deletion and complementation constructs are listed in Table S1.
Determination of the transcription initiation sites of yfgL gene. 5′-Rapid Amplification of cDNA Ends (RACE)-polymerase chain reaction (PCR) was performed using the SMARTer RACE cDNA Amplication Kit (Clontech Laboratories) following the manufacturer's instructions.
Bacterial growth assays. An 18-h culture of each strain was used to inoculate each 5-mL LB broth or M9 minimal medium (MM) aliquot to the same colony-forming units (CFU), respectively. Each culture was grown at 37°C and growth was monitored hourly by serial dilution and plating to LB agar with quantitation of CFU. Data are presented as the mean ± SEM from 3 independent trials.
Extraction of outer membrane proteins and analysis of YfgL. OM proteins were extracted from each strain grown in 20-mL LB medium or nutrient broth (NB) broth with sodium lauroyl sarcosinate (Sigma-Aldrich) and recovered by ultracentrifugation, as described previously.Citation44 Samples were separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie brilliant blue R250 (Gibco-BRL). For protein gel blots, proteins were separated using 12% SDS-PAGE and blotted to a Hybond-C membrane (Amersham), probed with polyclonal antibodies raised against YfgL, and revealed with peroxidase-labeled goat anti-rabbit antibody and the enhanced chemiluminescent substrate. Anti-YfgL antiserum was obtained from rabbits immunized with the recombinant YfgL protein, as described elsewhere (LTK BioLaboratories).
Adhesion and invasion assays. Human retinal pigment epithelial cells ARPE-19 were maintained in Dulbecco's modified Eagle's medium (DMEM)/F12 (1:1) supplemented with 10% heat-inactivated fetal bovine serum (FBS). Human colorectal epithelial cells Caco-2 and human lung epithelial cells A549 were maintained in DMEM supplemented with 10% heat-inactivated FBS and 1% nonessential amino acids (Gibco/BRL). Adhesion and invasion assays were performed mainly according to the methods described previously.Citation45 For adhesion assays, ARPE-19 cells in 24-well plates (∼1 × 106 cells per well) were prewashed with 1× phosphate-buffered saline (PBS). Mid-log-phase K. pneumoniae cells (A600, 0.4 to 0.6) in FBS-free DMEM/F12 medium were added to each well at a multiplicity of infection (MOI) of 10 bacteria/cell and incubated for 15 min in a humidified 5% CO2 atmosphere at 37°C. After incubation, wells were washed with 1× PBS 3 times, and bacteria were released by the addition of 0.1% Triton X-100 (Sigma-Aldrich). For invasion assays, K. pneumoniae cells in FBS-free DMEM/F12 medium were added to ARPE-19 cells. After centrifugation at 200× g for 5 min, plates were incubated for 1.5 h, and washed 3 times with 1× PBS, followed by a second incubation for 1.5 h in fresh medium containing 100 μg/mL gentamicin to kill extracellular bacteria. Finally, cells were incubated again and lysed with 0.1% Triton X-100. Recovered bacteria were quantified by plating appropriate dilutions on LB agar plates and counting CFU. These experiments are done in duplicate and repeated at least 3 times.
Immunofluorescence and microscopy. To detect whether the K. pneumoniae YfgL was surface exposed or not, dilutions of overnight cultured K. pneumoniae in 1× PBS and were added on slides. After drying, bacterial cells were incubated with 3% bovine serum albumin (BSA) (Sigma-Aldrich) in PBS as a blocking solution for 1 h, and then incubated with rabbit anti-K1 (1:1000) or anti-YfgL (1:1000) antiserum for 15 min, respectively. After incubation, cells were washed with 1× PBS 3 times, and Alexa Fluor 488 anti-rabbit IgG (Invitrogen) were added for 30 min at room temperature. To visualize cell adherence of K. pneumoniae, ARPE-19 cells (∼5 × 105 cells per well) grown on 12-mm round cover-glasses in 24-well plates. Overnight cultured K. pneumoniae in FBS-free DMEM/F12 medium were added to each well at a higher MOI of 104 and incubated for 15 min. Following infection, cells were washed with 1× PBS 3 times, fixed with 2% paraformaldehyde for 15 min, and permeabilized with 0.1% Triton X-100 for 15 min. After fixation and permeabilization, cells were incubated in 3% BSA in PBS for 1 h, and then incubated with rabbit anti-K1 antiserum (1:1000) for 15 min. After incubation, cells were washed with 1× PBS 3 times, and Alexa Fluor 488 anti-rabbit IgG (Invitrogen) were added for 30 min at room temperature. The actin cytoskeleton was stained with rhodamine-labeled phalloidin (Invitrogen) for 15 min, and nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (AppliChem GmbH) for 5 min at room temperature. To visualize potential cell internalization of the NTUH-K2044, ARPE-19 cells (∼5 × 105 cells per well) grown on 12-mm round cover-glasses in 24-well plates were infected for 2 h with the NTUH-K2044 carrying a plasmid encoding green fluorescent protein (GFP) at a high MOI of 10.Citation4 41 Following infection, cells were washed with 1× PBS, fixed with 2% paraformaldehyde for 15 min, and permeabilized with 0.1% Triton X-100 for 15 min. The actin cytoskeleton was stained with rhodamine-labeled phalloidin for 15 min, and nuclei were stained with DAPI for 5 min at room temperature. Images were initially observed using a Zeiss Axioplan 2 fluorescence microscope. To determine the potential location of the NTUH-K2044, confocal microscopy and 3-dimensional (3D) image analysis were performed. Images were captured by a Leica SP5 confocal microscope with 405-, 488-, and 543-nm lasers. Serial confocal z-stacks were taken with steps of 0.4 μm, and images were combined to observe cell-associated bacteria. When necessary, 3D images were analyzed by XYZ sections using the software Volocity 6.1 (PerkinElmer).
Microarray hybridization. Total RNA was purified with the RNeasy Mini Kit (Qiagen) from log-phase cultures of the NTUH-K2044 wild-type and yfgL mutant strains grown in LB medium. Total RNA (40 μg) labeling, microarray hybridization, colorimetric detection, and densitometry were performed and analyzed as described previously.Citation11,46 Expression level of 23S rRNA on each membrane was used as an internal standard for normalization. Processed microarray data files have been deposited in the National Center for Biotechnology Information under GEO accession number GSE69327.
Microarray validation. A randomly selected subgroup of genes that were highly affected by YfgL, as determined by microarray hybridization, was analyzed by real-time reverse-transcription quantitative PCR (qPCR), as described previously Citation31.
Fimbrial switch orientation assay. A modification of a previously described method was used to determine the orientation of the K. pneumoniae fim switch.Citation47 To investigate whether type-1 fimbria phase switching varies between the NTUH-K2044 and its derivative strains, the switch region was PCR amplified from overnight bacterial cultures cultivated either in static or shaking LB broth. The orientation of the phase switch, “on” or “off,” can be determined by PCR amplification of the switch region, followed by digestion with HincII and analyzed on 2% TAE-acrylamide-gels.
Quantitative real-time RT-PCR analysis. To measure the influence of the yfgL-gene deletion on the expression of the fim operon, total RNA from appropriate cultures of K. pneumoniae strains was first treated with DNase I (Qiagen) to remove any contaminating genomic DNA. A 400-ng sample of total RNA was reverse-transcribed and amplified by PCR monitored with SYBR green dye (Invitrogen) in an ABI7900 thermocycler (Applied Biosystems 7900). For each gene, the calculated threshold cycle (Ct) was normalized to the Ct of the 23S rRNA gene from the same cDNA sample before calculating the fold change using the ΔΔCt method.Citation48
Mannose-sensitive yeast agglutination (MSYA). Yeast aggregation assays were performed and monitored visually as described previously.Citation49 The titer was recorded as the highest dilution of bacteria yielding a positive aggregation reaction. These experiments are done in duplicate and repeated 3 times.
Sensitivity to antimicrobial agents. The minimum inhibitory concentrations (MICs) of colistin, polymyxin B, vancomycin, erythromycin and imipenem were determined using an agar dilution method according to the recommendations of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID).Citation50 These experiments are done in duplicate and repeated 3 times.
Serum killing assays. The survival of exponential-phase bacteria in nonimmune human serum was measured as previously described.Citation51 Briefly, a log-phase inoculum of 2.5 × 104 colony-forming units (CFU) was mixed at a 1:3 vol/vol ratio with mixed nonimmune human serum donated by 5 healthy volunteers. The final mixture, comprising 75% nonimmune serum by volume, was incubated at 37°C for 3 h. The colony count was determined by plating of serial dilutions on LB agar, and the mean survival ratio was plotted. A mean survival ratio ≥1 corresponds to serum resistance.
Phagocytosis and killing assays by human neutrophils. Human neutrophils were freshly isolated from peripheral blood donated by healthy volunteers.Citation41 For phagocytosis assay, plasmid pCRII-TOPO with a gene encoding green fluorescence protein (GFP) was electroporated into the wild-type, mutants and their respective complementation strains. An inoculum containing 108 CFU of bacteria was opsonized with 25% normal human serum for 15 min on ice and incubated with 106 human neutrophils at 37°C for 45 min. Cells were washed, fixed and stained with rhodamine-phalloidin as previously described.Citation52 After preparation, the cells were observed by confocal microscopy under × 630 image magnification, and the numbers of intracellular bacteria in 5 fields were counted (20–40 cells in each field). The sum of the intracellular bacteria was divided by the total number of cells in these fields and to calculate the number of intracellular bacteria in 100 cells. For killing assay, an inoculum containing 103 CFU of bacteria was opsonized with 25% normal human serum for 15 min on ice and incubated with or without 105 human neutrophils in 1× PBS at 37°C for 45 min. Percent survival of wild-type and mutant strains was calculated on the basis of the viable counts relative to those for no-neutrophil controls.
Mouse inoculation experiments. Virulence was evaluated by mortality in a murine model of septicemia generated by intraperitoneal (IP) injection according to the methods described previously.Citation30 The 50% lethal dose (LD50) was calculated as described by Reed and Muench.Citation53 To investigate the contribution of YfgL protein or type-1 fimbriae in bacteremia and proinflammatory response, 5-week-old BALB/c mice were administered IP with the same inoculation dose (1 × 103 CFU) of the wild-type, yfgL mutant, the yfgL complementation or fim mutant strain (4 mice for each strain), respectively. Surviving animals were sacrificed at 20 hours after challenge; organ homogenates (including liver and spleen) were cultured for quantification of CFU. Sera were collected at 20 hrs and IL-6 levels were measured by ELISA (R&D Systems, Minneapolis, MN).
Statistical Analyses. Data are presented as means ±standard error of the mean (SEM). Statistical significance was assessed by a 2-tailed Student's t test using Prism 5 (Graphpad) software and indicated as * P < 0.05 and ** P < 0.01 (compared to the wild-type); other comparisons were not statistically significant (P ≥ 0.05). Survival was analyzed by Kaplan-Meier analysis with a log-rank test. P values of <0.05 were considered significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
Conceived and designed the experiments: PFH CRH TLL JTW. Performed the experiments: PFH CTC. Analyzed the data: PFH. Wrote the paper: PFH JTW.
KVIR_S_1171435.zip
Download Zip (108.2 KB)Acknowledgments
We thank the staff of the imaging core at the First Core Labs, the National Taiwan University College of Medicine, for assistance with microscopy.
Funding
This work was supported by the Ministry of Science and Technology, the Excellent Research Projects of National Taiwan University, the National Taiwan University Hospital and the Liver Disease Prevention and Treatment Research Foundation of Taiwan.
References
- Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 1998; 11:589-603; PMID:9767057
- Ramphal R, Ambrose PG. Extended-spectrum beta-lactamases and clinical outcomes: current data. Clin Infect Dis 2006; 42 Suppl 4:S164-72; PMID:16544267; http://dx.doi.org/10.1086/500663
- Ko WC, Paterson DL, Sagnimeni AJ, Hansen DS, Von Gottberg A, Mohapatra S, Casellas JM, Goossens H, Mulazimoglu L, Trenholme G, et al. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerging infectious diseases 2002; 8:160-6; PMID:11897067; http://dx.doi.org/10.3201/eid0802.010025
- Tsai FC, Huang YT, Chang LY, Wang JT. Pyogenic liver abscess as endemic disease, Taiwan. Emerging infectious diseases 2008; 14:1592-600; PMID:18826824; http://dx.doi.org/10.3201/eid1410.071254
- Cheng DL, Liu YC, Yen MY, Liu CY, Wang RS. Septic metastatic lesions of pyogenic liver abscess. Their association with Klebsiella pneumoniae bacteremia in diabetic patients. Arch Intern Med 1991; 151:1557-9; PMID:1872659; http://dx.doi.org/10.1001/archinte.1991.00400080059010
- Chiu CT, Lin DY, Liaw YF. Metastatic septic endophthalmitis in pyogenic liver abscess. J Clin Gastroenterol 1988; 10:524-7; PMID:3053874; http://dx.doi.org/10.1097/00004836-198810000-00009
- Fung CP, Chang FY, Lee SC, Hu BS, Kuo BI, Liu CY, Ho M, Siu LK. A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut 2002; 50:420-4; PMID:11839725; http://dx.doi.org/10.1136/gut.50.3.420
- Liu YC, Cheng DL, Lin CL. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch Intern Med 1986; 146:1913-6; PMID:3532983; http://dx.doi.org/10.1001/archinte.1986.00360220057011
- Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL, Chang SC. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis 2007; 45:284-93; PMID:17599305; http://dx.doi.org/10.1086/519262
- Chung DR, Lee SS, Lee HR, Kim HB, Choi HJ, Eom JS, Kim JS, Choi YH, Lee JS, Chung MH, et al. Emerging invasive liver abscess caused by K1 serotype Klebsiella pneumoniae in Korea. J Infect 2007; 54:578-83; PMID:17175028; http://dx.doi.org/10.1016/j.jinf.2006.11.008
- Chou HC, Lee CZ, Ma LC, Fang CT, Chang SC, Wang JT. Isolation of a chromosomal region of Klebsiella pneumoniae associated with allantoin metabolism and liver infection. Infect Immun 2004; 72:3783-92; PMID:15213119; http://dx.doi.org/10.1128/IAI.72.7.3783-3792.2004
- Chung DR, Lee H, Park MH, Jung SI, Chang HH, Kim YS, Son JS, Moon C, Kwon KT, Ryu SY, et al. Fecal carriage of serotype K1 Klebsiella pneumoniae ST23 strains closely related to liver abscess isolates in Koreans living in Korea. Eur J Clin Microbiol Infect Dis 2012; 31:481-6; http://dx.doi.org/10.1007/s10096-011-1334-7
- Fung CP, Lin YT, Lin JC, Chen TL, Yeh KM, Chang FY, Chuang HC, Wu HS, Tseng CP, Siu LK. Klebsiella pneumoniae in gastrointestinal tract and pyogenic liver abscess. Emerging infectious diseases 2012; 18:1322-5; PMID:22840473; http://dx.doi.org/10.3201/eid1808.111053
- Hsu CR, Pan YJ, Liu JY, Chen CT, Lin TL, Wang JT. Klebsiella pneumoniae Translocates across the Intestinal Epithelium via Rho GTPase- and Phosphatidylinositol 3-Kinase/Akt-Dependent Cell Invasion. Infect Immun 2015; 83:769-79; PMID:25452552; http://dx.doi.org/10.1128/IAI.02345-14
- Fader RC, Gondesen K, Tolley B, Ritchie DG, Moller P. Evidence that in vitro adherence of Klebsiella pneumoniae to ciliated hamster tracheal cells is mediated by type 1 fimbriae. Infect Immun 1988; 56:3011-3; PMID:2902014
- Hornick DB, Allen BL, Horn MA, Clegg S. Adherence to respiratory epithelia by recombinant Escherichia coli expressing Klebsiella pneumoniae type 3 fimbrial gene products. Infect Immun 1992; 60:1577-88; PMID:1312518
- Tarkkanen AM, Virkola R, Clegg S, Korhonen TK. Binding of the type 3 fimbriae of Klebsiella pneumoniae to human endothelial and urinary bladder cells. Infect Immun 1997; 65:1546-9; PMID:9119502
- Di Martino P, Livrelli V, Sirot D, Joly B, Darfeuille-Michaud A. A new fimbrial antigen harbored by CAZ-5/SHV-4-producing Klebsiella pneumoniae strains involved in nosocomial infections. Infect Immun 1996; 64:2266-73; PMID:8675336
- Di Martino P, Bertin Y, Girardeau JP, Livrelli V, Joly B, Darfeuille-Michaud A. Molecular characterization and adhesive properties of CF29K, an adhesin of Klebsiella pneumoniae strains involved in nosocomial infections. Infect Immun 1995; 63:4336-44; PMID:7591068
- Struve C, Bojer M, Krogfelt KA. Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect Immun 2008; 76:4055-65; PMID:18559432; http://dx.doi.org/10.1128/IAI.00494-08
- Bolla JM, Lazdunski C, Pages JM. The assembly of the major outer membrane protein OmpF of Escherichia coli depends on lipid synthesis. EMBO J 1988; 7:3595-9; PMID:3061802
- Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 2005; 121:235-45; PMID:15851030; http://dx.doi.org/10.1016/j.cell.2005.02.015
- Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol 2006; 61:151-64; PMID:16824102; http://dx.doi.org/10.1111/j.1365-2958.2006.05211.x
- Gatsos X, Perry AJ, Anwari K, Dolezal P, Wolynec PP, Likic VA, Purcell AW, Buchanan SK, Lithgow T. Protein secretion and outer membrane assembly in Alphaproteobacteria. FEMS Microbiol Rev 2008; 32:995-1009; PMID:18759741; http://dx.doi.org/10.1111/j.1574-6976.2008.00130.x
- Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat Rev Microbiol 2009; 7:206-14; PMID:19182809; http://dx.doi.org/10.1038/nrmicro2069
- Rolhion N, Barnich N, Claret L, Darfeuille-Michaud A. Strong decrease in invasive ability and outer membrane vesicle release in Crohn's disease-associated adherent-invasive Escherichia coli strain LF82 with the yfgL gene deleted. J Bacteriol 2005; 187:2286-96; PMID:15774871; http://dx.doi.org/10.1128/JB.187.7.2286-2296.2005
- Fardini Y, Chettab K, Grepinet O, Rochereau S, Trotereau J, Harvey P, Amy M, Bottreau E, Bumstead N, Barrow PA, Virlogeux-Payant I. The YfgL lipoprotein is essential for type III secretion system expression and virulence of Salmonella enterica Serovar Enteritidis. Infect Immun 2007; 75:358-70; PMID:17060472; http://dx.doi.org/10.1128/IAI.00716-06
- Sarris PF, Zoumadakis C, Panopoulos NJ, Scoulica EV. Distribution of the putative type VI secretion system core genes in Klebsiella spp. Infect Genet Evolution 2011; 11:157-66; PMID:20932940; http://dx.doi.org/10.1016/j.meegid.2010.09.006
- Merino S, Camprubi S, Alberti S, Benedi VJ, Tomas JM. Mechanisms of Klebsiella pneumoniae resistance to complement-mediated killing. Infect Immun 1992; 60:2529-35; PMID:1587619
- Hsieh PF, Liu JY, Pan YJ, Wu MC, Lin TL, Huang YT, Wang JT. Klebsiella pneumoniae peptidoglycan-associated lipoprotein and murein lipoprotein contribute to serum resistance, antiphagocytosis, and proinflammatory cytokine stimulation. J Infect Dis 2013; 208:1580-9; PMID:23911714; http://dx.doi.org/10.1093/infdis/jit384
- Hsieh PF, Lin TL, Lee CZ, Tsai SF, Wang JT. Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis 2008; 197:1717-27; PMID:18433330; http://dx.doi.org/10.1086/588383
- Abraham JM, Freitag CS, Clements JR, Eisenstein BI. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci U S A 1985; 82:5724-7; PMID:2863818; http://dx.doi.org/10.1073/pnas.82.17.5724
- Rouquet G, Porcheron G, Barra C, Reperant M, Chanteloup NK, Schouler C, Gilot P. A metabolic operon in extraintestinal pathogenic Escherichia coli promotes fitness under stressful conditions and invasion of eukaryotic cells. J Bacteriol 2009; 191:4427-40; PMID:19376853; http://dx.doi.org/10.1128/JB.00103-09
- Amy M, Velge P, Senocq D, Bottreau E, Mompart F, Virlogeux-Payant I. Identification of a new Salmonella enterica serovar Enteritidis locus involved in cell invasion and in the colonisation of chicks. Res Microbiol 2004; 155:543-52; PMID:15313254; http://dx.doi.org/10.1016/j.resmic.2004.03.005
- Khairnar NP, Kamble VA, Mangoli SH, Apte SK, Misra HS. Involvement of a periplasmic protein kinase in DNA strand break repair and homologous recombination in Escherichia coli. Mol Microbiol 2007; 65:294-304; PMID:17630970; http://dx.doi.org/10.1111/j.1365-2958.2007.05779.x
- Fernandez L, Breidenstein EB, Song D, Hancock RE. Role of intracellular proteases in the antibiotic resistance, motility, and biofilm formation of Pseudomonas aeruginosa. Antimicrob Agents Chemother 2012; 56:1128-32; PMID:22123702; http://dx.doi.org/10.1128/AAC.05336-11
- Palomino C, Marin E, Fernandez LA. The fimbrial usher FimD follows the SurA-BamB pathway for its assembly in the outer membrane of Escherichia coli. J Bacteriol 2011; 193:5222-30; PMID:21784935; http://dx.doi.org/10.1128/JB.05585-11
- Alcantar-Curiel MD, Blackburn D, Saldana Z, Gayosso-Vazquez C, Iovine NM, De la Cruz MA, Girón JA. Multi-functional analysis of Klebsiella pneumoniae fimbrial types in adherence and biofilm formation. Virulence 2013; 4:129-38; PMID:23302788; http://dx.doi.org/10.4161/viru.22974
- Tsai YK, Fung CP, Lin JC, Chen JH, Chang FY, Chen TL, Siu LK. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob Agents Chemother 2011; 55:1485-93; PMID:21282452; http://dx.doi.org/10.1128/AAC.01275-10
- Chen JH, Siu LK, Fung CP, Lin JC, Yeh KM, Chen TL, Tsai YK, Chang FY. Contribution of outer membrane protein K36 to antimicrobial resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother 2010; 65:986-90; PMID:20211860; http://dx.doi.org/10.1093/jac/dkq056
- Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 2004; 199:697-705; PMID:14993253; http://dx.doi.org/10.1084/jem.20030857
- Kortman GA, Boleij A, Swinkels DW, Tjalsma H. Iron availability increases the pathogenic potential of Salmonella typhimurium and other enteric pathogens at the intestinal epithelial interface. PloS one 2012; 7:e29968; PMID:22272265; http://dx.doi.org/10.1371/journal.pone.0029968
- Hsieh PF, Lin HH, Lin TL, Wang JT. CadC regulates cad and tdc operons in response to gastrointestinal stresses and enhances intestinal colonization of Klebsiella pneumoniae. J Infect Dis 2010; 202:52-64; PMID:20497056; http://dx.doi.org/10.1086/653079
- Hernandez-Alles S, Alberti S, Alvarez D, Domenech-Sanchez A, Martinez-Martinez L, Gil J, Tomás JM, Benedí VJ. Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology 1999; 145(Pt 3):673-9; PMID:10217501; http://dx.doi.org/10.1099/13500872-145-3-673
- Sahly H, Podschun R, Oelschlaeger TA, Greiwe M, Parolis H, Hasty D, Kekow J, Ullmann U, Ofek I, Sela S. Capsule impedes adhesion to and invasion of epithelial cells by Klebsiella pneumoniae. Infect Immun 2000; 68:6744-9; PMID:11083790; http://dx.doi.org/10.1128/IAI.68.12.6744-6749.2000
- Ang S, Lee CZ, Peck K, Sindici M, Matrubutham U, Gleeson MA, Wang JT. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect Immun 2001; 69:1679-86; PMID:11179343; http://dx.doi.org/10.1128/IAI.69.3.1679-1686.2001
- Struve C, Forestier C, Krogfelt KA. Application of a novel multi-screening signature-tagged mutagenesis assay for identification of Klebsiella pneumoniae genes essential in colonization and infection. Microbiology 2003; 149:167-76; PMID:12576590; http://dx.doi.org/10.1099/mic.0.25833-0
- ABI UBe. Relative Quantitation of Gene Expression: Applied Biosystems. Foster City, CA: Applied Biosystems. 2001
- Muller CM, Aberg A, Straseviciene J, Emody L, Uhlin BE, Balsalobre C. Type 1 fimbriae, a colonization factor of uropathogenic Escherichia coli, are controlled by the metabolic sensor CRP-cAMP. PLoS pathogens 2009; 5:e1000303; PMID:19229313; http://dx.doi.org/10.1371/journal.ppat.1000303
- EUCAST Definitive Document E.DEF 3.1, June 2000: Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by agar dilution. Clin Microbiol Infect 2000; 6:509-15; PMID:11168187; http://dx.doi.org/10.1046/j.1469-0691.2000.00142.x
- Hsieh PF, Lin TL, Yang FL, Wu MC, Pan YJ, Wu SH, Wang JT. Lipopolysaccharide O1 antigen contributes to the virulence in Klebsiella pneumoniae causing pyogenic liver abscess. PloS one 2012; 7:e33155; PMID:22427976; http://dx.doi.org/10.1371/journal.pone.0033155
- Pan YJ, Lin TL, Hsu CR, Wang JT. Use of a Dictyostelium model for isolation of genetic loci associated with phagocytosis and virulence in Klebsiella pneumoniae. Infect Immun 2011; 79:997-1006; PMID:21173313; http://dx.doi.org/10.1128/IAI.00906-10
- Reed LJ MH. A simple method of estimating fifty percent endpoints. Am J Hyg 1938; 27:493-7
