ABSTRACT
Rat bite fever (RBF), a worldwide occurring and most likely under-diagnosed zoonosis caused by Streptobacillus moniliformis, represents the most prominent disease of Streptobacillus infections. Recently, novel members have been described, from which a reservoir in rats and other animal species and a zoonotic potential can be assumed. Despite regularly published case reports, diagnostics of RBF continues to represent a ‘diagnostic dilemma’, because the mostly applied 16S rRNA sequence analysis may be uncertain for proper pathogen identification. Virtually nothing is known regarding prevalence in humans and animal reservoirs. For a realistic assessment of the pathogen's spread, epidemiology and virulence traits, future studies should focus on the genomic background of Streptobacillus. Full genome sequence analyses of a representative collection of strains might facilitate to unequivocally identify and type isolates. Prevalence studies using selective enrichment mechanisms may also enable the isolation of novel strains and candidate species of this neglected group of microorganisms.
Introduction
For almost a century, Streptobacillus moniliformis represented a monotypic species within the genus StreptobacillusCitation1 (Streptobacillus, Fusobacteriales) causing streptobacillary rat bite fever (RBF) and Haverhill fever (HF).Citation2 RBF was first noted by Wagabhatt some 2,300 y ago in IndiaCitation3 and describes 2 similar yet distinct syndromes, from which the other – albeit less often – is caused by Spirillum minus (due to lack of a type strain not listed in the Approved List of Bacterial Names).Citation2,4 Spirillum minus infection, also known as sudoku, has been reported in Asia and is not further discussed here. The acute disease symptoms of the bacterial zoonosis streptobacillary RBF or food-borne HF include fever, malaise, muscle pain, arthritis and abscess formation, endocarditis, bacteremia, and maculopapular, petechial or pustular rash as well as vomiting and pharyngitis.Citation4 Most likely under-reported worldwide, streptobacillary RBF is predominantly transmitted through rat bites and scratches,Citation4 whereas HF is transmitted directly or indirectly by contact with rat urine.Citation5,6 In untreated cases RBF mortality ranges from 7 to 13%.Citation7-9 Approximately 50–100% of wild rats usually asymptomatically carry S. moniliformis in their oro- or nasopharynx and shed the organism with saliva and urine,Citation2,10 but abscess formation has also been described in rats and mice.Citation11,12 Other rodents as well as companion and exotic animal species and livestock are principally reported to be susceptible to clinical infection besides rats and mice, but mice may strain-dependently develop disease.Citation4,11,13-22
Detection of streptobacillosis due to S. moniliformis is referred to as a “diagnostic dilemma”Citation23 because reasons for under-diagnosing in susceptible host species are missing notice of a rodent bite or contact, non-specific clinical symptoms,Citation6 fastidious growth of the widely unknown microorganism and a lack of reliable diagnostics, non-notifiable disease and broad chemotherapeutic susceptibility. With respect to known diagnostic difficulties with this microorganism this review summarizes diagnostic approaches to detect streptobacillary infection in humans and animals.
Properties of the agent
Host spectrum
S. moniliformis has been isolated from various animal species. It is frequently found in wild rats (Rattus norvegicus),Citation10 but also in rats housed as pets,Citation12,24 and has also been found in laboratory rats.Citation25-27 Isolates exist also from laboratory mice (Mus musculus)Citation11,28-30 and from turkeys (Meleagris gallopavo).Citation13,16,18,22 Isolates from rats, mice, turkeys and humans were shown to belong to the same species. Citation31 Reports on possible infections in other livestock species date decades back into the last century.Citation17,20 However, as these strains are physically not available, reports are not fully in congruence with the genotypic and phenotypic properties of S. moniliformis. Moreover, as recent studies have even found large quantities also in genital tracts from cows and ewes,Citation32 there is some doubt if streptobacilli in livestock are identical with the RBF organism. Anecdotally, streptobacilli have been described from exotic host species like gerbils and squirrels that were occasionally named “Streptothrix paraxeri cepapi” after Smith's bush squirrel (Paraxerus cepapi),Citation33-35 partially with involvement of human infection resembling RBF,Citation33,36 but these isolates have also not been stored. Based on metagenomics data from cotton ratsCitation37 it may be possible that streptobacilli in other rodent species might in fact represent separate species (). Further proof of S. moniliformis in exotic species was recorded from a koala and macaques.Citation19,21,38 Carnivores like dogs, cats, weasels and ferrets were occasionally found to be colonized or even suffer from Streptobacillus infection.Citation14,15,39 It remains unclear whether such findings really represent S. moniliformis, although identified after mouthing wild rats, or if Streptobacillus species other than S. moniliformis may be involved that belong to the mouth microbiota and occasionally cause also disease in dogs and cats. S. moniliformis is an important zoonotic agent and is usually transmitted to humans by direct contact with rats (rat bite fever), but infection of humans is also possible through contaminated food (HF).Citation40-42 Contact with carnivorous animals is, however, only rarely believed to lead to human RBF.Citation43-48
Figure 1. Maximum parsimony (MP) tree showing the phylogenetic relationship of cultured Leptotrichiaceae species and those only represented by environmental 16S rRNA gene sequences. The tree was calculated in ARB using DNAPARS and based on 16S rRNA gene sequences spanning at least gene termini 97 to 1356.Citation138 Shorter sequences were added after tree construction without changing the overall tree topologies. Large circles represent nodes that were at least also present with high bootstrap support in the Maximum likelihood (ML) tree. Small circles mark nodes that were also present in the MP and neighbor joining tree, but in the ML tree only supported by bootstrap values <70%. GenBank/EMBL/DDBJ accession numbers of the sequences are given in parentheses. Numbers at branch nodes refer to bootstrap values >70% (100 replicates). Fusobacterium nucleatum subsp. nucleatum ATCC 25586T was used as outgroup. Bar: 0.1 nucleotide substitutions per nucleotide position. T marks type strain sequences.

After the genus Streptobacillus was held monotypic for almost a century, a second species, S. hongkongensis, has been isolated from 2 humans with peritonsillar abscess and septic arthritis.Citation49 Recently, a third species was isolated from the lungs of a cat with pneumoniaCitation50 which has been described as S. felis.Citation51 Some other isolates formerly assigned to S. moniliformis exist, from which S. notomytis from a spinifex hopping mouse (Notomys alexis) and from black rats (Rattus rattus) and S. ratti from an asymptomatically colonized black rat were recently described.Citation52,53 Contrarily to Nolan et al.,Citation54 various potentially novel Streptobacillus species and phylotypes consistent with operational taxonomic units have been identified in the last few years from Atlantic salmonCitation55 and microbiomes of digestive tracts in dolphins and sea lions,Citation56-58 upper respiratory tracts in cotton rats,Citation37 digestive tracts in dogs,Citation39,59,60 intestinal tract of a ducks,Citation61 genital tracts in livestock,Citation32 and skin and gut microbiomes in humans ().Citation62,63 This fuels the assumption that Streptobacillus species are far more distributed in the environment aside from their natural hosts than previously thought. Contrarily, former Streptobacillus-like organisms from fishCitation55 and guinea pigsCitation64-67 are even more distantly related to classical Streptobacillus species and indeed represent novel genera, that have been recently described.Citation68,69
Virulence factors
Despite recent advances in decoding the complete genome of S. moniliformis and further Streptobacillus species no designated virulence associated genes have been described.Citation54 Concerning pathogenicity one might refer to possible virulence properties in an α-hemolytic strain.Citation27 Indeed, hemolytic strains of S. moniliformis, S. hongkongensis, and S. felis were involved in clinical disease in a rat,Citation70 a dog,Citation15 a cat,Citation50 and a human,Citation51 but other clinical isolates, especially those causing severe or even fatal disease turned out to be non-hemolytic so that other virulence factors apparently play a more important role. These might include the extraordinary high amount of DNase in all strains, which is released independently from bacterial growth.Citation71 Further reflections on virulence regard the lipopolysaccharideCitation27 and the agglutination of erythrocytes. However, as depicted in the chapter on hemagglutination the experiments unequivocally suggest the presence of adhesins, a mechanism involved in bacterial pathogenicity which is a prerequisite for the “successful” infection of a host. Indeed, there appear to remain other factors besides adhesins as can be concluded from the fact that hemagglutination could principally also be observed in non-host species for S. moniliformis. These might include not yet identified genetic factors at the host side which can be concluded from differences in susceptibility to infection like for instance the genetically diverse, highly susceptible C57BL/6 J mice compared to BALB/c mice.Citation11,30 C57BL/6 J mice are known to show an exacerbated release of IL-12 compared to BALB/c mice, if Toll-like receptor (TLR) 2 agonists on the surface of Listeria monocytogenes are stimulated.Citation72 This could also explain a more severe pro-inflammatory response in C57BL/6 J mice by TLR-mediated recognition of S. moniliformis.Citation73 However, although neutrophils seem to represent the predominating leukocyte cell fraction in RBF patients, mouse macrophages are known to be killed earlier in the presence of engulfed S. moniliformis.Citation74
Diagnostics
Direct techniques for detection of infection
Phenotypic identification
Bacterial cultivation from clinical samples
Streptobacillus infection is mostly diagnosed by isolation of the organism from blood, synovia, pus or other fluids, nevertheless the organism is difficult to grow in culture and requires specific media and incubation conditions. An anticoagulant in blood cultures, sodium polyanethol sulfonate (SPS; trade name “Liquoid”), used to grow bacteria from blood samples from patients suspected of bacteremia inhibits growth of the organism in concentrations as low as 0.0125%.Citation75 Therefore, other additives are necessary for isolation of S. moniliformis.Citation76 Good growth of all species can be achieved on Columbia agar supplemented with 5% sheep blood after 2–5 d of incubation at 37°C in the presence of 5–10% CO2, but initial culture of the organism from clinical specimens can be difficult due to overgrowth by faster growing and less fastidious bacteria. We had the best culturing results using tryptone soy agar or broth (containing 30 g tryptone soy (Oxoid), 5 g yeast extract (Merck, Darmstadt, Germany), 800 ml Aqua dest., optionally 12 g agar and the addition of 200 ml decomplemented horse serum (Oxoid) after autoclaving). Growth of streptobacilli can further be improved by a 5–20% supplementation of common media with serum or ascitic fluid. Citation70 Some authors have used media supplemented with colistin, sulfamethoxazole-trimethoprim and nalidixic acid for primary isolation from colonized mucosal sites by suppressing Gram-negative contaminating flora.Citation10,11,24 However, work in our lab revealed that S. hongkongensis DSM 26322T is the only member of the genus that is trimethoprim/sulfamethoxazol-sensitive as well as nalidixic acid-sensitive Streptobacillus strains occurred.Citation31,51 In liquid media (e.g. tryptone soy broth) with addition of serum, streptobacillary growth can be detected after 2–7 d as typical “puff-ball” or “bread crumb-like” appearance.Citation2,4
Suboptimal growth at least for some strains of S. moniliformis and S. hongkongensis can still be detected in an aerobic and anaerobic atmosphere.Citation26,49,70 Isolates from guinea pigs were historically associated with S. moniliformis and reported to grow under strictly anaerobic conditions.Citation64 Investigations in our lab have now shown that these strains from guinea pigs are indeed obligate anaerobes and now form a novel taxon, Caviibacter abscessus, within the family Leptotrichiaceae.Citation69
Growth characteristics
Colonies of Streptobacillus are tiny, drop-like, shiny, slightly convex, 0.1–0.4 mm in diameter after 48–72 h of incubation in a capnophilic atmosphere. Some of the colonies show a “fried-egg” appearance indicating the presence of L-forms.Citation2 Whereas L-form variants arise spontaneously on agar media, the formation of cell wall deficient bacteria is thought to be a consequence of specific immunity in vivo and thus are responsible for clinical relapses and resistance to antimicrobial agents that interfere with cell wall synthesis.Citation77 The L-forms, which are sometimes regarded as non-pathogenic variants in vivo Citation78 and which can be induced in vitro on sheep blood agar by addition of 2 IU/ml penicillin,Citation79 spontaneously revert into bacillary forms after subculturing.
The vast majority of strains are non-hemolytic, but strains with slightly α-hemolytic colonies are known to occur Citation15,27 and the type strains of S. felis 131000547T, S. ratti OGS16T and S. hongkongensis DSM 26322T all typically demonstrate α-hemolysis on sheep blood agar. Citation31,50,51,53
Morphologic features
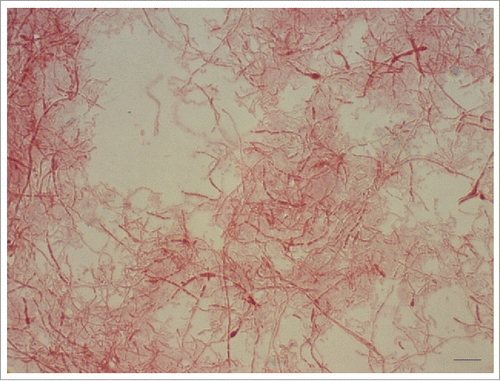
The microscopic features are consistent with Gram-negative, pleomorphic, fusiform to filamentous, non-spore forming, non-encapsulated, non-acid-fast rods which are arranged in chains and clumps. Sometimes, especially in aged cultures, irregular, lateral bulbar swellings can be seen, that resemble a “string of beads” or a necklace, which is the translation of the Latin word moniliformis. The 0.1–0.7 × 1–5 µm sized bacteria tend to pleomorphism and might form up to 150 µm unbranched filaments in stains from cultures compared to stains from infection sites ().Citation4
Figure 2. Gram-negative, pleomorphic cells of a 6-day-old culture of Streptobacillus moniliformis DSM 12112T are arranged in chains and clumps and display irregular, lateral bulbar swellings, that resemble a ‘string of beads’ or a necklace – the translation of the Latin word moniliformis. The 0.1–0.7 × 1–5 µm sized bacteria tend to pleomorphism and might form up to 150 µm unbranched filaments. Oil immersion, ×1000 magnification. Bar, 5 µm.
Electron microscopy was carried out with one strain of a ‘S. moniliformis-like organism’ isolated from a calve suffering from interstitial pneumonia.Citation17 The “bread crumb-like” floccules from liquid media appeared as “densely staining filaments and swollen bodies” which could also be appreciated by light microscopy. This isolate was not subjected to molecular analysis or sequencing and other key characteristics were missing, e.g., being dependent on a capnophilic atmosphere or pathogenicity for mice.
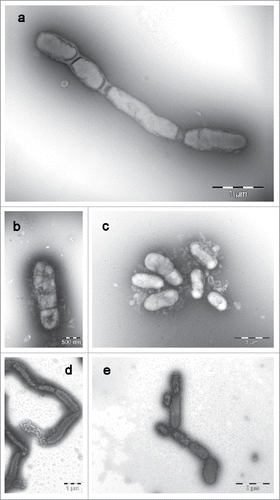
Own transmission electron micrographs (JEM-1011; JEOL, Freising, Germany) of cells of S. moniliformis DSM 12112T after growth on sheep blood agar at 37°C for 7 d show oval to elongated cells in a diameter range of 0.3–0.7 µm and lengths from 0.9 to 5.2 µm, without any flagella, but with a recognizable cell envelope, and partly an arrangement in chains (). The comparison of electron micrographs from the recently described species S. hongkongensis DSM 26322T, S. felis 131000547T, S. notomytis AHL307–1T, and S. ratti OGS16T grown under the same conditions and incubation times do not show discernible differences ().
Figure 3. Transmission electron micrographs (JEM-1011; JEOL, Freising, Germany) of cells of a: Streptobacillus moniliformis DSM 12112T, b: Streptobacillus felis 131000547T, c: Streptobacillus ratti OGS16T, d: Streptobacillus notomytis AHL 370–1T and e: Streptobacillus hongkongensis DSM 26322T. All strains were grown on sheep blood agar at 37°C for 7 d. Images were taken with negative contrast (PTA method) at ×3000 to ×10,000 magnifications. Bars, 500 nm, 1000 and 2000 nm, respectively.
Biochemical properties
For a review of biochemical tests that need to contain the addition of serum to the respective media see refs.Citation2,4,31 In our opinion the best conventional biochemical results were obtained with phenol red broth base (Difco, distributed by Becton Dickinson [Heidelberg, Germany]) supplemented with carbohydrates to be tested. The original recipe from Difco gives better results than the later modification by Becton Dickinson. All tests should be read after prolonged incubation at 37°C for up to 7 d. Some authors suggested to obtain biochemical profiles with commercially available systems, e.g. API-E (bioMeriéux, Nürtingen, Germany),Citation10 but use of those commercial biochemical platforms remains controversial.Citation70 However, a set of S. moniliformis field and reference strains in our laboratory was mostly in accordance with known patterns.Citation2,4,10,11,26,27,70,80 In contrast to biochemical assays, enzymatic pattern testing does not require proliferating bacteria. Most studies assessing enzymatic profiles of streptobacilli were using the API-ZYM system (bioMeriéux).Citation24,27,80 Positive reactions were recorded for alkaline phosphatase, butyrate esterase, caprylate esterase, myristate lipase, leucine arylamidase, chymotrypsin, acid phosphatase, and glucuronidase activities.Citation24 We have recently employed another commercially available biochemical system (Merlin Micronaut, Bornheim, Germany) that was specifically adapted to the growth of Streptobacillus.Citation31 A choice of suitable biochemical tests is given in . In summary, Streptobacillus species can be indicated by a combination of growth and micro-morphological characteristics together with some important, congruently negative resulting key reactions like cytochrome oxidase, catalase, urease, and nitrate reduction, Voges-Proskauer-reaction and indole production. Further biochemistry might be variable between strains of the same species and is not adequate to differentiate different Streptobacillus species.
Table 1. Physiological characteristics of Streptobacillus species known to date obtained by VITEK2-compact with the NHI card†, API-Zym‡Footnote# (both bioMeriéux) and classical reactions§ (modified afterCitation53); Taxa: 1, Streptobacillus moniliformis DSM 12112T; 2, results from 6 Streptobacillus moniliformis reference strains (ATCC 27747, ATCC 49567, ATCC 49940, NCTC 11194, CIP 55–48 and CIP 81–99); 3, Streptobacillus hongkongensis DSM 26322T; 4, Streptobacillus felis 131000547T; 5, Streptobacillus notomytis AHL 370–1T; 6, Streptobacillus ratti OGS16T; +, positive; -, negative; +/− variable.
Chemotaxonomic pattern
Fatty acid profiles obtained by gas-liquid chromatography, together with characteristic growth, have been used for rapid identification of S. moniliformis.Citation80-86 The major cellular fatty acid peaks are tetradecanoic acid (14:0), palmitic acid (16:0), octadecanoic acid with linoleic acid (18:2) and oleic acid (18:1), and stearic acid (18:0).Citation84,85 Fatty acid profiles obtained with use of gas chromatography coupled with mass spectrometry showed major peaks for C16:0, C18:2, C18:1, and C18:0 fatty acids, a profile characteristic of S. moniliformis.Citation82 In light of additional species of Streptobacillus this must be scrutinized, because these species cannot be differentiated by their fatty acid patterns alone. A comprehensive comparison of fatty acid profiles from all Streptobacillus species known to date is presented in .Citation53
Table 2. Cellular fatty acid pattern of Streptobacillus species known to date (modified after Citation53); Taxa: 1, Streptobacillus moniliformis DSM 12112T; 2, Streptobacillus hongkongensis DSM 26322T; 3, Streptobacillus felis 131000547T; 4, Streptobacillus notomytis AHL 370–1T; 5, Streptobacillus ratti OGS16T. Biomass for fatty acid analysis was harvested after 3 d of growth in capnophilic environment with 10% CO2 on Columbia sheep blood agar at 36°C.
Polar lipids are poorly understood in Streptobacillus species. A lack of quinones and a specific polyamine pattern different from those of the α-, β- and γ-subclasses of the proteobacteria was proposed by Hofmann and Wullenweber et al.Citation70,71 The protein profiling from whole cell preparations displayed a species specific pattern of 40–50 proteins ranging from 18 to 100 kDa. Four major protein bands in the region 60–67 kDa were formed that accounted for 20–30% of the total protein.Citation87 Earlier assumptions of protein-based strain differences between human and murine isolates as well as between Haverhill and rat bite fever strainsCitation87 were not confirmed by other authors.Citation80,88 On the other hand, especially differences of HF- versus RBF-strains are unlikely because rats represent the source of infection in both cases and disparities could better be explained by different gene expression following oral or parenteral infectionCitation4 or simply by too few HF-strains under study. Additionally, the time between infection and strain isolation from the host following rat exposure is usually too short to facilitate adaptation of strains and expression of a different phenotype.
Experimental infection
Historically, the classical foot pad test was performed for confirmation of S. moniliformis infection by injection into mice, which led to septic arthritis within few days.Citation70 This painful procedure is now obsolete as better in vitro diagnostics are available. A number of studies have, nevertheless, proven that S. moniliformis strains isolated from susceptible host species were able to cause infection in rodents, thereby partially fulfilling Koch's postulates.Citation11,22
Hemagglutination
Screening for adhesive properties was performed for 14 S. moniliformis strains and for S. notomytis AHL370-1T by hemagglutination experiments using erythrocytes from 11 different host donor species, i.e. red blood cells from humans, BALB/c and C57B1/6J mice, rats, turkeys, guinea-pigs, hamsters, chicken, sheep, horses, pigs and cattle were included.Citation31,71 Adhesive properties were detected in all S. moniliformis strains tested. Erythrocytes of the different vertebrate species were agglutinated with varying intensity. The strongest reactions could be observed in erythrocytes from turkeys, humans, guinea-pigs and pigs, followed by rats and chickens. C57BL/6J mice, known to represent a highly susceptible mouse strain toward streptobacillosis,Citation11 were less strongly agglutinated compared to erythrocytes from the more resistant BALB/c mice. By adding mannose, a known agonist of a common adhesin receptor, no significant differences could be observed indicating mannose-resistant agglutination in all cases. No differences were observed between agglutination of erythrocytes from ‘original’ host species (from which respective strains were originally isolated) and other host red blood cells, but susceptibility was generally highest in species of potential hosts compared to non-host species. There were no differences in the hemagglutinating behavior between RBF and HF strains of S. moniliformis.Citation71
Serum agglutination
For direct identification of S. moniliformis agglutination reactions with specific serum have been used in the past.Citation89 Direct immunofluorescence was also employed to achieve identification by staining S. moniliformis bacteria with a polyclonal antiserum ().Citation7 None of these tests is commercially available and specificity of such assays remains to be reviewed.
Mass spectrometry
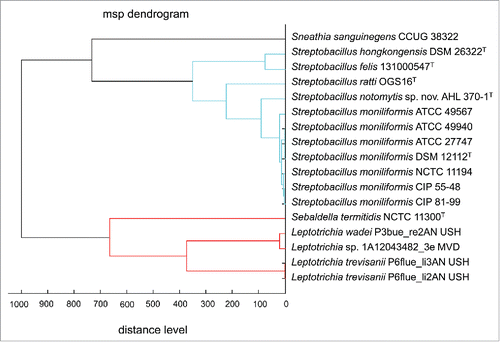
Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) was found to be a fast and reliable tool for species identification of S. moniliformis.Citation50 In the meantime, this technique has proven to contain enough discriminatory information to differentiate all currently known species of Streptobacillus.Citation31,53 Likewise, commercial databases do not contain spectra of all members, but respective spectra can be obtained via MALDI-UP, a user-to-user dedicated database platform.Citation90 A representative cluster analysis of MALDI-TOF spectra is depicted in . In a repeatedly culture-negative clinical case of RBF, employment of PCR and electrospray ionization followed by mass spectrometry (PCR/ESI-MS) proved to be a useful tool in detection of S. moniliformis from a synovial fluid but not from the patient's serum. The authors highlight that this technique was culture-independent and even successful in specimens obtained following initiation of antimicrobial therapy.Citation91
Figure 5. Dendrogram including all main spectra peak lists (MSP) of the family Leptotrichiaceae available in the Bruker Taxonomy Database; spectra of Streptobacillus ratti OGS16T, Streptobacillus notomytis AHL 370–1T, Streptobacillus hongkongensis DSM 26322T, Streptobacillus felis 131000547T, Streptobacillus moniliformis and Sebaldella termitidis NCTC 11300T reference strains were recorded using the direct transfer protocol. The dendrogram was generated using the BioTyper MSP Dendrogram Creation Standard Method (v1.4) of the MALDI BioTyper OC Software (v3.1, build 66). The database used (DB 5627, BrukerDaltonics) comprised solely 24 spectra from Streptobacillus moniliformis DSM 12112T; T type strain, ATCC: American Type Culture Collection, Rockville, USA, DSM: Deutsche Sammlung für Mikroorganismen, Braunschweig, Germany, CIP: Collection of Institute Pasteur, Paris, France, NCTC: National Collection of Type Cultures, London, UK.
Fourier transform-infrared spectroscopy (FT-IR)
As a vibration-spectroscopic technique, FT-IR is using the mid-infrared region of the electromagnetic spectrum to analyze the total composition of dried films of microorganism cells. In comparison with the protein-fingerprints obtained by MALDI-TOF MS, FT-infrared-spectra mirror information about the sum of biomolecules, like carbohydrates, lipids, proteins and other cell components.Citation92 This method had already been used as a fingerprint-based tool for rapid and reliable classification of a large number of clinically relevant pathogensCitation93-95 including recently described Streptobacillus species ().Citation31,51 However, at present, the lack of enough strains from the novel species makes it difficult to validate and test the performance of the species identifying methods.
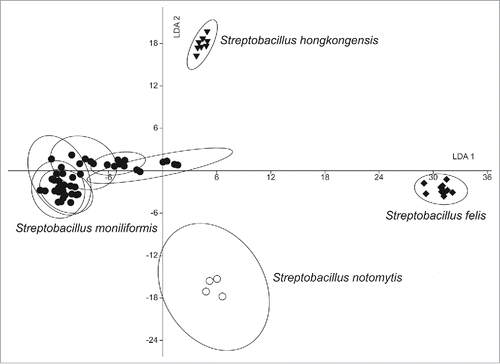
Figure 6. Linear discriminant analysis (LDA) analysis of 69 infrared spectra of 10 Streptobacillus isolates obtained by Fourier-transform infrared-spectroscopy (FT-IR) using OPUS Software (vers. 4.2, BrukerOptics, Ettlingen). The wave numbers 550–1800 and 2800–3200 cm−1 of second derivative spectra were selected and vector normalized. After a principal component analysis, the first 30 components were used for the LDA. In this LDA every isolate was defined as one group. Spectra of Streptobacillus notomytis AHL 370–1T are represented by circles, Streptobacillus moniliformis by dots, Streptobacillus hongkongensis DSM 26322T by triangles and Streptobacillus felis 131000547T by diamonds.
Antimicrobial properties
Despite its good response to various antimicrobial treatments one should attempt susceptibility testing for every isolate. Most studies employed disk diffusion testing where diameters of zones with growth inhibition are recorded according to a norm (e.g., according to the German standard DIN 58940).Citation11,85,96 Some authors have used minimum inhibitory concentration (MIC) testing with the agar incorporation testCitation80 or a breakpoint method,Citation70,71 that gave mostly congruent results compared with the disk diffusion method.Citation70 With respect to MIC testing with automatic systems the slow growth of Streptobacillus hinders a reliable end point measurement and can only be read by visual interpretation during off-label use. One has to take into account, that no official breakpoints specific for Streptobacillus have been published to date. Therapeutics of choice are generally penicillin, streptomycin and tetracycline.Citation70 Therapy in mice was successfully initiated with 1 g ampicillin/L drinking water given for 2 weeks, followed by chlortetracycline for 1 week.Citation11 Expectably, MIC testing results are too vague and – like biochemistry – not appropriate to differentiate species.
Storage of bacteria
To keep Streptobacillus in strain collections, freezing of fresh bacterial biomass and supplementation of respective medium with 20% serum or in pure cattle serum with 6% glucose at −70°C works well for re-cultivation even after several years. Deep freezing in brain heart infusion (BHI) broth supplemented with 10–20% (w/v) of glycerol can also be advised (own observations).Citation88 Lyophilization in fetal calf serum with 6% glucose is also a good option for long-term storage in our laboratory. Resuscitation of sub-lethally damaged strains was successfully achieved by centrifuging the previously frozen organism onto a human endothelial cell culture where regular growth could be initiated.Citation97
Molecular identification
Molecular properties
Determinations of guanine/cytosine (G/C) contents of 23 S. moniliformis as well as the novel species revealed a nearly identical G/C content of 25.7–28.9 mol% in all investigated Streptobacillus strains.Citation31
A high level of DNA-DNA-homology (DDH) between 14 strains of S. moniliformis could be shown by Hofmann.Citation71 As concluded from an earlier definition all of the investigated S. moniliformis had DDH levels above 70% thus indicating them as members of a single species.Citation98,99 According to this definition S. notomytis AHL370-1T revealed 68% homology to the S. moniliformis type strain, thereby at best justifying a separate subspecies.Citation98 We therefore believe that DDH of Streptobacillus species gives weak results.Citation31 Instead, average nucleotide identity (ANI) was carried out according to the method described by Goris et al.,Citation100 with which analogous results could be confirmed. They could demonstrate a close relationship between DDH values and ANI in that the recommended cut-off point of 70% for species discrimination corresponded to 95% ANI. The 95–96% species boundary is also supported by Richter & Rosselló-Móra,Citation101 who have developed an alignment free interface to calculate ANI also used in this study. In contrast to the highly homologous group of S. moniliformis strains all novel members could be unequivocally discriminated.
Species specific PCR for S. moniliformis
PCR protocols published to date for the detection of S. moniliformis in clinical samples and for identification focus on respective partial gene sequences of the 16S rRNA gene.Citation10,12,97,102-104 The method by Boot et al.Citation102 amplified a 296 bp fragment and showed considerable sensitivity but also some flaws in specificity due to amplicon sequence similarities with Leptotrichia sp., Fusobacterium necrogenes and Sebaldella termitidis.Citation102,103 Although these non-specificities could be solved by macro restriction with the endonuclease BfaI, Kimura et al.Citation10 advanced this PCR with respect to specificity by improving oligonucleotide primers according to . Thus, a 269 bp fragment was amplified and cross reactivity with the above mentioned bacterial species was no longer detected. We have further modified the method by Kimura et al.Citation10 slightly by changing the annealing temperature to 53°C for 1 min. We have calculated a diagnostic sensitivity of 2 × 102 bacteria with this PCR by detecting as few as 10 pg of serially diluted purified DNA lysate of S. moniliformis DSM 12112T endowed in homogenized rat lung tissue. Details on our PCR have been previously described.Citation50 A different PCR protocol employing primer pair SbmF/SbmR () yielded a significantly longer amplicon (1,222 bp),Citation12 thereby further improving specificity. Both PCR protocols were suitable to detect all S. moniliformis strains from humans, rats, mice and turkeys and also S. felis, S. notomytis and S. ratti, but S. hongkongensis was not detected in the PCR by Rohde et al.Citation12,51-53 Summarizing, all the mentioned PCR systems must presently be regarded rather genus than species specific. In an era of easy access to sequencing techniques it is always desirable to sequence amplicons. S. moniliformis and Leptotrichia sp. turned out to non-specifically cross-react in a fluorescence in situ hybridization assay (FISH) for rapid identification of Fusobacterium spp.Citation105 which in turn suggests its use also for the direct detection of S. moniliformis.
Table 3. Oligonucleotide primer sequences and PCR conditions of the target genes used for the detection of Streptobacillus species.
Marker gene sequencing
16S rRNA gene
A number of studies have used full length or partial 16S rRNA gene sequencing as a diagnostic tool for species determination of S. moniliformis in clinical samplesCitation10,39,97,106-109 as well as for laboratory confirmation of suspicious isolates.Citation24 Because of the extraordinary role of 16S rRNA gene sequencing in bacterial taxonomy this gene enables to compare isolates and phylotypes obtained in microbiome studies. On the other hand, 16S rRNA gene sequencing can be insufficient for definite species resolution.Citation110 For unequivocal identification of Streptobacillus species in particular, this gene should always – especially in the highly homologous species S. moniliformis, S. felis, S. notomytis and S. ratti – be confirmed by another gene locus or method such as those described below. Contrarily, in S. moniliformis with a sufficient choice of strains, intraspecies heterogeneity was too low to distinguish strains from different origins and hosts ().Citation31
Other housekeeping genes
Species specific gyrB primers were designed to amplify a 514 bp fragment of the gene for gyrase B in order to identify S. moniliformis.Citation24 Within the phylum Fusobacteria sequencing of 16S rRNA, 16S-23S rRNA internal transcribed spacer, gyrB, groEL, recA, rpoB, conserved indels and genes for group-specific proteins, 43 kDa outer membrane protein and zinc protease have been proposed for species identification or phylogenetic analysisCitation111-120 and more than 31 whole genome sequences have been released in GenBank. On the basis of next generation sequencing a number of functional genes was tested for their phylogenetic potential.Citation31 To overcome the above mentioned uncertainties, we have used groEL, recA and gyrB in addition to 16S rRNA, which unequivocally could discriminate Streptobacillus strains to species level.
Indirect techniques for detection of infection
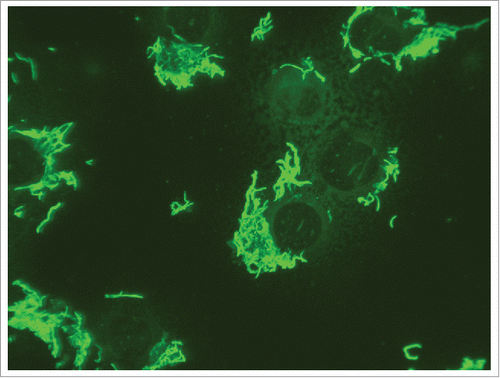
Although the authors are unaware of any seroprevalence surveillance studies in humans to assess the number of atypical or subclinical cases with the potentially lethal RBF microorganism, various serologic approaches exist to detect antibodies against S. moniliformis in serum. Among these are direct slide agglutination and complement fixation techniques with human or naturally or experimentally infected animal sera.Citation15,40,74,121-128 Compared to modern methods, these tests show a flaw in sensitivity and specificity. Nevertheless, agglutinating antibodies were also used early in taxonomic studies to type S. moniliformis strains.Citation125 A complement fixation testCitation74,129,130 and an indirect immunofluorescence assay (IFA)Citation11,27,75 have been employed in intravenously infected mice. The latter was prepared by using air-dried and heat fixed bacterial suspensions with an OD of 0.2 on IFA slides as antigen. Alternatively, antigen for serology can be prepared by adding a bacterial suspension to cells (e.g., L929, HeLa cells) grown on IFA slides (). This has the advantage that bacteria adhere to the cells so that unspecific reactions can easily be identified.
A protocol for an enzyme-linked immunosorbent assay (ELISA) was evaluatedCitation26,88 and propagated for testing mouse and rat colonies.Citation88 The authors used washed and merthiolate-inactivated bacteria cultured in broth. This test was found superior in detecting significantly more positive animals compared to culture.Citation26 Different immunoglobulin subclasses could be detected with the usual shift from IgM to IgG with duration of infection by using different secondary antibodies.Citation26 Unfortunately, this test failed to unequivocally detect true infection due to lack in specificity and therefore immune blotting (IB) with whole cell antigens was advised to confirm positive or doubtful reactions from ELISA.Citation25 Interestingly, antibodies in sera of guinea pigs were also detected by ELISA, but guinea pigs were resistant to oral or nasal infection with a rat strain of S. moniliformisCitation131 and strains infecting guinea pigs formerly assigned to S. moniliformis were found to represent a novel species,Citation69 thereby suggesting the possibility of cross reactivity of this ELISA with other species.Citation88
Use of membrane proteins of S. moniliformis instead of whole bacterial cells reduces the background reactions in ELISA and increases its specificity (Nicklas, unpublished data). Proteins can easily be prepared as described by Livingston et al. Citation132 for Helicobacter hepaticus. These proteins can also be coupled to polystyrene microspheres (Luminex Corporation distributed by Diamex, Heidelberg, Germany) and applied in bead-based multiplex serology. We use this test as primary test for health surveillance of rodent colonies and IFA with cells grown on IFA slides after infection with S. moniliformis as a confirmatory test ().
Employing SDS-PAGE of S. moniliformis whole cell preparations, a species specific pattern of 40–50 proteins was derived.Citation87 By IB, however, a number of approximately 10 different antibodies to respective immunogenic antigens of the 18–87 kDa range were demonstrated.Citation25 Rodent sera were considered positive if an antibody activity against at least 2 antigens of the 32–55 kDa range could be detected. With this assumption IB yielded a diagnostic sensitivity of 78% and a diagnostic specificity of 85%.Citation25 Interestingly, though S. notomytis AHL370-1T displayed a unique profile in electrophoretic protein patternsCitation87 no antigenic differences could be observed for this species compared to S. moniliformis.Citation88
In contrast to viral serology, there are few publications which emphasize serology for bacterial infections, especially to confirm the microbiological status of laboratory rodent colonies. Serology has the advantage that no animals have to be sacrificed and that serum samples can easily be shipped to external laboratories. On the other hand, the risk of false-positive and false-negative reactions must be considered. None of these serological tests is currently commercially available.
However, antibodies can routinely be found in infected rats after 2 to 4 weeks post infection.Citation26 Conversely, mice strain-dependently suffered from natural infection and even died before antibodies could be detected.Citation11 Especially susceptible were C57BL/6J mice usually displaying cervical lymphadenitis in contrast to BALB/cJ, C3H/He, CB6F1, B6D2F1 and DBA/2J mice. Seroconversion following oral infection was only observed in C57BL/6J, C57BL/10, AKR/N, B6D2F1 and DBA/2J and in all of the tested mouse strains after intravenous injection, despite severe clinical symptoms were only observed in all C57BL/6J and some DBA/2J mice.Citation11,70 After intranasal application, that represents the natural way of infection, antibodies were not reliably detected in mice and rats,Citation11,133 but experiments were terminated already 4–6 weeks after infection. We experimentally infected BALB/c intranasally and detected antibodies by ELISA and IFA usually after 8 weeks or later (Nicklas, unpublished data). Other authors came to the conclusion that genetic factors as well as individual resistance were responsible for different strain susceptibility in mice.Citation11,70 Intravenous and subcutaneous infection of non-specified mice led to a weak neutrophilia and maximum antibody titers not exceeding 1:640. Despite an effect that homologous antibodies existing prior to infection prolonged the incubation period, the authors concluded that the organism was in some fashion resistant to phagocytosis in vivo.Citation74
With respect to specificity (serological) cross reactions to most other rodent bacteria could be excluded for ELISA and IFA, except for some members of the order Mycoplasmatales.Citation70,88 An age-dependent effect was reported for the ELISA so that routine monitoring of rats should be done up to an age of 16 weeks to prevent false positive reactivity.Citation70
Discussion
RBF is occurring worldwide and is believed to be under-recognized and under-diagnosed in humans.Citation4 The risk of infection with any organism following a rat bite is 1–10%,Citation8 but the risk of developing streptobacillary RBF and the infectious dose are unknown. According to another survey 40,000 rat bites are noticed every yearCitation134 and approximately 2% of rat bites are followed by an infection.Citation135 Untreated RBF is associated with a case fatality rate of up to 13%.Citation70 The reasons for under-diagnosing streptobacillosis in man and animals include organism as well as host specific factors like unsuitable diagnostic tools, the fastidious growth and broad chemotherapeutic susceptibility of this microorganism, a non-notifiable disease as well as missing notice of a rodent bite, non-specific clinical symptoms, especially in chronic infections and a broad spectrum of differential diagnoses. Additionally, only very severe clinical causes will be diagnostically worked up and few laboratories and physicians are experienced with RBF or are even aware of the disease. In addition to the natural reservoir of rats, mice and other rodents, streptobacillary infections have also been reported to occur in livestock as well as zoo animals like calves, a pig, turkeys, non-human primates, and a koala.Citation4,13,16-22 Recently, various publications suggest that Streptobacillus species might be far more common and distributed in the environment or as commensal microbiota than previously thought.Citation32,37,59-63 It could recently be shown that the natural reservoir for the very rare cases of human S. hongkongensis infection known to date is indeed the human oropharynx* and presumably not an unidentified animal or environmental reservoir.Citation49 Despite a certain amount of annually published case reports, most of which have solely used 16S rRNA sequencing alone for definite diagnosis, there has not been much progress in the diagnosis of acute clinical cases of RBF in the last decades. Moreover, as pointed out earlier, 16S rRNA sequencing alone may be insufficient for unequivocally determining the involved pathogen to species level. In conclusion, the better knowledge of the global spread of Streptobacillus and its variability demands both a higher awareness as well as better diagnostic approaches in human as well as in animal diagnostics. Hence, the present review focused on a large spatiotemporal collection of S. moniliformis isolates from different host species and also included all currently known novel members of the genus. We aimed to analyze whether well-established diagnostic tools still fit the demands of an increasing diversity of Streptobacillus members and host species and also report our experiences with modern as well as little-known diagnostics.
Primary isolation by culture methods remains difficult as colonies are small and shiny and appear only after incubation for several days in an atmosphere enriched with CO2. Isolation from clinical samples on blood agar is unlikely in mixed cultures (e.g., oral cavity, nasopharynx, intestinal tract) due to overgrowth by less fastidious organisms but is easily possible from otherwise sterile environments (e.g., blood, joint fluid, abscesses). Colonies may differ in size as L-form variants arise spontaneously. They are usually non-hemolytic but strains with a weak α-hemolysis do occur. In liquid media containing serum or ascitis fluid bacteria show typical “puff-ball” or “bread-crumb-like” appearance as a sediment and a clear supernatant. The yield can be improved by adding antimicrobial agents like for instance colistin, sulfamethoxazole-trimethoprim and nalidixic acid to the culture medium, but even then the isolation rate can be low. Although Kimura et al.Citation10 could detect a prevalence of up to 92% by PCR they succeeded to culture only 7 isolates from more than 1000 suspicious colonies. We have earlier shown that physiological parameters are problematic for typing Streptobacillus. In general, carbohydrate fermentation tests and other tests requiring proliferating bacteria may be difficult to read as some strains grow poorly resulting in very weak reactions. Biochemistry is furthermore largely dependent on the test system itself, possible batch-to-batch variation and the person reading the tests and even with commercial test systems it was not unequivocally possible to get identical results for the same species or even the same strains in repeated experiments.Citation31 Nevertheless, it is possible to use information from biochemistry together with growth characteristics, when working with suspicious isolates, but the characters are generally too weak to differentiate species. If standardized test systems are employed, one should use tests determining end-point measurements (e.g. API ZYM®, VITEK2-compact® (NHI profile), Merlin Micronaut Streptobacillus profile) that are thus independent from bacterial growth compared to tests requiring viable bacteria. Important key reactions for all members of the genus Streptobacillus are cytochrome oxidase, catalase, urease, nitrate reduction, Voges-Proskauer and indole production (all negative). Chemotaxonomic analysis was slightly deviant from other studies for S. moniliformis that have found homologous major fatty acid profiles of tetradecanoic acid (14:0), palmitic acid (16:0), octadecanoic acid with linoleic acid (18:2) and oleic acid (18:1), and stearic acid (18:0).Citation82,84,85 In contrast, we have detected relatively uniform fatty acid patterns of C16:0, C18:0, C18:1ω9c, summed feature 5 C18:0 ANTE/C18:2ω6,9c and C18:1ω6c for all Streptobacillus species.Citation53 Antimicrobial resistance profiles – albeit also not suitable for species or genus discrimination – have revealed that S. moniliformis – since no β-lactamase activity could be demonstrated so farCitation54 – is susceptible to all β-lactam antibiotics.Citation31,80 Penicillin G was repeatedly reported being the most efficient antimicrobial substance in vitro and in vivo, which further supports its use as the drug of choice in the treatment of RBF and HF, followed by tetracycline.Citation2 The successful use of the combination of clindamycin with rifampin (for enhanced tissue concentration) has been described in a case of abscess formation.Citation136 Because of generally low MIC values the strains from our study confirmed a generally good therapeutic basis, but, nevertheless, some isolates were in vitro resistant or intermediate resistant to ciprofloxacin, erythromycin, gentamicin, nalidixic acid and streptomycin.Citation31
Hence, these very similar, non-discriminatory physiological results suggest that other traits should be propagated for the identification of Streptobacillus species. Based on spectral differences our group could recently show that 23 S. moniliformis strains from at least 5 different host species isolated over the past 90 y from almost all subcontinents as well as the type strains of S. hongkongensis and S. felis were unequivocally differentiated by MALDI-TOF MS and also by FT-IR, where the spectral information mirrors information from a broad variety of main component biomolecules.Citation31,137 This was also true for the 2 recently described novel species S. notomytis and S. ratti.Citation52,53 Together with the ease, expense and availability of these methods in the microbiological laboratory nowadays, at least MALDI-TOF MS should presently be regarded as the new gold standard in species discrimination, but – on the other hand – usually requires previous cultivation of the organism. Nevertheless, by the worldwide spread of the MALDI-TOF MS technology in clinical microbiological laboratories, we expect a significantly higher number of reliable diagnoses for Streptobacillus sp. This is particularly true, since relevant entries for a specific database extension are available.Citation90 The published PCR assays are genus rather than species specific. A diagnostic sensitivity of less than 2 × 102 cfu was calculated for homogenized rat lung tissue. Diagnostic flaws concerning specificity toward other genera were successfully corrected in the meantime. Based on the novel members, a real species specific real-time PCR remains to be developed, thereby further improving also sensitivity. Phylogenetic results largely confirmed the findings with spectroscopy. Molecular data derived from 16S rRNA gene sequencing as well as multiple protein-coding phylogenies of groEL, gyrB and recA resulted – independent of the treeing method – in almost identical phylogenetic trees for respective nucleotide and amino acid alignments (data not shown). Intraspecies homology is high as can be concluded from G/C contents and average nucleotide identities. With respect to epidemiology and virulence factor analysis there is still a great demand for a broader insight into multiple Streptobacillus genomes. Studies using selective enrichment steps like for instance antibody enhanced isolations may facilitate the acquisition of novel strains from different host species in order to fill these gaps.
Conclusion
Rat bite fever represents a significant public health threat that is under-diagnosed in humans and animal species. Novel species of the genus made it necessary to critically review diagnostic tools with respect to species specificity. We have provided an update in diagnostics to improve detection and isolation of these neglected microorganisms. All members of the recently extended genus can be reliably differentiated from a pure culture by MALDI-TOF MS, FT-IR spectroscopy and also by sequence analysis of selected functional genes. 16S rRNA sequencing alone is adequate to allocate the pathogen to the correct genus, but may be insufficient for definite species diagnosis since also other Streptobacillus species except S. moniliformis are adapted to the rat oropharynx. Contrarily, growth characteristics and classical phenotypic methods and also standardized biochemistry are laborious and also only suitable for genus determination, but do not possess enough discriminatory power to sufficiently differentiate Streptobacillus on the species level. Based on additional full genome sequences, the detection of further housekeeping genes will enable the development of new tools like multilocus sequence typing (MLST) or multilocus sequence analysis (MLSA) for both, molecular epidemiology as well as in-depth infra-species resolution and the determination of clonality.Citation110 Further genetic studies on Streptobacillus should also include investigations on possible virulence determinants and differences in pathogenic mechanisms between strains.
Abbreviations
| ANI | = | average nucleotide identity |
| API | = | analytical profile index |
| ATCC | = | American Type Culture Collection, Rockville, USA |
| BHI | = | brain heart infusion |
| bp | = | base pair |
| CIP | = | Collection of Institute Pasteur, Paris, France |
| DDH | = | DNA-DNA homology |
| DIN | = | Deutsche Industrienorm (German standard) |
| DSM | = | Deutsche Sammlung für Mikroorganismen, Braunschweig, Germany |
| ELISA | = | enzyme-linked immunosorbent assay |
| FISH | = | fluorescence in situ hybridization |
| FT-IR | = | Fourier transform-infrared spectroscopy |
| G/C | = | guanine/cytosine (contents) |
| HF | = | Haverhill fever |
| IB | = | immunoblotting |
| IFA | = | immunofluorescence assay |
| IU | = | international units |
| kDa | = | kilo Dalton |
| L-form | = | cell wall deficient variant, derived from Lister (L) institute |
| MALDI-TOF MS | = | Matrix-assisted laser desorption ionization time-of-flight mass spectrometry |
| MLSA | = | multilocus sequence analysis |
| MLST | = | multilocus sequence typing |
| MIC | = | minimum inhibitory concentration |
| NCTC | = | National Collection of Type Cultures, London, UK |
| OD | = | optical density |
| PCR | = | polymerase chain reaction |
| RBF | = | Rat bite fever |
| S. | = | Streptobacillus |
| SDS-PAGE | = | sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| SPS | = | sodium polyanethol sulfonate |
| T | = | type strain |
| TLR | = | Toll-like receptor |
| w/v | = | mass/volume |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Ethical statements
The authors state that we complied with all of the legal requirements pertaining to the German Cancer Research Center (Deutsches Krebsforschungszentrum) in which animal experiments were done. The procedures were approved by the Ethics Committee of Animal Experimentation in Germany (notification no. A10-02 [Regierungspräsidium Karlsruhe]).
Acknowledgments
For excellent technical assistance we thank Ulrike Kling, Anna Mohr, Asmahan Omar, Katharina Engel, Mersiha Curić, Barbara Depner and Jens Heinbächer, Annegret Männig for proof-reading and Barbara Gamb for making even the most exotic manuscripts available. Stefanie P. Glaeser and Norman Mauder are acknowledged for their contribution to one of the figures.
Funding
The Hessian State Laboratory (Hessisches Landeslabor) is supported by Hessian Ministry for the Environment, Climate Change, Agriculture and Consumer Protection (HMUKLV).
References
- Levaditi C, Nicolau S, Poincloux P. Sur le rôle étiologique de Streptobacillus moniliformis (nov. spec.) dans l'érythème polymorphe aigu septicémique. C R Acad Sci 1925; 180:1188-90.
- Elliott SP. Rat bite fever and Streptobacillus moniliformis. Clin Microbiol Rev 2007; 20:13-22; PMID:17223620; http://dx.doi.org/10.1128/CMR.00016-06
- Row R. Cutaneous spirochetosis produced by rat bite in Bombay. Bulletin de la Societe´ de Pathologie Exotique 1918; 11:188-95.
- Gaastra W, Boot R, Ho HT, Lipman LJ. Rat bite fever. Vet Microbiol 2009; 133:211-28; PMID:19008054; http://dx.doi.org/10.1016/j.vetmic.2008.09.079
- Bleich A, Nicklas W. Zoonoses transmitted by mouse and rat maintained as laboratory or pet animals [in German]. Berl Münch Tierärztl Wochenschr 2008; 121:241-55.
- Regnath T, Kurb N, Wolf M, Ignatius R. Rat-bite fever – two cases of infection with Streptobacillus moniliformis within two months [in German]. Dtsch Med Wochenschr 2015; 140:741-3; PMID:25970414; http://dx.doi.org/10.1055/s-0041-102114
- Graves MH, Janda JM. Rat-bite fever (Streptobacillus moniliformis): a potential emerging disease. Int J Infect Dis 2001; 5:151-5; PMID:11724672; http://dx.doi.org/10.1016/S1201-9712(01)90090-6
- Hagelskjaer L, Sorensen I, Randers E. Streptobacillus moniliformis infection: 2 cases and a literature review. Scand J Infect Dis 1998; 30:309-11; PMID:9790145; http://dx.doi.org/10.1080/00365549850161016
- Washburn RG. Spirillum minus (rat bite fever). In: Mandell GL, Bennett JE, Dolin R, eds. Principles and Practice of Infectious Diseases. Philadelphia, PA: Elsevier Churchill Livingstone, 2005; 2810.
- Kimura M, Tanikawa T, Suzuki M, Koizumi N, Kamiyama T, Imaoka K, Yamada A. Detection of Streptobacillus spp. in feral rats by specific polymerase chain reaction. Microbiol Immunol 2008; 52:9-15; PMID:18352907; http://dx.doi.org/10.1111/j.1348-0421.2008.00005.x
- Wullenweber M, Kaspareit-Rittinghausen J, Farouq M. Streptobacillus moniliformis epizootic in barrier-maintained C57BL/6 J mice and susceptibility to infection of different strains of mice. Lab Anim Sci 1990; 40:608-12; PMID:2172624
- Rohde J, Rapsch C, Fehr M. Case report: Abscessation due to Streptobacillus moniliformis in a rat [in German]. Prakt Tierarzt 2008; 89:466-73.
- Boyer CIJ, Bruner DW, Brown JA. A Streptobacillus, the cause of tendon-sheath infection in turkeys. Avian Dis 1958; 2:418-27; http://dx.doi.org/10.2307/1587482
- Das AM. Streptobacillus moniliformis isolated from an abcess of a dog. Ind J Comp Microbiol Immunol Infect Dis 1986; 7:115.
- Ditchfield J, Lord LH, McKay KA. Streptobacillus moniliformis infection in a dog. Can Vet J 1961; 2:457-9; PMID:17421433
- Glünder G, Hinz KH, Stiburek B. Joint disease in turkeys caused by Streptobacillus moniliformis in Germany [in German]. Dtsch Tierärztl Wochenschr 1982; 89:367-70.
- Gourlay RN, Flanagan BF, Wyld SG. Streptobacillus actinoides (Bacillus actinoides): isolation from pneumonic lungs of calves and pathogenicity studies in gnotobiotic calves. Res Vet Sci 1982; 32:27-34; PMID:7089379
- Mohamed YS, Moorhead PD, Bohl EH. Natural Streptobacillus moniliformis infection of turkeys, and attempts to infect turkeys, sheep, and pigs. Avian Dis 1969; 13:379-85; PMID:5816049; http://dx.doi.org/10.2307/1588506
- Russell EG, Straube EF. Streptobacillary pleuritis in a koala (Phascolarctos cinereus). J Wildl Dis 1979; 15:391-4; PMID:501842; http://dx.doi.org/10.7589/0090-3558-15.3.391
- Smallwood RP. Rat bite fever from the bite of a pig. Brit Med J 1929; 29:1159.
- Valverde CR, Lowenstine LJ, Young CE, Tarara RP, Roberts JA. Spontaneous rat bite fever in non-human primates: a review of two cases. J Med Primatol 2002; 31:345-9; PMID:12519213; http://dx.doi.org/10.1034/j.1600-0684.2002.01036.x
- Yamamoto R, Clark GT. Streptobacillus moniliformis infection in turkeys. Vet Rec 1966; 79:95-100; PMID:5968147; http://dx.doi.org/10.1136/vr.79.4.95
- Rumley RL, Patrone NA, White L. Rat-bite fever as a cause of septic arthritis: a diagnostic dilemma. Ann Rheum Dis 1987; 46:793-5; PMID:3689005; http://dx.doi.org/10.1136/ard.46.10.793
- Hayashimoto N, Yoshida H, Goto K, Takakura A. Isolation of Streptobacillus moniliformis from a pet rat. J Vet Med Sci 2008; 70:493-5; PMID:18525173; http://dx.doi.org/10.1292/jvms.70.493
- Boot R, Van de Berg L, Vlemminx MJ. Detection of antibodies to Streptobacillus moniliformis in rats by an immunoblot procedure. Lab Anim 2006; 40:447-55; PMID:17018215; http://dx.doi.org/10.1258/002367706778476442
- Koopman JP, Van den Brink ME, Vennix PP, Kuypers W, Boot R, Bakker RH. Isolation of Streptobacillus moniliformis from the middle ear of rats. Lab Anim 1991; 25:35-9; PMID:1826334; http://dx.doi.org/10.1258/002367791780808211
- Wullenweber M, Jonas C, Kunstyr I. Streptobacillus moniliformis isolated from otitis media of conventionally kept laboratory rats. J Exp Anim Sci 1992; 35:49-57; PMID:1534999
- Glastonbury JR, Morton JG, Matthews LM. Streptobacillus moniliformis infection in Swiss white mice. J Vet Diagn Invest 1996; 8:202-9; PMID:8744742; http://dx.doi.org/10.1177/104063879600800210
- Kaspareit-Rittinghausen J, Wullenweber M, Deerberg F, Farouq M. [Pathological changes in Streptobacillus moniliformis infection of C57bl/6 J mice]. Berl Münch Tierärztl Wochenschr 1990; 103:84-7.
- Wullenweber M, Hedrich HJ, Reetz IC. Susceptibility to streptobacillosis of mice is highly influenced by genetic factors. AALAS Bulletin 1991; 30:43.
- Eisenberg T, Nicklas W, Mauder N, Rau J, Contzen M, Semmler T, Hofmann N, Aledelbi K, Ewers C. Phenotypic and genotypic characteristics of members of the genus Streptobacillus. PLoS One 2015; 10:e0134312; PMID:26252790; http://dx.doi.org/10.1371/journal.pone.0134312
- Swartz JD, Lachman M, Westveer K, O'Neill T, Geary T, Kott RW, Berardinelli JG, Hatfield PG, Thomson JM, Roberts A, et al. Characterization of the vaginal microbiota of ewes and cows reveals a unique microbiota with low levels of lactobacilli and near-neutral pH. Front Vet Sci 2014; 1:19; PMID:26664918; http://dx.doi.org/10.3389/fvets.2014.00019
- Wilkins EG, Millar JG, Cockcroft PM, Okubadejo OA. Rat-bite fever in a gerbil breeder. J Infect 1988; 16:177-80; PMID:3351317; http://dx.doi.org/10.1016/S0163-4453(88)94047-9
- McMillan B, Boulger LR. Squirrel-bite fever. Trans R Soc Trop Med Hyg 1968; 62:567; PMID:5691463; http://dx.doi.org/10.1016/0035-9203(68)90146-6
- Schottmüller H. On the etiology and clinical cause of the 'bite disease' (rat-, cat-, squirrel-bite diesase) [in German]. Dermatol Wochenschr Ergänzungsh 1914; 58:77-103.
- Gray HH. Squirrel bite fever. Trans R Soc Trop Med Hyg 1967; 61:857; http://dx.doi.org/10.1016/0035-9203(67)90047-8
- Chaves-Moreno D, Plumeier I, Kahl S, Krismer B, Peschel A, Oxley AP, Jauregui R, Pieper DH. The microbial community structure of the cotton rat nose. Environ Microbiol Rep 2015; PMID:26306992
- Iyer MAK. Spirillum fever caused by a monkey bite. Indian Med Gaz 1936; 71:462.
- Wouters EG, Ho HT, Lipman LJ, Gaastra W. Dogs as vectors of Streptobacillus moniliformis infection? Vet Microbiol 2008; 128:419-22; PMID:18061376; http://dx.doi.org/10.1016/j.vetmic.2007.10.019
- Parker F, Hudson NP. The etiology of Haverhill fever (Erythema arthriticum epidemicum). Am J Pathol 1926; 2:357-807; PMID:19969709
- Shanson DC, Gazzard BG, Midgley J, Dixey J, Gibson GL, Stevenson J, Finch RG, Cheesbrough J. Streptobacillus moniliformis isolated from blood in four cases of Haverhill fever. Lancet 1983; 2:92-4; PMID:6134972; http://dx.doi.org/10.1016/S0140-6736(83)90072-7
- Sprecher MH, Copeland JR. Haverhill fever due to Streptobacillus moniliformis treated with streptomycin. J Am Med Assoc 1947; 134:1014-6; PMID:20251984; http://dx.doi.org/10.1001/jama.1947.72880290001008
- Faro S, Walker C, Pierson RL. Amnionitis with intact amniotic membranes involving Streptobacillus moniliformis. Obstet Gynecol 1980; 55:9S-11S; PMID:7360458; http://dx.doi.org/10.1097/00006250-198003001-00003
- Peel MM. Dog-associated bacterial infections in humans: isolates submitted to an Australian reference laboratory, 1981–1992. Pathol 1993; 25:379-84; http://dx.doi.org/10.3109/00313029309090863
- Maynard JH, McNaughton WM, Travis T. Streptobacillus moniliformis cellulitis and bacteraemia following a dog bite. Commun Dis Intell 1986; 10:2-3.
- Nixon JH. ‘‘Rat bite fever’’ caused by a ferret. Bri Med J 1914; 2:629; http://dx.doi.org/10.1136/bmj.2.3379.629
- Dick GE, Tunnicliff R. Streptothrix isolated from blood of a patient bitten by weasel. J Infect Dis 1918; 23:183-7; http://dx.doi.org/10.1086/infdis/23.2.183
- Gilbert GL, Cassidy JF, Bennett NM. Rat-bite fever. Med J Aust 1971; 2:1131-4; PMID:5167177
- Woo PC, Wu AK, Tsang CC, Leung KW, Ngan AH, Curreem SO, Lam KW, Chen JH, Chan JF, Lau SK. Streptobacillus hongkongensis sp. nov., isolated from patients with quinsy and septic arthritis, and emended descriptions of the genus Streptobacillus and the species Streptobacillus moniliformis. Int J Syst Evol Microbiol 2014; 64:3034-9; PMID:24912824; http://dx.doi.org/10.1099/ijs.0.061242-0
- Eisenberg T, Nesseler A, Nicklas W, Spamer V, Seeger H, Zschöck M. Streptobacillus sp. isolated from a cat with pneumonia. J Clin Microbiol Case Reports 2014; 2014:1-7.
- Eisenberg T, Glaeser S, Nicklas W, Mauder N, Contzen M, Aledelbi K, Kämpfer P. Streptobacillus felis sp. nov. isolated from a cat with pneumonia. Int J Syst Evol Microbiol 2015; 65:2172-8; PMID:25858245; http://dx.doi.org/10.1099/ijs.0.000238
- Eisenberg T, Glaeser SP, Ewers C, Semmler T, Nicklas W, Rau J, Mauder N, Hofmann N, Imaoka K, Kimura M, Kämpfer P. Streptobacillus notomytis sp. nov. isolated from a spinifex hopping mouse (Notomys alexis) THOMAS, 1922 and emended description of Streptobacillus Levaditi et al. 1925, Eisenberg et al. 2015 emend. Int J Syst Evol Microbiol. 2015 Oct 5; PMID: 26438009; http://dx.doi.org/10.1099/ijsem.0.000654
- Eisenberg T, Imaoka K, Kimura M, Glaeser SP, Ewers C, Semmler T, Rau J, Nicklas W, Tanikawa T, Kämpfer P. Streptobacillus. ratti sp. nov., isolated from a black rat (Rattus rattus). Int J Syst Evol Microbiol. 2016 Apr;66(4):1620-6; PMID: 26705259; http://dx.doi.org/10.1099/ijsem.0.000869.
- Nolan M, Gronow S, Lapidus A, Ivanova N, Copeland A, Lucas S, Del Rio TG, Chen F, Tice H, Pitluck S, et al. Complete genome sequence of Streptobacillus moniliformis type strain (9901). Stand Genomic Sci 2009; 1:300-7; PMID:21304670
- Palmer R, Drinan E, Murphy T. A previously unknown disease of farmed Atlantic salmon: pathology and establishment of bacterial aetiology. Dis Aquat Org 1994; 19:7-14; http://dx.doi.org/10.3354/dao019007
- Bik EM, Chow E, Carlin KP, Jensen ED, Venn-Watson S, Relman DA. Indigenous microbiota of the bottlenose dolphin. 2nd ASM Conference on beneficial microbes: beneficial host-microbial interactions. San Diego, California, 2008.
- Bik EM, Rohlik CM, Chow E, Carlin KP, Jensen ED, Venn-Watson S, et al. Indigenous microbiota of marine mammals. 13th International Symposium on Microbial Ecology. Seattle, Washington, 2010.
- Bik EM, Costello EK, Switzer AD, Callahan BJ, Holmes SP, Wells RS, Carlin KP, Jensen ED, Venn-Watson S, Relman DA. Marine mammals harbor unique microbiotas shaped by and yet distinct from the sea. Nature communications 2016; 7:10516; PMID:26839246; http://dx.doi.org/10.1038/ncomms10516
- Dewhirst FE, Klein EA, Thompson EC, Blanton JM, Chen T, Milella L, Buckley CM, Davis IJ, Bennett ML, Marshall-Jones ZV. The canine oral microbiome. PLoS One 2012; 7:e36067; PMID:22558330; http://dx.doi.org/10.1371/journal.pone.0036067
- Xenoulis PG, Palculict B, Allenspach K, Steiner JM, Van House AM, Suchodolski JS. Molecular-phylogenetic characterization of microbial communities imbalances in the small intestine of dogs with inflammatory bowel disease. FEMS Microbiol Ecol 2008; 66:579-89; PMID:18647355; http://dx.doi.org/10.1111/j.1574-6941.2008.00556.x
- Strong T, Dowd S, Gutierrez AF, Coffman J. Amplicon pyrosequencing of wild duck eubacterial microbiome from a fecal sample reveals numerous species linked to human and animal diseases [v1; ref status: awaiting peer review, http://f1000r.es/1yy]. F1000Research 2013; 2:1-7; PMID:24358860
- Hullar MA, Lancaster SM, Li F, Tseng E, Beer K, Atkinson C, Wähälä K, Copeland WK, Randolph TW, Newton KM, et al. Enterolignan-producing phenotypes are associated with increased gut microbial diversity and altered composition in premenopausal women in the United States. Cancer Epidemiol Biomarkers Prev 2015; 24:546-54; PMID:25542830; http://dx.doi.org/10.1158/1055-9965.EPI-14-0262
- Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, Nomicos E, Polley EC, Komarow HD, Murray PR, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res 2012; 22:850-9; PMID:22310478; http://dx.doi.org/10.1101/gr.131029.111
- Aldred P, Hill AC, Young C. The isolation of Streptobacillus moniliformis from cervical abscesses of guinea-pigs. Lab Anim 1974; 8:275-7; PMID:4421530; http://dx.doi.org/10.1258/002367774780943670
- Fleming MP. Streptobacillus moniliformis isolations from cervical abscesses of guinea-pigs. Vet Rec 1976; 99:256; PMID:790752; http://dx.doi.org/10.1136/vr.99.13.256-b
- Kirchner BK, Lake SG, Wightman SR. Isolation of Streptobacillus moniliformis from a guinea pig with granulomatous pneumonia. Lab Anim Sci 1992; 42:519-21; PMID:1460856
- Smith W. Cervical abscesses of guinea-pigs. Joumal of Pathology and Bacteriology 1941; 37:29-37; http://dx.doi.org/10.1002/path.1700530104
- Eisenberg T, Kämpfer P, Ewers C, Semmler T, Glaeser SP, Collins E, Ruttledge M, Palmer R. Oceanivirga salmonicida gen. nov. sp. nov., a novel member from the Leptotrichiaceae isolated from Atlantic salmon (Salmo salar) International Journal of Systematic and Evolutionary Microbiology 2016; in press.
- Eisenberg T, Glaeser SP, Ewers C, Semmler T, Drescher B, Kämpfer P. Caviibacter abscessus gen. nov. sp. nov., a novel member from the Leptotrichiaceae isolated from guinea pigs (Cavia porcellus) International Journal of Systematic and Evolutionary Microbiology 2016; in press.
- Wullenweber M. Streptobacillus moniliformis - a zoonotic pathogen. Taxonomic considerations, host species, diagnosis, therapy, geographical distribution. Lab Animal 1995; 29:1-15; http://dx.doi.org/10.1258/002367795780740375
- Hofmann N. Phenotypical and molecular taxonomic investigations on the systematic status of Streptobacillus moniliformis, the agent of rat-bite-fever [in German]. Faculty of Biology, Leibniz Universität Hannover. Thesis Dr. rer. nat., Faculty of Biology, Leibniz Universität Hannover 1994; 105 pp.
- Liu T, Matsuguchi T, Tsuboi N, Yajima T, Yoshikai Y. Differences in expression of toll-like receptors and their reactivities in dendritic cells in BALB/c and C57BL/6 mice. Infect Immun 2002; 70:6638-45; PMID:12438336; http://dx.doi.org/10.1128/IAI.70.12.6638-6645.2002
- Irvine L, Wills T. Streptobacillus moniliformis: a mouse trying to become a rat. Clin Microbiol Newslett 2006; 28:118-20; http://dx.doi.org/10.1016/j.clinmicnews.2006.07.002
- Savage NL. Host-parasite relationships in experimental Streptobacillus moniliformis arthritis in mice. Infect Immun 1972; 5:183-90; PMID:4635498
- Lambe DW, Jr., McPhedran AM, Mertz JA, Stewart P. Streptobacillus moniliformis isolated from a case of Haverhill fever: biochemical characterization and inhibitory effect of sodium polyanethol sulfonate. Am J Clin Pathol 1973; 60:854-60; PMID:4586017; http://dx.doi.org/10.1093/ajcp/60.6.854
- Shanson DC, Pratt J, Greene P. Comparison of media with and without 'Panmede' for the isolation of Streptobacillus moniliformis from blood cultures and observations on the inhibitory effect of sodium polyanethol sulphonate. J Med Microbiol 1985; 19:181-6; PMID:2984425; http://dx.doi.org/10.1099/00222615-19-2-181
- Domingue GJ, Sr., Woody HB. Bacterial persistence and expression of disease. Clin Microbiol Rev 1997; 10:320-44; PMID:9105757
- Freundt EA. Experimental investigations into the pathogenicity of the L-phase variant of Streptobacillus moniliformis. Acta Pathol Microbiol Scand 1956; 38:246-58; PMID:13326444; http://dx.doi.org/10.1111/j.1699-0463.1956.tb03172.x
- Freundt EA. Streptobacillus moniliformis infection in mice. Acta Pathol Microbiol Scand 1956; 38:231-45; PMID:13326443; http://dx.doi.org/10.1111/j.1699-0463.1956.tb03171.x
- Edwards R, Finch RG. Characterisation and antibiotic susceptibilities of Streptobacillus moniliformis. J Med Microbiol 1986; 21:39-42; PMID:3950962; http://dx.doi.org/10.1099/00222615-21-1-39
- Anglada A, Comas L, Euras JM, Sanmarti R, Vilaro J, Brugues J. [Arthritis caused by Streptobacillus moniliformis: a case of fever induced by a rat bite]. Med Clin (Barc) 1990; 94:535-7; PMID:2192206
- Pins MR, Holden JM, Yang JM, Madoff S, Ferraro MJ. Isolation of presumptive Streptobacillus moniliformis from abscesses associated with the female genital tract. Clin Infect Dis 1996; 22:471-6; PMID:8852965; http://dx.doi.org/10.1093/clinids/22.3.471
- Razin S, Boschwitz C. The membrane of the Streptobacillus moniliformis L-phase. J Gen Microbiol 1968; 54:21-32; PMID:5729609; http://dx.doi.org/10.1099/00221287-54-1-21
- Rowbotham TJ. Rapid identification of Streptobacillus moniliformis. Lancet 1983; 2:567; PMID:6136713; http://dx.doi.org/10.1016/S0140-6736(83)90591-3
- Rygg M, Bruun CF. Rat bite fever (Streptobacillus moniliformis) with septicemia in a child. Scand J Infect Dis 1992; 24:535-40; PMID:1411321; http://dx.doi.org/10.3109/00365549209052641
- Torres L, Lopez AI, Escobar S, Marne C, Marco ML, Perez M, Verhaegen J. Bacteremia by Streptobacillus moniliformis: first case described in Spain. Eur J Clin Microbiol Infect Dis 2003; 22:258-60; PMID:12709841
- Costas M, Owen RJ. Numerical analysis of electrophoretic protein patterns of Streptobacillus moniliformis strains from human, murine and avian infections. J Med Microbiol 1987; 23:303-11; PMID:3585963; http://dx.doi.org/10.1099/00222615-23-4-303
- Boot R, Bakker RH, Thuis H, Veenema JL, De Hoog H. An enzyme-linked immunosorbent assay (ELISA) for monitoring rodent colonies for Streptobacillus moniliformis antibodies. Lab Anim 1993; 27:350-7; PMID:8277708; http://dx.doi.org/10.1258/002367793780745516
- Burke WA, Kwong O, Halpern R. Ratbite fever due to Streptobacillus moniliformis: a report of two cases. Calif Med 1959; 91:356-8; PMID:13806142
- Rau J, Eisenberg T, Wind C, Lasch P, Sting R. MALDI-TOF MS user platform (MALDI-UP) @ http://maldi-up.ua-bw.de/ and http://maldi-tof-ms-user-platform.ua-bw.de/ – An information-forum for exchange of user-made database extensions 2015.
- Mackey JR, Melendez EL, Farrell JJ, Lowery KS, Rounds MA, Sampath R, Bonomo RA. Direct detection of indirect transmission of Streptobacillus moniliformis rat bite fever infection. J Clin Microbiol 2014; 52:2259-61; PMID:24719439; http://dx.doi.org/10.1128/JCM.00259-14
- Helm D, Labischinski H, Schallehn G, Naumann D. Classification and identification of bacteria by Fourier-transform infrared spectroscopy. J Gen Microbiol 1991; 137:69-79; PMID:1710644; http://dx.doi.org/10.1099/00221287-137-1-69
- Kuhm AE, Suter D, Felleisen R, Rau J. Identification of Yersinia enterocolitica at the species and subspecies levels by Fourier transform infrared spectroscopy. Appl Environ Microbiol 2009; 75:5809-13; PMID:19617388; http://dx.doi.org/10.1128/AEM.00206-09
- Preisner O, Lopes JA, Guiomar R, Machado J, Menezes JC. Fourier transform infrared (FT-IR) spectroscopy in bacteriology: towards a reference method for bacteria discrimination. Anal Bioanal Chem 2007; 387:1739-48; PMID:17086390; http://dx.doi.org/10.1007/s00216-006-0851-1
- Wenning M, Scherer S. Identification of microorganisms by FTIR spectroscopy: perspectives and limitations of the method. Appl Microbiol Biotechnol 2013; 97:7111-20; PMID:23860713; http://dx.doi.org/10.1007/s00253-013-5087-3
- Roughgarden JW. Antimicrobial therapy of ratbite fever. A review. Archives of Internal Medicine 1965; 116(1):39-54; http://dx.doi.org/10.1001/archinte.1965.03870010041007
- Loridant S, Jaffar-Bandjee MC, La Scola B. Shell vial cell culture as a tool for Streptobacillus moniliformis “resuscitation”. Am J Trop Med Hyg 2011; 84:306-7; PMID:21292904; http://dx.doi.org/10.4269/ajtmh.2011.10-0466
- Johnson JL. Bacterial Classification III. Nucleic acids in bacterial classification. In: Krieg NR, Holt JG, eds; The Williams & Wilkins Co.: Baltimore, London, 1984;pp. 8-11.
- Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 1987; 37:463-4; http://dx.doi.org/10.1099/00207713-37-4-463
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 2007; 57:81-91; PMID:17220447; http://dx.doi.org/10.1099/ijs.0.64483-0
- Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 2009; 106:19126-31; PMID:19855009; http://dx.doi.org/10.1073/pnas.0906412106
- Boot R, Oosterhuis A, Thuis HC. PCR for the detection of Streptobacillus moniliformis. Lab Anim 2002; 36:200-8; PMID:11943086; http://dx.doi.org/10.1258/0023677021912352
- Boot R, Van de Berg L, Reubsaet FA, Vlemminx MJ. Positive Streptobacillus moniliformis PCR in guinea pigs likely due to Leptotrichia spp. Vet Microbiol 2008; 128:395-9; PMID:18023543; http://dx.doi.org/10.1016/j.vetmic.2007.10.007
- Dubois D, Robin F, Bouvier D, Delmas J, Bonnet R, Lesens O, Hennequin C. Streptobacillus moniliformis as the causative agent in spondylodiscitis and psoas abscess after rooster scratches. J Clin Microbiol 2008; 46:2820-1; PMID:18562588; http://dx.doi.org/10.1128/JCM.00744-08
- Sigge A, Essig A, Wirths B, Fickweiler K, Kaestner N, Wellinghausen N, Poppert S. Rapid identification of Fusobacterium nucleatum and Fusobacterium necrophorum by fluorescence in situ hybridization. Diagn Microbiol Infect Dis 2007; 58:255-9; PMID:17350209; http://dx.doi.org/10.1016/j.diagmicrobio.2007.01.001
- Berger C, Altwegg M, Meyer A, Nadal D. Broad range polymerase chain reaction for diagnosis of rat-bite fever caused by Streptobacillus moniliformis. Pediatr Infect Dis J 2001; 20:1181-2; PMID:11740332; http://dx.doi.org/10.1097/00006454-200112000-00021
- Glasman PJ, Thuraisingam A. Rat bite fever: a misnomer? BMJ Case Rep 2009; 2009; PMID:22180758
- Nakagomi D, Deguchi N, Yagasaki A, Harada K, Shibagaki N, Kimura M, Imaoka K, Shimada S. Rat-bite fever identified by polymerase chain reaction detection of Streptobacillus moniliformis DNA. J Dermatol 2008; 35:667-70; PMID:19017047; http://dx.doi.org/10.1111/j.1346-8138.2008.00541.x
- Wallet F, Savage C, Loiez C, Renaux E, Pischedda P, Courcol RJ. Molecular diagnosis of arthritis due to Streptobacillus moniliformis. Diagn Microbiol Infect Dis 2003; 47:623-4; PMID:14711486; http://dx.doi.org/10.1016/S0732-8893(03)00167-6
- Glaeser SP, Kämpfer P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst Appl Microbiol 2015; 38:237-45; PMID:25959541; http://dx.doi.org/10.1016/j.syapm.2015.03.007
- Woo PC, Wong SS, Teng JL, Leung KW, Ngan AH, Zhao DQ, Tse H, Lau SK, Yuen KY. Leptotrichia hongkongensis sp. nov., a novel Leptotrichia species with the oral cavity as its natural reservoir. J Zhejiang Univ Sci B 2010; 11:391-401; PMID:20506569; http://dx.doi.org/10.1631/jzus.B1000056
- Conrads G, Claros MC, Citron DM, Tyrrell KL, Merriam V, Goldstein EJ. 16S-23S rDNA internal transcribed spacer sequences for analysis of the phylogenetic relationships among species of the genus Fusobacterium. Int J Syst Evol Microbiol 2002; 52:493-9; PMID:11931161; http://dx.doi.org/10.1099/00207713-52-2-493
- Sun D, Zhang H, Lv S, Wang H, Guo D. Identification of a 43-kDa outer membrane protein of Fusobacterium necrophorum that exhibits similarity with pore-forming proteins of other Fusobacterium species. Res Vet Sci 2013; 95:27-33; PMID:23433684; http://dx.doi.org/10.1016/j.rvsc.2013.01.016
- Kim HS, Lee DS, Chang YH, Kim MJ, Koh S, Kim J, Seong JH, Song SK, Shin HS, Son JB, et al. Application of rpoB and zinc protease gene for use in molecular discrimination of Fusobacterium nucleatum subspecies. J Clin Microbiol 2010; 48:545-53; PMID:19955278; http://dx.doi.org/10.1128/JCM.01631-09
- Shah HN, Olsen I, Bernard K, Finegold SM, Gharbia S, Gupta RS. Approaches to the study of the systematics of anaerobic, gram-negative, non-sporeforming rods: current status and perspectives. Anaerobe 2009; 15:179-94; PMID:19695337; http://dx.doi.org/10.1016/j.anaerobe.2009.08.003
- Strauss J, White A, Ambrose C, McDonald J, Allen-Vercoe E. Phenotypic and genotypic analyses of clinical Fusobacterium nucleatum and Fusobacterium periodonticum isolates from the human gut. Anaerobe 2008; 14:301-9; PMID:19114111; http://dx.doi.org/10.1016/j.anaerobe.2008.12.003
- Jin J, Haga T, Shinjo T, Goto Y. Phylogenetic analysis of Fusobacterium necrophorum, Fusobacterium varium and Fusobacterium nucleatum based on gyrB gene sequences. J Vet Med Sci 2004; 66:1243-5; PMID:15528856; http://dx.doi.org/10.1292/jvms.66.1243
- Jalava J, Eerola E. Phylogenetic analysis of Fusobacterium alocis and Fusobacterium sulci based on 16S rRNA gene sequences: proposal of Filifactor alocis (Cato, Moore and Moore) comb. nov. and Eubacterium sulci (Cato, Moore and Moore) comb. nov. Int J Syst Bacteriol 1999; 49 Pt 4:1375-9; PMID:10555315; http://dx.doi.org/10.1099/00207713-49-4-1375
- Lawson PA, Gharbia SE, Shah HN, Clark DR, Collins MD. Intrageneric relationships of members of the genus Fusobacterium as determined by reverse transcriptase sequencing of small-subunit rRNA. Int J Syst Bacteriol 1991; 41:347-54; PMID:1715737; http://dx.doi.org/10.1099/00207713-41-3-347
- Gupta RS, Sethi M. Phylogeny and molecular signatures for the phylum Fusobacteria and its distinct subclades. Anaerobe 2014; 28:182-98; PMID:24969840; http://dx.doi.org/10.1016/j.anaerobe.2014.06.007
- Anonym. From the Centers for Disease Control and Prevention. Rat-bite fever–New Mexico, 1996. JAMA 1998; 279:740-1; PMID:9508137; http://dx.doi.org/10.1001/jama.279.10.740
- Brown TMP, Nunemaker JC. Rat-bite fever. A review of the American cases with reevaluation of etiology; report of cases. Bulletin Johns Hopkins Hospital 1942; 70:201-36.
- Raffin BJ, Freemark M. Streptobacillary rat-bite fever: a pediatric problem. Pediatrics 1979; 64:214-7; PMID:112571
- Savage NL, Joiner GN, Florey DW. Clinical microbiological, and histological manifestations of Streptobacillus moniliformis-induced arthritis in mice. Infect Immun 1981; 34:605-9; PMID:7309242
- van Rooyen CE. The biology, pathogenesis and classification of Streptobacillus moniliformis. Joumal of Pathology and Bacteriology 1936; 43:455-72; http://dx.doi.org/10.1002/path.1700430303
- Heilman FR. A study of Asterococcus muris (Streptobacillus moniliformis). I. Morphologic aspects and nomenclature. Journal of Infectious Diseases 1941; 69(1):32-44; http://dx.doi.org/10.1093/infdis/69.1.32
- Heilman FR. A study of Asterococcus muris (Streptobacillus moniliformis). II. Cultivations and biochemical activities. Journal of Infectious Diseases 1941; 69(1):45-51.
- Nelson JB. The reaction of antisera for B. actinoides. Journal of Bacteriology 1933; 26(3):321-7; PMID:16559658
- Bell DP, Elmes PC. Effects of certain organisms associated with chronic respiratory disease on SPF and conventional rats. J Med Microbiol 1969; 2:511-9; PMID:5404805; http://dx.doi.org/10.1099/00222615-2-4-511
- Gay FW, Maguire ME, Baskerville A. Etiology of chronic pneumonia in rats and a study of the experimental disease in mice. Infect Immun 1972; 6:83-91; PMID:4634460
- Boot R, Van de Berg L, Koedam MA, Veenema JL, Vlemminx MJ. Resistance to infection of guinea pigs with a rat Streptobacillus moniliformis. Scand J Lab Anim Sci 2007; 34:1-5.
- Livingston RS, Riley LK, Steffen EK, Besch-Williford CL, Hook RR, Jr., Franklin CL. Serodiagnosis of Helicobacter hepaticus infection in mice by an enzyme-linked immunosorbent assay. J Clin Microbiol 1997; 35:1236-8; PMID:9114413
- Boot R, van Herck H, van der Logt J. Mutual viral and bacterial infections after housing rats of various breeders within an experimental unit. Lab Anim 1996; 30:42-5; PMID:8709572; http://dx.doi.org/10.1258/002367796780744929
- Anonym. Urban pest management. Washinton: Committee on Urban Pest Management, 1980.
- Ordog GJ, Balasubramanium S, Wasserberger J. Rat bites: fifty cases. Ann Emerg Med 1985; 14:126-30; PMID:3970397; http://dx.doi.org/10.1016/S0196-0644(85)81073-8
- Legout L, Senneville E, Mulleman D, Solau-Gervais E, Flipo RM, Mouton Y. Rat bite fever mimicking rheumatoid arthritis. Scand J Infect Dis 2005; 37:532-3; PMID:16012023; http://dx.doi.org/10.1080/00365540510032114
- Naumann D. Infrared spectroscopy in microbiology. In: Meyers RA, ed. Encyclopedia of Analytical Chemistry. Chichester: John Wiley & Sons Ltd., 2000; 102-31.
- Brosius J, Palmer ML, Kennedy PJ, Noller HF. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A 1978; 75:4801-5; PMID:368799; http://dx.doi.org/10.1073/pnas.75.10.4801
- Chen PL, Lee NY, Yan JJ, Yang YJ, Chen HM, Chang CM, Lee HC, Ko NY, Lai CH, Ko WC. Prosthetic valve endocarditis caused by Streptobacillus moniliformis: a case of rat bite fever. J Clin Microbiol 2007; 45:3125-6; PMID:17652475; http://dx.doi.org/10.1128/JCM.01169-07