ABSTRACT
The Influenza virus is a leading cause of respiratory disease in the United States each year. While the virus normally causes mild to moderate disease, hospitalization and death can occur in many cases. There are several methodologies that are used for detection; however problems such as decreased sensitivity and high rates of false-negative results may arise. There is a crucial need for an effective sample preparation technology that concentrates viruses at low abundance while excluding resident analytes that may interfere with detection. Nanotrap particles are hydrogel particles that are coupled to chemical dye affinity baits that bind a broad range of proteins and virions. Within minutes (<30 minutes), Nanotrap particles concentrate low abundant proteins and viruses from clinically complex matrices. Nanotrap particles with reactive red baits concentrated numerous respiratory viruses including various strains and subtypes of Influenza virus, Coronavirus, and Respiratory Syncytial Virus from saliva, nasal fluid swab specimens, and nasal aspirates. Detection was enhanced more than 10-fold when coupled to plaque assays and qRT-PCR. Importantly, Nanotrap particle can efficiently capture and concentrate multiple viral pathogens during a coinfection scenario. These results collectively demonstrate that Nanotrap particles are an important tool that can easily be integrated into various detection methodologies.
Introduction
Influenza (commonly referred to as the “flu”) is a serious respiratory illness that is caused by the Influenza virus, a negative-sense RNA virus belonging to the family Orthomyxoviridae.Citation1 The virus leads to seasonal epidemics as well as global pandemics. In the United States alone, the virus is responsible for more than 35,000 deaths and 200,000 hospitalizations each year.Citation2,3 While seasonal epidemics occur each year, there have been several pandemics that have occurred during the last century. The emergence of a novel H1N1 strain in 1918 led to 500 million infections and over 50 million deaths worldwide.Citation4 Almost a century later, there was the appearance of the novel H1N1 strain of Influenza in 2009Citation5 that was antigenically different than any previous H1N1 epidemic strains yet striking similar to the 1918 H1N1 strain.Citation6 There is still concern that another pandemic, similar to the “1918 Spanish flu,” can emerge that would kill millions worldwide. The ability of Influenza virus to rapidly evolve results in vaccines and diagnostic assays becoming potentially ineffective.
Current Influenza diagnostic capabilities have limitations, especially in the detection of the virus at low viral genomic copies.Citation4 While Rapid Influenza Diagnostic Tests (RIDTs) can detect the virus in less than thirty minutes, specimens must be collected as early in the illness as possible (by 72 hours after infection) so that high viral loads and viral antigen levels are present.Citation7 Studies conducted by CDC have shown that RIDTs are only 50–70% sensitive compared to reverse transcription polymerase chain reaction (RT-PCR) assays and viral culture, leading to many false negative results.Citation8 This is largely due to the use of a small volume of sample in these assays as well as the inclusion and subsequent interference of high abundant proteins in complex solutions such as nasopharyngeal aspirates and swab samples. Therefore, a definitive diagnosis must be confirmed by molecular assays or viral culture. Molecular assays such as RT-PCR are able to identify viral RNA from various strains and subtypes of Influenza. However, one limitation is that the detection of viral RNA by these assays is not indicative of viable virus or on-going Influenza viral replication in the respiratory specimen, which is only possible with viral culturing.Citation6 Propagation of the virus through culturing facilitates analysis of the virus by additional methods that can be used to compare the virulence and antiviral resistance of novel, circulating, and vaccine strains of the virus.Citation6 Therefore, there is a substantial need for an accurate and reliable sample preparation methodology that concentrates whole virus from clinically complex specimens and increases the sensitivity of various diagnostic assays for Influenza and other respiratory pathogens.
Nanotrap particles are a versatile technology that can address these critical diagnostic challenges. Importantly, while most concentrators require cold-chain methodologies, Nanotrap particles can be utilized at both ambient and elevated temperatures. Furthermore, the particles are capable of concentrating analytes of interest from large volumes of complex biological fluids (such as nasal aspirates) into a significantly smaller volume (as low as 25 µL).Citation9,10 At the heart of Nanotrap technology is a base particle consisting of a network of crosslinked environmentally-responsive polymer chains that have been functionalized with affinity baits to facilitate analyte capture and retention.Citation11 The versatility of the Nanotrap particles stems from the variety of affinity baits that can be immobilized onto the polymer matrix. To date, Nanotrap particles with affinity dyes as well as ligates containing cationic, carboxylic acid, or sulfonic acid groups have been generated and utilized to target a wide range of small proteins and peptides. The analyte capture performance of the Nanotrap particles can be further altered by customizing the particle architecture (such as addition of a charged or inert polymer shell) to suit the intended application.Citation12-14
This novel sample preparation methodology has been utilized for the concentration and enhanced detection of infectious disease proteins, including a low-abundant bacterial antigen Outer surface protein A (OspA) used to diagnose Lyme diseaseCitation10 and the nucleoprotein of Rift Valley fever virus (RVFV).Citation9 While the Nanotrap particles were originally designed to specifically harvest proteins and other small molecules, recent findings by Shafagati et al have shown that the Nanotrap particles can also be used in the capture and detection of virions.Citation15 The ability of the Nanotrap particles to capture virions allows samples to be analyzed through numerous downstream analytical techniques (both protein and nucleic acid based) including standard sandwich ELISAs, lateral flow immunoassays, mass spectroscopy (MS) techniques, or q-RT-PCR assays. In addition to RVFV, Nanotrap particles have been shown to capture other viruses such as human immunodeficiency virus (HIV), Venezuelan equine encephalitis virus (VEEV), Influenza, human Coronavirus, and Adenovirus.Citation11,15 Here these findings were expanded to determine the utility of Nanotrap particles for respiratory pathogen detection, focusing primarily on Influenza virus. Data within demonstrate that this novel sample preparation tool can be coupled to both plaque assays and qRT-PCR, to enhance viral detection at varying concentrations. Importantly, the Nanotrap particles captured virus in various clinically relevant matrices commonly used in influenza diagnostics, including nasal aspirates, nasal swabs, and saliva. Lastly, Nanotrap capture was not curtailed in a coinfection scenario with various subtypes and strains of Influenza as well as other respiratory viruses.
Results
Nanotrap particles captured influenza A and B viruses
Previous published findings demonstrated that several Nanotrap particles were capable of capturing different viruses, including respiratory pathogens such as Influenza.Citation11 In order to quantitatively determine which Nanotrap particles were able to enhance detection of Influenza A, 5 different types of Nanotrap particles (NT45, NT46, NT53, NT69, and NT120) were screened for virus capture and enrichment performance (). The Nanotrap particles were incubated with one milliliter of Influenza A/California/04/09 (IA H1N1) at 5E+05 pfu/mL. Following capture, viral RNA was extracted and the amount of virus captured was determined using qRT-PCR. While all 5 Nanotrap particles successfully captured and enriched the virus, NT46, a Nanotrap particle with an immobilized reactive red dye as the bait, provided a 10-fold increase in detection compared to samples without Nanotrap particles, which were processed in parallel (). The enrichment capability of NT46 was then confirmed in other strains and subtypes of Influenza. Another Influenza A strain, Brisbane/10/2007 (H3N2) (IA H3N2), and Influenza B were tested with and without NT46. For IA H3N2, a 9-fold increase in detection was obtained with qRT-PCR (). For Influenza B, there was a 7-fold increase in detection ().
Figure 1. Nanotrap particles captured Influenza A virions. One milliliter of Influenza A/California/4/2009 (IA H1N1), Influenza A/Brisbane/10/2007 (IA H3N2), or Influenza B/Taiwan/2/62 (Influenza B) were diluted to 1E+05 pfu/mL in PBS and incubated with 100 µL NT45, NT46, NT53, NT55, or NT69 for 30 minutes at room temperature. Samples were centrifuged, unbound material removed, and the pellet was washed one time with 200 µL distilled water and incubated for an additional 15 minutes. Samples were then centrifuged and unbound material was discarded. Samples were processed for qRT-PCR (panels A-C) or plaque assays (panels D-F). No NT samples (at 100 µL volumes) were processed in parallel.
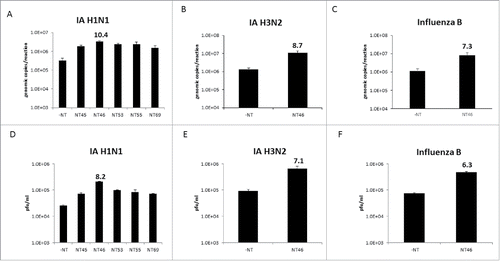
Table 1. Nanotrap particle bait and shell chemistries.
To ensure that whole infectious virus was being captured, plaque assays were performed. As in panel A, five Nanotrap particles were incubated with Influenza A, the pellet resuspended in media, and plaque assays were performed directly on the Nanotrap-viral mixture. Once again, NT46 was the best performer, providing an 8-fold increase in detection compared to the no Nanotrap control (). With NT46, a 7-fold increase in detection for IA H3N2 () and 6-fold increase in detection with Influenza B () were obtained. Collectively, these results demonstrate that infectious virions can be captured and enriched using NT46 and increased detection, up to 10-fold and 8-fold in qRT-PCR and plaque assay, respectively can be achieved. While Influenza capture can vary depending on the strain and subtype, NT46 consistently enhanced the detection of various Influenza viruses with plaque assays and qRT-PCR. Therefore subsequent experiments were focused on evaluating the enrichment performance of NT46.
Nanotrap particles bind and captured influenza A hemagglutinin proteins
It was hypothesized that the Nanotrap particles capture virus via the surface glycoproteins. To test this hypothesis, hemagglutinin (HA) protein was incubated with Nanotrap particles and captured assessed by western blot. In agreement with the results presented in , NT46 was the most efficient at capturing HA protein from IA H1N1 (HA1) (). NT46 was able to capture as little as 10 ng/mL of HA1 protein, yielding approximately 100 fold enrichment compared to the no NT control (). There are a variety of Influenza virus subtypes (currently 16 HA subtypes) and mutations in the HA protein occur quite frequently.Citation16 To determine if Nanotrap particles can broadly bind to HA proteins, HA5 and HA7 proteins were also tested. NT46 was also capable of capturing and enriching HA5 and HA7 proteins (). These results indicate that Nanotrap particles can bind to multiple HA proteins and suggest that they would be capable of capturing Influenza viruses even if they were to mutate.
Figure 2. Nanotrap particles enhanced detection of Influenza hemagglutinin proteins. (A) One mL of histidine-tagged hemagglutinin protein (His-HA) from IA H1N1 (1 µg/mL) was incubated with 100 µL NT45, NT46, NT53, NT55, or NT69 for 30 minutes at room temperature. (B) IA H1N1 His-HA (1 µg/mL, 100 ng/mL, or 10 ng/mL) was incubated with 100 µL NT46 for 30 minutes at room temperature. (C) His-HA proteins (1 µg/mL) from H1N1, H5N1, or H5N8 were incubated with 100 µL NT46 for 30 minutes at room temperature. (D) His-HA proteins (1 µg/mL) from H1N1, H7N2, and H7N7 were incubated with 100 µL NT46 for 30 minutes at room temperature. Samples in panels A-D were analyzed for the presence of Influenza HA by western blot using antibodies directed against the histidine tag. Control samples are His-HA at 100 μg/mL and/or 10 μg/mL (panels C and D) and a no NT (1 μg/mL of His-HA) sample at a volume of 10 µL processed in parallel.
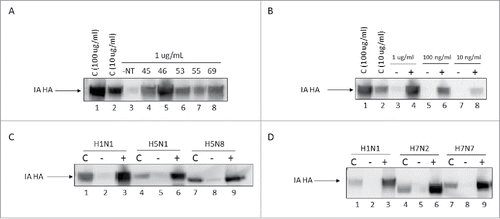
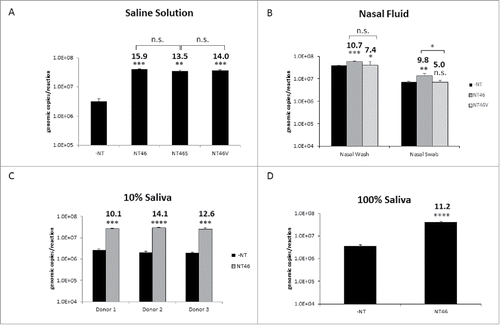
Figure 3. Nanotrap particles enhanced detection of Influenza in clinically relevant matrices. (A) IA H1N1 was diluted to 1E+05 pfu/mL in PBS and incubated with NT46, NT46 with an outer shell (NT46S), or NT46 with an outer shell coated with VSA (NT46V). Samples were processed for qRT- PCR as described in . No NT samples (at 100 µL volumes) were processed in parallel. (B) For the nasal wash samples, IA H1N1 was diluted to 1E+05 pfu/mL in 10% human nasal fluid and incubated with NT46 or NT46V. For the nasal swab samples, Influenza A/California/4/2009 was diluted to 1E+06 pfu/mL in 100% human nasal fluid and spiked onto a flocked swab. The swab was swirled in 1 mL PBS, the swab head was cut and placed in a spin basket on top of the microcentrifuge tube containing the sample, and centrifuged at 14,000 rpm for 5 minutes. The swab head and spin basket were removed, the sample transferred to a new microcentrifuge tube, and 100 µL of NT46 or NT46V was added to each sample. Samples were processed for qRT-PCR as described in . No NT samples (at 100 µL volumes) were processed in parallel. Statistical significance for panels A and B were determined through one-way ANOVA followed by Tukey's multiple comparisons test: *p-value < 0 .05, **p-value ≤ 0 .01, and ***p-value ≤ 0 .001. n.s. is not significant. Samples were compared to the –NT sample (significant of comparison indicated without brackets) and to each other (significance of comparison indicated with brackets). C) IA H1N1 was diluted to 1E+05 pfu/mL in 10% human saliva from three different donors and incubated with NT46. Samples were processed for qRT-PCR as described in . No NT samples (at 100 µL volumes) were processed in parallel. D) IA H1N1 was diluted to 1E+05 pfu/mL in 100% human saliva from a single donor and incubated with NT46. Samples were processed for qRT-PCR as described in . No NT samples (at 100 µL volumes) were processed in parallel. Statistical significance for panels C and D were determined through student's t-tests: ***p-value ≤ 0 .001, ****p-value < 0 .0001 (compared to the –NT sample).
Reactive red nanotrap particles enhanced viral detection in clinically relevant matrices
There are several specimens that can be used for Influenza diagnostics. These include nasopharyngeal swabs, nasopharyngeal aspirates and washes, deep nasal swabs, throat swabs, and saliva swabs and aspirates. The next set of experiments set out to demonstrate that reactive red particles can enrich virus in these clinically relevant matrices. Nanotrap particles with an outer shell surrounding the particle core have previously been shown to function well in serum.Citation9,15 It was speculated that the incorporation of an outer shell would deter viral and host proteins in nasal fluid and saliva from binding to the Nanotrap particles. Therefore, in parallel to NT46, two additional red particles with different architectures were tested for Influenza capture (). NT46S contains an inert polymer shell surrounding the dye-functionalized Nanotrap particle core, while the NT46V particle core is encapsulated within a charged polymer shell containing sulfonic acid groups. Capture of Influenza with NT46, NT46S, and NT46V were compared to one another in a saline solution containing no additional host proteins. In this scenario, there was no significant difference in viral RNA detection among the three reactive red particle types (). Since there was no difference in capture for the two core-shell particles, only NT46 and NT46V were utilized in subsequent experiments.
NT46 and NT46V were next utilized in a nasal aspirate scenario. IA H1N1 was spiked in 10% nasal fluid in a saline solution. The Nanotrap particles were then incubated with 1ml of the spiked sample. Interestingly, the results showed a near 11-fold increase in detection for NT46 and a 7-fold increase in detection for NT46V (), suggesting that the addition of a sulfonic acid outer shell was not necessary to enhance Influenza capture. However, the capture differences between the two particles were not statistically different. IA H1N1 capture by NT46 was also demonstrated with nasal fluid from a second donor (Fig. S1). The capture efficiency of NT46 and NT46V was then tested in a swab scenario. IA H1N1 was spiked into nasal fluid and 50μl of the spiked samples was added onto a swab. The swab was suspended in 950μl saline solution, the swab heads spun down to remove all fluid, and the Nanotrap particles added to the sample. While both Nanotrap particles were able to concentrate IA H1N1, there was a statistically significance difference in capture with the two Nanotrap particles. The use of NT46V resulted in a 5-fold increase in detection compared to nearly 10-fold increase with the use of NT46 ().
Lastly, Nanotrap particle capture in saliva was performed. Sputum and saliva samples are less frequently used in viral diagnostics as the viral yield is significantly lower for these types of specimens.Citation17,18 However, saliva is a good tool for detection as it is less invasive and easier to collect from patients. It was speculated that since the components of saliva are less viscous compared to nasal fluid samples, saliva samples would be easier to couple with Nanotrap particles. IA H1N1 was spiked into 10% saliva. To evaluate the effect of individual saliva protein composition on Nanotrap particle binding to Influenza A virus, saliva from three different donors was tested. Samples were briefly sonicated and incubated with NT46. NT46 provided an increase in detection (10.1–14.1-fold) compared to the no NT control (). Importantly, three different batches of NT46 tested in parallel demonstrated similar enrichment capabilities in both saline and 10% saliva (Fig. S2), demonstrating that the Nanotrap particles perform consistently. NT46 was also able to capture and enrich IA H1N1 in neat saliva (). Collectively, these results demonstrate that a core-only reactive red particle, NT46, can be utilized to dramatically concentrate virus and increase viral detection in various clinical matrices containing numerous host and viral proteins.
NT46 enhanced influenza A virus detection at both high and low titers
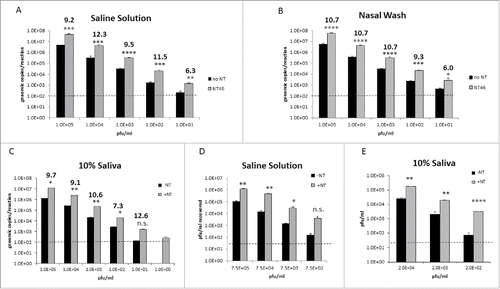
As previously mentioned, viral shedding is highest by 48 hours after infection with Influenza and dramatically increase thereafter.Citation19 If samples are not collected during the peak times, false negative results are likely to occur. The goal of the next set of experiments was to utilize the Nanotrap particles to concentrate virus and increase the lower limit of detection (LOD) when viral titers are virtually undetectable. IA H1N1 was spiked into saline solution, 10% nasal fluid, or 10% saliva at various concentrations and the ability of NT46 to capture IA H1N1 was analyzed with qRT-PCR or plaque assays (). At every concentration of virus tested and in all three fluids, NT46 was capable of capturing and enriching IA H1N1. Notably, at 1.0E+01 pfu/ml, viral RNA is nearing the LOD of our qRT-PCR assay. However with NT46 incubation, IA H1N1 is well above the LOD (panels A-C). Likewise IA H1N1 at 1.0E+00 pfu/ml in saliva was undetectable without Nanotrap particles, but could be readily detected with the use of NT46 (panel C). Plaque assays confirmed these results, demonstrating a consistent enrichment of IA H1N1 virions down to 7.5E+02 pfu/ml in saline (panel D) or 2.0E+02 pfu/ml in saliva (panel E). These results demonstrate that NT46 can increase detection of IA H1N1 at both high and low viral titers.
Figure 4. Nanotrap particles enhanced detection of Influenza at both high and low titers. A-C) IA H1N1 was diluted from 1E+05 pfu/mL to 1E+01 pfu/ml in saline (panel A), 1E+05 pfu/mL to 1E+01 pfu/ml in nasal wash (panel B), or 1E+05 pfu/mL to 1E+00 pfu/ml in 10% human saliva (panel C) and incubated with NT46. Samples were processed for qRT-PCR as described in . No NT samples (at 100 µL volumes) were processed in parallel. D and E) IA H1N1 was diluted from 7.5E+05 pfu/ml to 7.5E+02 pfu/ml in saline solution (panel D) or diluted from 2.0E+04 pfu/mL to 2.0E+02 pfu/ml in 10% human saliva (panel E) and incubated with NT46. Samples were processed for plaque assay as described in . No NT samples (at 100 µL volumes) were processed in parallel. Statistical significance for all panels was determined through student's t-tests: *p-value ≤ 0 .05, **p-value ≤ 0 .01, **p-value ≤ 0 .01, ****p-value ≤ 0 .0001, and n.s is not significant (compared to the –NT sample). Dashed lines represent the lower LOD for the assays, which is 100 genomic copies/reaction for IA H1N1 qRT-PCR and 2E+01 pfu/ml for the plaque assays.
Nanotrap particles enhanced detection of multiple respiratory pathogens
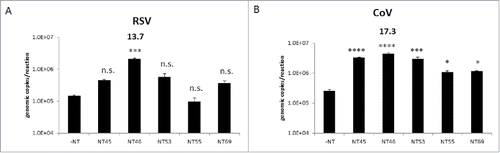
It had previously been demonstrated that Nanotrap particles can capture other respiratory pathogens such as Adenovirus (a DNA virus) and Coronavirus (another RNA virus).Citation11 The enrichment capabilities of the Nanotrap particles were further investigated with various viruses causing respiratory ailments. Respiratory syncytial virus A2001/3–12 strain (RSV) and Coronavirus 229E strain (CoV), two enveloped RNA viruses were tested with and without Nanotrap particles. For both RSV and CoV, the five Nanotrap particles used for the initial Influenza A screenings were utilized (). With the exception of NT55, four out of the five Nanotrap particles captured and enriched RSV (). All five Nanotrap particles enriched CoV (). However, NT46 once again was the best performer for both viruses, providing a 14-fold and 17-fold increase in detection for RSV and CoV, respectively. Collectively, the results demonstrated that one Nanotrap particle, NT46, can be used in the capture and enrichment of multiple respiratory pathogens.
Figure 5. Nanotrap particles captured other respiratory viral pathogens. One ml of respiratory syncytial virus (RSV) diluted to 1E+05 genomic copies/5 µL (panel A) or coronavirus (CoV) strain 229E diluted to 1E+05 genomic copies/5 µL (panel B) were added to 100 µL NT45, NT46, NT53, NT55, or NT69. Samples were processed for qRT-PCR as described in . No NT samples (at 100 µL volumes) were processed in parallel. Statistical significance was determined through one-way ANOVA followed by Tukey's multiple comparisons test: *p-value < 0 .05, ***p-value ≤ 0 .001, and ****p-value < 0 .0001 (compared to the –NT sample). n.s. is not significant.
Nanotrap particles enhanced detection of respiratory viruses in a coinfection scenario
Coinfections of Influenza with other respiratory pathogens such as RSV and CoV can occur (especially in children and immunocompromised adults).Citation20-22 Therefore, it is important for a Nanotrap particle to enrich virus not only in the presence of other host and viral analytes, but also in the presence of other infectious pathogens. In order to test the enrichment capability of the Nanotrap particles, NT46 was utilized in a dual infection scenario with multiple respiratory viruses. It was hypothesized that two virus types will compete for binding of NT46, and detection with Nanotrap particles for one or both viruses may therefore be decreased. To test this hypothesis, a mock coinfection scenario was performed by incubating NT46 with a sample containing both IA H1N1 and IA H3N2 at 1E+05 pfu/mL in saline solution. For the NT46-incubated samples, there was no significant change in the detection of IA H1N1 in the presence of IA H3N2. Likewise there was no statistically significant drop in IA H3N2 detection in the presence of IA H1N1 (). Next, a mock infection scenario with IA H1N1 and Influenza B (IB) viruses in saline solution was performed. Again, for the samples incubated with NT46, there was no significant change in either IA H1N1 or IB detection (). There was no significant change in detection for the no Nanotrap samples in the presence of multiple viruses (data not shown).
Figure 6. Nanotrap particles captured multiple viruses in one sample. (A) Influenza A/California/4/2009 (IA H1N1), Influenza A/Brisbane/10/2007 (IA H3N2), or a combination of the 2 viruses were diluted to 1E+05 pfu/mL in PBS and incubated with NT46. B) IA H1N1, Influenza B/Taiwan/2/62 (IB), or a combination of the two viruses were diluted to 1E+05 pfu/mL in PBS and incubated with NT46. (C) IA H1N1, CoV 229E strain, or a combination of the two viruses were diluted to 1E+05 pfu/mL (for IA) or 1E+05 genomic copies/5µL (for CoV) in PBS and incubated with NT46. (D) IA H1N1, RSV A2 strain, or a combination of the two viruses were diluted to 1E+05 pfu/mL (for IA H1N1) or 1E+05 genomic copies/5µL (for RSV) in PBS and incubated with NT46. Samples were processed and analyzed for qRT PCR with viral-specific primers as described in . No NT samples (at 100 µL volumes) were processed in parallel. Fold enrichment values for NT46 samples are shown. Statistical significance was determined through student's t- tests: **p-value ≤ 0 .01 (compared to the –NT sample), n.s. is not significant.
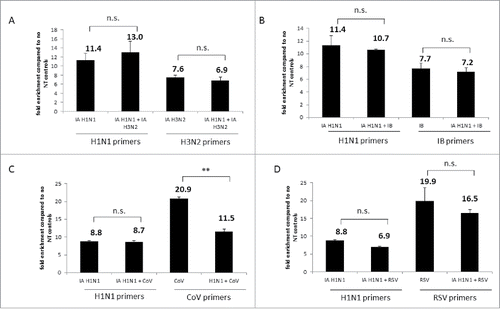
Coinfection scenarios with Influenza and two other respiratory viruses, CoV and RSV, were then performed in 10% saliva. In the coinfection scenario with IA H1N1 and CoV incubated with NT46, there was no change in detection for IA H1N1. However, there was a very significant drop in CoV detection from 21-fold with CoV alone to 11-fold in the presence of IA H1N1 (). In contrast to the previous experiments, there was a decrease in IA H1N1 detection in the presence of RSV, falling from 9-fold for IA H1N1 alone to 7-fold in the coinfection scenario. Although not deemed statistically significant, a decrease in RSV detection from 20-fold to 16-fold was seen in the presence of IA H1N1 (). Once again, there was no significant change in detection for the no NT samples (data not shown). Collectively, these results indicate that NT46 can capture and concentrate a variety of different viruses in a coinfection scenario. However, while enrichment with NT46 remained high, various viruses may be competing with each other for NT46 binding.
Nanotrap particles rapidly capture influenza A virus
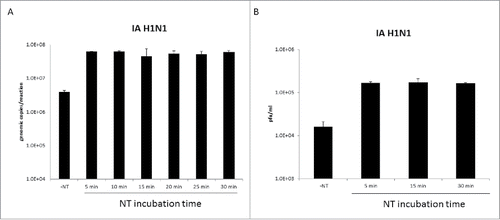
One of the primary benefits of the Nanotrap particles is their ability to capture and concentrate viruses. However, this does add additional sample processing steps to the diagnostic workflow. Therefore to determine if the capture time could be shortened, Nanotrap particles were incubated with IA H1N1 for various periods of time (5 to 30 minutes) and viral capture assayed by qRT-PCR () or plaque assays (). Five minutes of incubation time provided similar levels of enrichment as those observed with even the longest incubation time tested (30 minutes). These results demonstrate that Nanotrap particles can rapidly capture Influenza virus and suggest that the Nanotrap particle workflow could be reduced by at least 25 minutes.
Figure 7. Nanotrap particles enriched Influenza A after five minutes of incubation. One milliliter of IA H1N1 was diluted to 1E+05 pfu/mL in PBS and incubated with 100 µL NT46 from 5 minutes to 30 minutes at room temperature. Samples were processed for qRT-PCR (panel A) or plaque assays (panel B) as described in . No NT samples (at 100 µL volumes) were processed in parallel.
Discussion
Nanotrap particles have traditionally been used for the capture and enrichment of proteins.Citation10,12,14,23 Several papers have shown that these hydrogel particles enrich numerous proteins such as insulin, myoglobin, and PDGF.Citation13,14 A 2011 paper by Douglas et al demonstrated that the Nanotrap particles can dramatically concentrate Lyme disease antigens in urine and enhance detection at previously undetectable concentrations.Citation10 Most recently, a paper published by Shafagati et al demonstrated that the Nanotrap particles can also be utilized for the enrichment and protection of viral antigens such as RVFV NP.Citation9 The Nanotrap particles can also be used for the capture of virions. In 2013, the capture and enrichment of virions from RVFV and other viral pathogens was shown with several Nanotrap particles.Citation15 Importantly, the virus can be inactivated with heat or detergent after Nanotrap particle incubation and viral nucleic acid is still detectable with qRT-PCR. Here, the previous findings have been expanded upon to demonstrate that the Nanotrap particles can be utilized as a sample preparation tool to concentrate Influenza and other respiratory pathogens.
The first goal was to determine the compatibility of the Nanotrap particles with the Influenza virus. From the initial Nanotrap particle screenings, NT46, a reactive red particle, was identified as the top candidate for capture of the H1N1 strain of Influenza. The enrichment capability of NT46 was confirmed in two other types of Influenza, an H3N2 Influenza A strain and Influenza B. The results suggested that despite variations in the HA and NA viral glycoproteins that could change binding to the Nanotrap core and/or bait, NT46 dramatically concentrates various strains and subtypes of Influenza. While the exact mechanism by which the Nanotrap particles, specifically NT46, bind to the Influenza virus is unknown, it is hypothesized that the Nanotrap particles bind to the virus via the virus' surface glycoproteins. Data demonstrating that NT46 captures HA1, HA5, and HA7 proteins support this hypothesis. As previously mentioned, NT46 contains a reactive red dye. This triazine-derived textile dye has a negative charge and has commonly been used for protein purification via affinity chromatography.Citation24 The structure of the triazine dyes mimics the structure of substrates that bind to the active sites of different proteins, and is therefore able to bind a broad class of proteins and enzymes.Citation25 It is possible that triazine dyes may bind to molecules involved in the infection mediated by viral envelope proteins, or more specifically, the extracellular portion of viral glycoproteins that is involved in virus-cell fusion.Citation26 It is further speculated that a charge-based interaction could a play a role in NT46 binding with the virus.Citation12 The host cell membrane and sialic acid receptors have a net negative charge while HA1 subunit of human Influenza A typically has a positive charge.Citation27 The negative charge of the anionic NT46 may mimic the interaction that the host has with the viral glycoproteins and therefore facilitate binding to the positive charged portion of HA. Alternately, other positively-charged Nanotrap particles could be binding to the negatively charged zone in the neuraminidase active site.Citation16 NT46 was previously shown to capture RVFV and HIV to varying degrees.Citation15 However a Nanotrap particle containing cibracon blue was the most effective at capturing RVFV and HIV, suggesting that RVFV and HIV glycoproteins have a greater affinity for cibracon blue as compared to the reactive red affinity bait. In order to investigate these theories, future studies will utilize electron microscopy to determine the exact mechanism of Nanotrap particle binding to the virion.
A common problem in diagnostics is the interference of high abundant host proteins. Nasopharyngeal and saliva samples contain viscous analytes that non-specifically bind to antibodies and can significantly decrease detection. Furthermore, nasal wash collection dramatically dilutes the sample as a total of 3–8 mL can be collected from one patient.Citation28,29 However, molecular assays and RIDTs allow testing of only 50–120 μL of sample. The nasal aspirate experiments performed here utilized 1 mL of sample with Nanotrap particles. While there is a slight drop in detection in clinically relevant matrices (nasal aspirate, nasal fluid, and saliva) compared to saline solution, there is significant enrichment of both saliva and nasal aspirate samples with NT46. NT46V, the core-shell variety of NT46, was tested to determine if the presence of a polymeric shell could improve Influenza detection in clinically relevant matrices. NT46V contains an outer polymer shell functionalized with sulfonic acid groups, which provides the particle with a negative charge. Since there was no significant change in capture with particles that contained a shell versus core-only particles, the presence of the polymer shell was not necessary for analyte capture for this specific application. Virions potentially bind to host proteins such as mucin that are found in nasal fluid and saliva. The more promiscuous nature of core-only particles may allow these protein-bound virions to be captured by the Nanotrap particles, whereas the presence of a polymer shell would make any affinity baits within the core less accessible, thereby excluding the virus-protein complexes and only binding individual virions. Interestingly, when using the core-only particles masking of binding sites was not observed. One possible explanation for this observation may be due to the negative charge of the NT46 particles. Results of this study suggest that the negative charge of the NT46 particle is capable of repelling some types of matrix proteins from binding, allowing for target analytes to interact with the affinity dye immobilized onto the core particles.
The enrichment capability of NT46 was further tested with other viruses causing upper and lower respiratory illnesses. For RSV and CoV, which are both enveloped RNA viruses, there was up to a 21-fold increase in detection with NT46. These results strongly suggests that the Nanotrap particle core-shell and/or bait interacts with the viral glycoproteins such as the HA protein of Influenza and the spike protein of CoV. These results demonstrated that one Nanotrap particle, NT46, can be coupled to a multiplex assay that allows for the testing of various pathogens in one panel. Therefore, the data within provide critical information that lays the groundwork for future studies coupling the Nanotrap particles with clinical samples.
It has previously been shown that Nanotrap particles work in a mixed infection scenario with RVFV and HIV coinfection.Citation15 Here, the findings were extended to respiratory pathogens, specifically with Influenza, CoV, and RSV. While the promiscuity of the Nanotrap particles allows for more than one virus or analyte to be captured, the results demonstrate that this feature does not interfere with the Nanotrap particle's enrichment capability in a dual infection scenario. This feature of the Nanotrap particles is favorable when the cause of infection is unknown as several viruses can result in the same symptoms. Moreover, patients may be coinfected with more than one pathogen at a time. While infectivity with multiple pathogens is rarely tested by clinicians, several studies have shown that dual respiratory virus infection occurs in almost 20% of cases of lower respiratory tract infections. This is especially common in children and in those with immunocompromised systems.Citation30,31 In one study, 20 out of 75 children were infected with more than one type of respiratory virus. In most cases with children, coinfection commonly occurs with RSV and another respiratory pathogen such as Influenza.Citation31 Although rare, coinfection with both Influenza A and B types have also been seen in patients.Citation32 It is also important to note that secondary bacterial infections with Streptococcus pneumoniae oftentimes arise after infection with Influenza and may lead to serious “superinfections.”Citation33 The results show that the Nanotrap can successfully capture and detect multiple viral pathogens in one sample. While the Nanotrap particles have been utilized for the capture of virus, there is no published data on the capture of bacteria with Nanotrap particles. Since bacteria are significantly larger than viruses, Nanotrap particles with larger surface areas may be optimal for the capture of bacteria. Future studies will investigate the enrichment capability of the Nanotrap particles with bacteria causing respiratory disease (such as Streptococcus pneumoniae) and couple the Nanotrap particle technology to a multiplex assay that can test for multiple pathogens, both viral and bacterial, in one sample and with one Nanotrap particle (NT46).
While data presented herein clearly demonstrated significant sample enrichment and detection of respiratory pathogens in complex biological fluids, there are possible limitations of the Nanotrap technology. Additional sample processing time and the need for specialized equipment (e.g. centrifuges) is a concern. One step to limit the additional processing time is to decrease the initial incubation period of the sample with the Nanotrap particles. As little as 5 minutes of incubation with NT46 captured and enriched Influenza A virus. In addition, magnetic versions of the Nanotrap particles are currently in development in order to improve the Nanotrap recovery process following sample incubation as well as shorten the sample processing time. By incorporating magnetic nanoparticles into the polymeric network of the Nanotrap particles, the Nanotraps can be collected and separated from the biofluid sample via magnetic decantation, thereby negating the need for specialized equipment such as benchtop centrifuges and allowing for the Nanotraps to be used in a resource-limited setting. Using magnetic decantation to separate the particles from the biofluid solution will also decrease the sample processing time by 30–60 minutes, depending on the workflow requirements, thereby allowing target analytes to be captured, recovered, and detected within 30 minutes or less. The second limitation relates to the availability of the affinity baits. In the event that an affinity bait is discontinued by the supplier, this would result in limited supply of the specific Nanotrap type, decreasing the availability of the Nanotrap, and increasing the cost of that Nanotrap type. As a measure to counter this, new affinity baits are being consistently tested to determine if there are any baits that can be substituted to generate a particle with capture and recovery performance to the particles that are in limited supply.
Future projects will also investigate the feasibility of extending this concept to other downstream methodologies, which will include lateral flow assays (LFA), enzyme-linked immunoassays (ELISA), and bead-based immunoassay platforms which are used to detect the presence of viral antigens such as Influenza NP. The Nanotrap particle technology may resolve the sensitivity issues that are a common problem during both LFAs and ELISA diagnostics as viral titers often fall below the threshold of detection. Lastly, the Nanotrap particles will be coupled to point-of-care diagnostic devices (RIDTs) that can rapidly (<30 minutes) detect various respiratory pathogens. This concept can be expanded to numerous emerging infectious diseases and provide a quick, safe, and easy-to-use tool for diagnostics in a field and clinical setting.
Materials and methods
Nanotrap particles
Nanotrap particles were provided by Ceres Nanosciences, Inc., Manassas, VA. Nanotraps are environmentally-responsive hydrogel particles that are based on crosslinked poly(N-isopropylacrylamide) chains. The Nanotrap particles are easily customizable and can be engineered to have the following features in addition to the main core particle: 1) a charged or inert polymer shell, 2) charged functional groups, such as carboxylic acid or amine functional groups, and/or 3) affinity dye functional groups, such as Cibacron Blue F3G-A. For the purposes of this paper, five different types of Nanotraps were engineered to suit the capture and enrichment of the target analyte. A series of QA/QC tests are conducted on each newly synthesized lot/batch of Nanotrap particles including comparing their analyte capture and recovery performance with existing lots of the same type of Nanotrap in order to ensure comparable performance and reduce batch-to-batch variability. Prior to sample incubation, 500 µL of particles were centrifuged at 14,000 rpm for 10 minutes. The Nanotrap storage solution was discarded and the pellet was resuspended in 500 µL distilled ultrapure water. The samples were centrifuged again at the same speed and time. The supernatant was once again discarded, and the pellet was resuspended in 400 µL distilled ultrapure water at a final concentration of 5 mg/mL.
Cell culture
The Madine-Darby canine kidney (MDCK) cell line was obtained from American Type Culture Collection (ATCC). The cells were grown in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FBS, 1% penicillin/streptomycin, and 1% glutamax (supplemented DMEM) and cultured in a humidified environment containing 5% CO2 at 37°C.
Viruses
Influenza A/California/4/09(H1N1), Influenza B/Taiwan/2/62, and Respiratory Syncytial Virus (A2001/3-12 strain) were obtained from BEI Resources (catalog numbers NR-13658, NR-3181, and NR-28526, respectively). Influenza A/Brisbane/10/2007 (H3N2) and Coronavirus strain 229E were obtained from Influenza Reagent Resource (catalog numbers FR-8 and FR-303, respectively). The Influenza viruses were propagated by infecting MDCK cells at 80–90% confluency at an MOI of 0.1 in Influenza Growth Media (IGM; DMEM supplemented with 1% bovine serum albumin, 1% non-essential amino acids, 1% L-glutamine, and 1% penicillin/streptomycin). Cell culture medium was collected from the cells when ∼80% cytopathic effect was observed [typically 24–48 hours post-infection (hpi)]. Cell culture medium was centrifuged at 10,000 rpm for 10 minutes to pellet the cellular debris. Cell free-viral supernatants were then filtered using a 0.22 µM filter and viral titers determined by plaque assays. The genome copies of the viruses were determined by extracting viral RNA from cell free viral supernatants and performing qRT- PCR with viral specific primers (described below).
Specimen samples
Nasal fluid was obtained from Lee Biosolutions (991-13-5). Human saliva was obtained from BioreclamationIVT (HMSALIVA). Samples were sonicated using the MISONIX Ultrasonic Liquid Processor (XL-2000 Series model) for 3 10-second pulses at 600 W. Between pulses, the samples were incubated on ice for 10 seconds. Nasal swabs obtained from BinaxNOW® Influenza A and B kits were used in the mock-swab scenarios.
Standard nanotrap particle incubation
According to a protocol standardized by Ceres Nanosciences, 1000 µL of sample (containing virus spiked in either IGM or phosphate buffered saline) was incubated with 100 µL of Nanotrap particles at 4 mg/mL for 30 minutes at room temperature. The sample was centrifuged at 14,000 rpm for 10 minutes and the supernatant containing the unbound material was discarded. The pellet was then washed one time with 200 µL distilled ultrapure water and incubated for an additional 20 minutes. The samples were then centrifuged at 14,000 rpm for 10 minutes, after which the unbound material was discarded and the pellet was resuspended in the appropriate buffer.
Plaque assays
MDCK cells were plated in 6 well plates at 1.0E+06 cells/ml in order to achieve 100% confluency. After Nanotrap particle incubation, the pellet was resuspended in 100 µL of IGM and serial dilutions were performed directly from the resuspended sample. For the –NT samples (control viral samples not incubated with Nanotrap particles), serial dilutions were performed on a starting volume of 100 µL. A volume of 400 µL of the serial dilution was added to each well in duplicate and incubated for 50 minutes. The primary overlay (known as the CV mixture) consisted of equal parts 0.9% agarose in distilled water and media containing 2X EMEM, 5% BSA, 2% non-essential amino acids, 2% penicillin/streptomycin, 2% sodium pyruvate, and 2% L-glutamine. The CV mixture was treated with 0.011% of 2 mg/mL Trypsin-TPCK (obtained from Sigma-Aldrich). A volume of 300 µL of the CV mixture was added directly to each well. The cells were fixed with 10% formaldehyde in water after 48–72 hpi. The cells were stained with 1% Crystal Violet in 20% ethanol and water. After two hours, the crystal violet stain was washed off and the plaques formed were counted to determine the plaque forming units per milliliter (pfu/mL).
RNA extraction and quantitative reverse transcription PCR
After Nanotrap particle incubation, the pellet was resuspended in 100 µL of distilled ultrapure water containing carrier RNA (Life Technologies) at a 1:128 ratio. For the –NT samples, a volume of 100 µL was processed in parallel. A volume of 375 µL TRIzol LS® Reagent (Life Technologies) was added to the +NT and –NT samples, vortexed for 10 seconds, and allowed to incubate at room temperature for 5 minutes. A volume of 100 µL chloroform was then added to the sample and vortexed for 10 seconds. After a five minute incubation, the samples were centrifuged at 4°C and at a speed of 12,000 × g for 15 minutes. The colorless upper aqueous phase (∼200 µL) was removed and saved for downstream analysis. A volume of 50 µL of sample was transferred to a 96-well plate and RNA extraction was performed with Ambion's MagMax 96-well Viral RNA extraction kit according to manufacturer's instructions.
In order to determine the number of viral genomic copies produced, qRT-PCR was performed using either the Superscript® III Platinum® SYBR® green One Step qRT-PCR Kit (Thermo Fisher Scientific) with viral specific primers or RNA UltraSense One-Step Quantitative RT-PCR System (Thermo Fisher Scientific) with viral specific primers and probe. IA H1N1, RSV and CoV were detected with SYBR® green PCR according to a standardized protocol using 15 µL of master mix containing enzyme mix, 2X reaction mix, 10 µM forward primer and 10 µM reverse primer. Primers used include: IA H1N1 forward primer – ATT ACT GGA CAC TAG TAG AGC, IA H1N1 reverse primer - GCA TTT CTT TCC ATT GCG AA, CoV, forward primer- CGC AAG AAT TCA GAA CCA GAG; CoV reverse primer - GGC AGT CAG GTT CTT CAA CAA, RSV forward primer – CAT CCA GCA AAT ACA CCA TCC A, RSV reverse primer – TTC TGC ACA TCA TAA TTA GGA GTA TCA A. The samples were heated at 50°C for 3 minutes hold, 95°C for 5 minutes hold, and 40 cycles of 95°C for 15 seconds and 49–52°C for 30 seconds. A melting curve was added at the end of the reaction.
For IA H3N2 and Influenza B detection, the RNA UltraSense kit was used according to a standardized protocol using 20 µL of master mix containing enzyme mix, 5X reaction mix, ROX reference dye, 10 µM TaqMan fluorogenic probe, 40 µM forward primer and 40 uM reverse primer added to 5 µL of extracted RNA. The primer and probe set was obtained from BEI resources (NR-15592 and NR-15592). The samples were heated at 50°C for 30 minutes, 95°C for 2 minutes, and at 95°C and 55°C for 45 cycles.
qRT-PCR analysis was performed using the StepOne Plus Real Time PCR System (Applied biosystems). Viral genomic copies were determined by comparing the unknowns to a standard curve containing known RNA quantities and extrapolating the value. RNA standards were made by extracting viral RNA from IA H1N1, IA H3N2, Influenza B, RSV, and CoV viral supernatants using TRIzol LS® Reagent and quantifying the viral RNA with Quant-iT Ribogreen RNA assay kit (ThermoFisher Scientific). Representative standard curves for each qRT-PCR assay are shown in Figure S3.
Western blot analysis
Histidine-tagged hemagglutinin (HA) proteins from Influenza A/California/07/2009 (H1N1), Influenza A/Indonesia/5/2005 (H5N1), Influenza A/gyrfalcon/Washington/41088-6/2014 (H5N8), Influenza A/New York/107/2003 (H7N2), and Influenza A/Netherlands/219/2003 (H7N7) were obtained from Influenza Reagent Resource (catalog numbers FR-559, FR-59, FR-1418, FR-69, and FR-71, respectively). Nanotrap pellets containing captured Influenza HA proteins were resuspended in 25 µL of blue lysis buffer (containing 1:1 mixture of T-PER reagent (Pierce, IL), 2× Tris-glycine SDS sample buffer (Novex, Invitrogen), 33 mM DTT, and protease and phosphatase inhibitor cocktail (1× Halt cocktail, Pierce)). No Nanotrap control samples were resuspended in 10 µL of lysis buffer. All samples were boiled for 10 min. The Nanotrap samples were then centrifuged at 14,000 rpm. The supernatant was saved into a new microcentrifuge tube containing 2 µL NuPAGE® 4X LDS buffer (Invitrogen) and samples centrifuged for a second time to ensure that all Nanotrap particles were pelleted. The supernatants were separated on NuPAGE® 4–12% Bis-Tris gels (Invitrogen) and transferred to nitrocellulose or PVDF membranes at 80 mA at 4°C overnight. The membranes were blocked with 3% milk in 1× PBS +0.1% Tween (PBS-T) for 1 hour at room temperature. His-tag antibody (Cell Signaling Technology) was diluted in 3% milk in PBS-T at a 1:1000 dilution. The membranes were then washed 3 times with PBS-T and incubated with secondary HRP-coupled anti-rabbit antibody (Cell Signaling Technology) diluted 1:2,000 in 3% milk for 1 hour and then washed 4 times with PBS-T for 5 minutes. The protein gel blots were visualized by chemiluminescence using SuperSignal West Femto Maximum Sensitivity Substrate kit (ThermoScientific) and a Bio-Rad Molecular Imager ChemiDoc XRS system (Bio-Rad).
Statistics
Statistical significance was determined using either student's unpaired t test to compare the mean or fold enrichment values of the two sample sets (no NT controls and NT46) or one-way ANOVA followed by Tukey's multiple comparisons test. Differences were deemed statistically significant if the p-value was ≤0 .05.
Disclosure of potential conflicts of interest
BL and AP are employed by Ceres Nanosciences. Co-author KKH is a member of the Scientific Advisory board at Ceres Nanosciences. BL is a shareholder at Ceres Nanosciences. The Nanotrap particles used in this study were research grade and provided by Ceres Nanosciences which are not commercially available products. This does not alter the authors' adherence to all of the policies on sharing data and materials.
KVIR_S_1185585_Supplemental.zip
Download Zip (155 KB)Funding
The following reagents were obtained through BEI Resources, NIAID, NIH: Human Respiratory Syncytial Virus, A2001/3–12 (NR-28526), Influenza A Virus, A/California/04/2009 (H1N1)pdm09, Cell Isolate (NR-13658) and Influenza B Virus, B/Taiwan/2/1962 (NR-3181). Histidine-tagged hemagglutinin (HA) proteins from Influenza A/California/07/2009 (H1N1), Influenza A/Indonesia/5/2005 (H5N1), Influenza A/gyrfalcon/Washington/41088-6/2014 (H5N8), Influenza A/New York/107/2003 (H7N2), and Influenza A/Netherlands/219/2003 (H7N7) were obtained from Influenza Reagent Resource, Influenza Division, WHO Collaborating Center for Surveillance, Epidemiology and Control of Influenza, Centers for Disease Control and Prevention, Atlanta, GA, USA (catalog numbers FR-559, FR-59, FR-1418, FR-69, and FR-71, respectively). Influenza A /Brisbane/10/2007 (H3N2) (FR-8) and Human Coronavirus, Strain 229E (ATCC® VR-740™) (FR-303) were also obtained through the Influenza Reagent Resource. NS was supported through a graduate student fellowship provided through the IIAD, HS-STEM Career Development Program. This material is based upon work supported by the US. Department of Homeland Security under Grant Award Number 20011-ST-104-000002. The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the US. Department of Homeland Security. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Labella AM, Merel SE. Influenza. Med Clin North Am 2013; 97:621-45; PMID:23809717; http://dx.doi.org/10.1016/j.mcna.2013.03.001
- Rothberg MB, Haessler SD, Brown RB. Complications of Viral Influenza. Am J Med 2008; 121:258-64; PMID:18374680; http://dx.doi.org/10.1016/j.amjmed.2007.10.040
- Baigent SJ, McCauley JW. Influenza type A in humans, mammals and birds: Determinants of virus virulence, host-range and interspecies transmission. BioEssays 2003; 25:657-71; PMID:12815721; http://dx.doi.org/10.1002/bies.10303
- Johnson NPAS, Mueller J. Updating the Accounts: Global Mortality of the 1918–1920 “Spanish” Influenza Pandemic. Bull Hist Med 2002; 76:105-15; PMID:11875246; http://dx.doi.org/10.1353/bhm.2002.0022
- Centers for Disease Control and Prevention. CDC Novel H1N1 Flu | The 2009 H1N1 Pandemic: Summary Highlights, April 2009-April 2010 [Internet]. [cited 2014 Apr 7]; Available from: http://www.cdc.gov/h1n1flu/cdcresponse.htm
- Cohen J. What's Old Is New: 1918 Virus Matches 2009 H1N1 Strain. Science 2010; 327:1563-4; PMID:20339037; http://dx.doi.org/10.1126/science.327.5973.1563
- Nonpharmaceutical Interventions for Pandemic Influenza, International Measures. Emerg Infect Dis 2006; 12:81-7; PMID:16494722; http://dx.doi.org/10.3201/eid1201.051370
- Centers for Disease Control and Prevention. CDC - Rapid Diagnostic Testing for Influenza: Information for Health Care Professionals | Health Professionals | Seasonal Influenza (Flu) [Internet]. [cited 2014 Apr 7]; Available from: http://www.cdc.gov/flu/professionals/diagnosis/rapidclin.htm
- Shafagati N, Lundberg L, Baer A, Patanarut A, Fite K, Lepene B, Kehn-Hall K. The use of Nanotrap particles in the enhanced detection of Rift Valley fever virus nucleoprotein. PloS One 2015; 10:e0128215; PMID:26020252; http://dx.doi.org/10.1371/journal.pone.0128215
- Douglas T, Tamburro D, Fredolini C, Espina B, Lepene BS, Ilag L, Espina V, Petricoin EF, Liotta LA, Luchini A. The Use of Hydrogel Microparticles to Sequester and Concentrate Bacterial Antigens in a Urine Test for Lyme Disease. Biomaterials 2011; 32:1157-66; PMID:21035184; http://dx.doi.org/10.1016/j.biomaterials.2010.10.004
- Shafagati N, Patanarut A, Luchini A, Lundberg L, Bailey C, Petricoin E, Liotta L, Narayanan A, Lepene B, Kehn-Hall K. The use of Nanotrap particles for biodefense and emerging infectious disease diagnostics. Pathog Dis 2014; 71(2):164-76; PMID:24449537; http://dx.doi.org/10.1111/2049-632X.12136
- Luchini A, Fredolini C, Espina BH, Meani F, Reeder A, Rucker S, Petricoin EF, Liotta LA. Nanoparticle Technology: Addressing the fundamental roadblocks to protein biomarker discovery. Curr Mol Med 2010; 10:133-41; PMID:20196732; http://dx.doi.org/10.2174/156652410790963268
- Luchini A, Geho DH, Bishop B, Tran D, Xia C, Dufour R, Jones C, Espina V, Patanarut A, Zhu W, et al. Smart Hydrogel Particles: Biomarker Harvesting: One-step affinity purification, size exclusion, and protection against degradation. Nano Lett 2008; 8:350-61; PMID:18076201; http://dx.doi.org/10.1021/nl072174l
- Patanarut A, Luchini A, Botterell PJ, Mohan A, Longo C, Vorster P, Petricoin EF, Liotta LA, Bishop B. Synthesis and characterization of hydrogel particles containing Cibacron Blue F3G-A. Colloids Surf Physicochem Eng Asp 2010; 362:8-19; http://dx.doi.org/10.1016/j.colsurfa.2010.03.023
- Shafagati N, Narayanan A, Baer A, Fite K, Pinkham C, Bailey C, Kashanchi F, Lepene B, Kehn-Hall K. The Use of NanoTrap Particles as a Sample Enrichment Method to Enhance the Detection of Rift Valley Fever Virus. PLoS Negl Trop Dis 2013; 7:e2296; PMID:23861988; http://dx.doi.org/10.1371/journal.pntd.0002296
- Covalciuc KA, Webb KH, Carlson CA. Comparison of Four Clinical Specimen Types for Detection of Influenza A and B Viruses by Optical Immunoassay (FLU OIA Test) and Cell Culture Methods. J Clin Microbiol 1999; 37:3971-4; PMID:10565916
- Ortiz de la Tabla V, Masiá M, Antequera P, Martin C, Gazquez G, Buñuel F, Gutiérrez F. Comparison of combined nose-throat swabs with nasopharyngeal aspirates for detection of pandemic influenza A/H1N1 2009 virus by real-time reverse transcriptase PCR. J Clin Microbiol 2010; 48:3492-5; PMID:20702662; http://dx.doi.org/10.1128/JCM.01105-10
- Lau LLH, Cowling BJ, Fang VJ, Chan KH, Lau EHY, Lipsitch M, Cheng CKY, Houck PM, Uyeki TM, Peiris JSM, et al. Viral shedding and clinical illness in naturally acquired influenza virus infections. J Infect Dis 2010; 201:1509-16; PMID:20377412; http://dx.doi.org/10.1086/652241
- Stefanska I, Romanowska M, Donevski S, Gawryluk D, Brydak LB. Co-infections with influenza and other respiratory viruses. Adv Exp Med Biol 2013; 756:291-301; PMID:22836647; http://dx.doi.org/10.1007/978-94-007-4549-0_36
- Yoshida LM, Suzuki M, Nguyen HA, Le MN, Vu TD, Yoshino H, Schmidt WP, Nguyen TTA, Le HT, Morimoto K, et al. Respiratory syncytial virus: co-infection and paediatric lower respiratory tract infections. Eur Respir J 2013; 42:461-9; PMID:23645407; http://dx.doi.org/10.1183/09031936.00101812
- Debiaggi M, Canducci F, Ceresola ER, Clementi M. The role of infections and coinfections with newly identified and emerging respiratory viruses in children. Virol J 2012; 9:247; PMID:23102237; http://dx.doi.org/10.1186/1743-422X-9-247
- Tamburro D, Fredolini C, Espina V, Douglas TA, Ranganathan A, Ilag L, Zhou W, Russo P, Espina BH, Muto G, et al. Multifunctional core-shell nanoparticles: discovery of previously invisible biomarkers. J Am Chem Soc 2011; 133:19178-88; PMID:21999289; http://dx.doi.org/10.1021/ja207515j
- Stellwagen E. Dye Affinity Chromatography [Internet]. In: Current Protocols in Protein Science. John Wiley & Sons, Inc; 2001 [cited 2014 Aug 7]. Available from: http://onlinelibrary.wiley.com/doi/10.1002/0471140864.ps0902s00/abstract
- Hattori T, Zhang X, Weiss C, Xu Y, Kubo T, Sato Y, Nishikawa S, Sakaida H, Uchiyama T. Triazine dyes inhibit HIV-1 entry by binding to envelope glycoproteins. Microbiol Immunol 1997; 41:717-24; PMID:9343823; http://dx.doi.org/10.1111/j.1348-0421.1997.tb01916.x
- Perez LG, O'Donnell MA, Stephens EB. The transmembrane glycoprotein of human immunodeficiency virus type 1 induces syncytium formation in the absence of the receptor binding glycoprotein. J Virol 1992; 66:4134-43; PMID:1602536
- Kobayashi Y, Suzuki Y. Compensatory Evolution of Net-Charge in Influenza A Virus Hemagglutinin. PLoS One 2012; 7:e40422; PMID:22808159; http://dx.doi.org/10.1371/journal.pone.0040422
- Garman E, Laver G. The Structure, Function, and Inhibition of Influenza Virus Neuraminidase [Internet]. In: Fischer WB, editor. Viral Membrane Proteins: Structure, Function, and Drug Design. Springer: US; 2005 [cited 2015 Mar 16]. page 247-67. Available from: http://link.springer.com/chapter/10.1007/0-387-28146-0_17
- Aldona Z, Wos MD. Available from: http://www.flu.nc.gov/providers/documents/FluTestingGuidanceSLPHFeb2013.pdf
- Massachusettes General Hospital. MGH Microbiology Specimen Collection Procedure for Rapid RSV Antigen Detection [Internet]. 2012; Available from: http://mghlabtest.partners.org/RSV_spec_collect_2010.pdf
- Drews AL, Atmar RL, Glezen WP, Baxter BD, Piedra PA, Greenberg SB. Dual Respiratory Virus Infections. Clin Infect Dis 1997; 25:1421-9; PMID:9431390; http://dx.doi.org/10.1086/516137
- Jennings LC, Anderson TP, Werno AM, Beynon KA, Murdoch DR. Viral etiology of acute respiratory tract infections in children presenting to hospital: role of polymerase chain reaction and demonstration of multiple infections. Pediatr Infect Dis J 2004; 23:1003-7; PMID:15545854; http://dx.doi.org/10.1097/01.inf.0000143648.04673.6c
- Calistri A, Salata C, Cosentino M, Asnicar S, Franchin E, Cusinato R, Pacenti M, Donatelli I, Palù G. Report of two cases of influenza virus A/H1N1v and B co-infection during the 2010/2011 epidemics in the Italian Veneto Region. Virol J 2011; 8:502; PMID:22050693; http://dx.doi.org/10.1186/1743-422X-8-502
- Rynda-Apple A, Robinson KM, Alcorn JF. Influenza and Bacterial Superinfection: Illuminating the Immunologic Mechanisms of Disease. Infect Immun 2015; 83:3764-70; PMID:26216421; http://dx.doi.org/10.1128/IAI.00298-15
