ABSTRACT
Aspergillus fumigatus is an opportunistic fungal pathogen and the most important species causing pulmonary fungal infections. The signaling by calcium is very important for A. fumigatus pathogenicity and it is regulated by the transcription factor CrzA. We have previously used used ChIP-seq (Chromatin Immunoprecipitation DNA sequencing) aiming to identify gene targets regulated by CrzA. We have identified among several genes regulated by calcium stress, the putative flavin transporter, flcA. This transporter belongs to a small protein family composed of FlcA, B, and C. The ΔflcA null mutant showed several phenotypes, such as morphological defects, increased sensitivity to calcium chelating-agent ethylene glycol tetraacetic acid (EGTA), cell wall or oxidative damaging agents and metals, repre-sentative of deficiencies in calcium signaling and iron homeostasis. Increasing calcium concentrations improved significantly the ΔflcA growth and conidiation, indicating that ΔflcA mutant has calcium insufficiency. Finally, ΔflcA-C mutants showed reduced flavin adenine dinucleotide (FAD) and were avirulent in a low dose murine infection model.
Introduction
Calcium signaling is very important for fungal morphology and metabolism.Citation1-5 The calcineurin phosphatase and the transcription factor Crz1p1/CrzA are essential for fungal calcium signaling.Citation1,2 In Saccharomyces cerevisiae, several environmental stresses, for instance osmotic, extreme pH, high temperature, ER stress and prolonged incubation with mating pheromone α-factor, are regulated by calcineurin.Citation5,6 Calcineurin also connects many stress response signaling pathways.Citation7 Cyclosporin A, an immunosuppressant, reduces calcineurin activity and causes morphological changes and growth reduction in A. nidulans, A. oryzae, Magnaporthe oryzae and Neurospora crassa.Citation8-12 Calcineurin activates the transcription factor Crz1 (Calcineurin Responsive Zinc Finger 1) transcription factor by dephosphorylating it when there in an increase cytosolic calcium and permitting its translocation to the nuclei.Citation1,13 Crz1 has a C2H2 zinc finger motif responsible for the binding to a CDRE (calcineurin-dependent response element) sequence in the promoter regions of the Crz1-regulated genes.Citation1,14
Fungal infections are becoming very important considering that presently there is a larger number of people dying from fungal infections than malaria and tuberculosis, what is due to the increase of the number of patients with immunosuppression.Citation15 The signaling by calcium is very important for fungal virulence and drug resistance.Citation1,2 Calcineurin has been demonstrated to be required for virulence in human fungal pathogens, such as Cryptococcus spp, Candida spp, Paracoccidioides brasiliensis, and Aspergillus fumigatus;Citation16-23 and fungal plant pathogens, such as Sclerotinia scleotiorum, Botrytis cinerea, Magnaporthe oryza, and Ustilago spp.Citation24-28 In all these fungal pathogens, calcineurin is important for growth, morphology, state transitions, cation homeostasis, stress responses, and cell membrane and cell wall integrity pathways.Citation1,2,29-33
Aspergillus fumigatus is an opportunistic fungal pathogen and the most important species causing pulmonary fungal infections.Citation15,34-38 Although A. fumigatus is able to cause many clinical forms, the most important is the invasive pulmonary aspergillosis (IPA), that has mortality rates as high as 80% in immunosuppressed patients.Citation15,36-38 We have used used ChIP-seq (Chromatin Immunoprecipitation DNA sequencing) aiming to identify gene targets regulated by CrzA. We have identified among several genes regulated by calcium stress, flcA, encoding the putative flavin adenine dinucleotide (FAD) transmembrane transporter (the predicted homolog of the S. cerevisiae flavin carrier FLC2).Citation39,40 The Candida albicans homolog of this gene was identified when expressed in S. cerevisiae because it increased the heme transport. Since S. cerevisiae cannot grow very well on media with heme as a single iron source, the overexpression of the C. albicans homolog, CaFLC1, improved considerably its growth on heme iron as single iron source.Citation40 In S. cerevisiae Flc mutants have deficiencies in the cell wall, are resident in the endoplasmic reticulum, and cannot transport FAD into the lumen of the endoplasmic reticulum.Citation40 Here, we characterized in more detail FlcA, which belongs to a small protein family of putative flavin transporters composed of 3 members, FlcA, B, and C. The flcA null mutant had several important phenotypes, such as morphogenetic defects, sensitivity to calcium, cell wall and oxidative damaging agents, and metals. Finally, ΔflcA-C mutants were avirulent in a low dose murine infection model.
Results
FlcA is a member of a small protein family
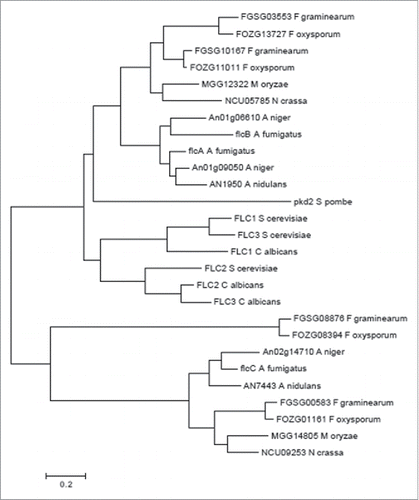
By using ChIP-seq to reveal CrzA targets in A. fumigatus, we identified the A. fumigatus homolog of the S. cerevisiae FAD transmembrane transporter FLC2 (Afu4g13340, flcA). BLASTp analysis revealed 2 putative paralogues of FlcA, FlcB (Afu2g17650, 61.1 % identity, 77.3 % similarity, e-value 0.0) and FlcC (Afu2g06100, 28.9 % identity, 46.8 % similarity, e-value 7e-59). The flcA-C gene models are supported by RNA-seq data (available at www.aspgd.org). The hypothetical proteins encoded by flcA-C were predicted to be 721, 612, and 723 amino acids in length and possessed a mass of 78.6, 66.3, and 79.4 kDa, respectively. We have compared the protein structures and organizations between FlcA-C by using the SMART interface (http://smart.embl-heidelberg.de/). The organization of the protein FlcA-C domains was conserved with 2 important Pfam domains (Fig. S1): (i) a TRP_N (PF14558) that might be involved in lipid binding and (ii) A TRP (PF06011) that represents a family of transient receptor protein channel-like proteins, which may be responsible for FAD transport into the endoplasmic reticulum lumen where it is important for oxidative protein folding. The predicted FlcA-C amino acid sequences have between 9 and 7 hydrophobic transmembrane domains, respectively (Fig. S2). FlcA-C contained N-terminal signal peptides putatively assigned by several prediction programs as endoplasmic reticulum resident proteins (SignalP, www.cbs.dtu.dk/services/SignalP; Euk-mPLoc 2.0, http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/; Predotar, htpps://urgi.versailles.inra.fr/predotar/predotar.html). The FlcA-C have several homologues in other filamentous fungi (protein identity greater than 70%) (). However, the phylogenetic analyses clearly demonstrated 2 different branches from yeasts and molds, and that FlcA and FlcB homologues are much closer than FlcC homologues ().
Figure 1. Phylogeny of A. fumigatus FlcA-C. The optimal tree for the A. fumigatus is represented. The tree was inferred using the Neighbor-Joining Method. Sequences were aligned with ClustalW and the tree was constructed by using MEGA6.
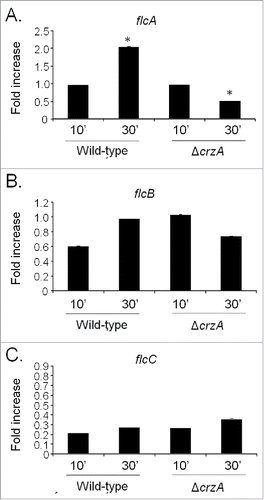
Next step, we compared flcA-C mRNA accumulation in the wild-type and ΔcrzA strains in response to a short exposure (10 or 30 min) to calcium (200 mM CaCl2) by using qRT-PCR (). These conditions are absolutely identical to the experimental conditions used for identifying flcA by ChIP-seq (de Castro et al., 2014). The flcA showed a twice increase in mRNA accumulation post calcium exposure in the wild-type strain, while its mRNA accumulation was decreased also twice after 30 minutes in the post calcium exposure ΔcrzA mutant (). The flcB and flcC do not show significant modulation post calcium exposure in the wild-type and ΔcrzA strains (). These results suggest that upon calcium exposure, CrzA influences flcA mRNA accumulation.
Figure 2. The A. fumigatus flcA expression is dependent on CrzA. The qRT-PCR for the A. fumigatus (A) flcA, (B) flcB, and (C) flcC genes. The strains were grown for 16 hours at 37 °C and transferred to 200 mM CaCl2 for 10 and 30 min. The results are expressed as fold increase of the control (in the absence of CaCl2) and the results were normalized with the βtub expression (*, p < 0.001).
Phenotypic characterization of an A. fumigatus putative flavin flcA-C transporter family
To a better understanding of the function of FlcA-C in A. fumigatus, the flcA-C genes were deleted with the pyrG marker (Fig. S3A to C). To exclude the possibility that undesired mutations throughout the creation of the deletion strains contributed to the phenotypes observed (Fig. S4), we have complemented the null mutants with the equivalent wild-type genes. The promoters of the flcA, B, and C genes are not affecting the pyrG promoter because the deletion strains grow to the same extent on minimal medium and minimal medium supplemented with uridine and uracil (Figs. S4). That provided strong evidence that the lack of uridine and uracil into the host is not affecting the pyrG promoter and consequently the reduced viability of these strains into the host is not due to a marker effect. The ΔflcA-C mutants were exhaustively investigated for phenotypes that could be affected by their absence (Figs. S6–8). However, we were only able to identify phenotypes for ΔflcA as described here. The ΔflcA mutant showed a dramatic reduction in radial growth in solid minimal and complete media when compared to the wild-type and ΔflcB-C strains, but surprisingly exhibited about the same dry weight in liquid minimal media ( and ). The viability of the ΔflcA-C conidia is similar to the wild-type strain (Fig. S9). Accordingly, conidial germination of the ΔflcA and wild-type strains in liquid minimal media showed the same germination and nuclear kinetics, while the apical tip of the ΔflcA strain showed bipolar elongation (). The compact morphology and reduced radial growth of the ΔflcA mutant on solid media () was attributed to an increase in apical branching in comparison to the wild-type strain (). We also investigated a possible transcriptional compensatory mechanism for the absence of each flc gene by measuring the flcA-C mRNA accumulation in ΔflcA-C mutant strains in response to a short pulse (10 or 30 min) of calcium (200 mM CaCl2) via qRT-PCR (). There is a significant increased flcA expression in ΔflcB and ΔflcC (about 3-fold at 0 and 30 and 10 and 30 min post calcium exposure, respectively; ). In ΔflcC and ΔflcA mutant strains, there are significant increases of about 6- and 3-fold in the flcB mRNA accumulation at 0 and 10 min, respectively (). There is significant increase in the flcC expression (about twice and 5-fold) at time 0 for both ΔflcA and ΔflcB mutant strains (). These results suggest that there are compensatory transcriptional mechanisms affecting increased flcA-C mRNA accumulation in the ΔflcA-C mutant strains.
Figure 3. The A. fumigatus ΔflcA has morphogenetic defects. The wild-type, ΔflcA-C, and their corresponding complementing strains were grown for 48 h at 37 °C on solid (A) or liquid MM (B). A. fumigatus wild-type and ΔflcA germlings were grown in liquid MM for 12 h and stained or not with calcofluor white (C, top panels, bars 5 µM) or for 20 h at 30 °C (C, lower panels, bars, 10 µM). (D) The edge of the colonies represented in the plates of (A). Bars, 50 µM. (E) The qRT-PCR for the A. fumigatus flcA-C genes in the wild-type, ΔflcA, ΔflcB, and ΔflcC strain. The strains were grown for 16 hours at 37 °C (time 0) and transferred to 200 mM CaCl2 for 10 and 30 min. The results are expressed as the number of cDNA copies of a specific flc gene divided by the number of copies of the cDNA of the normalizer βtub (p < 0.001).
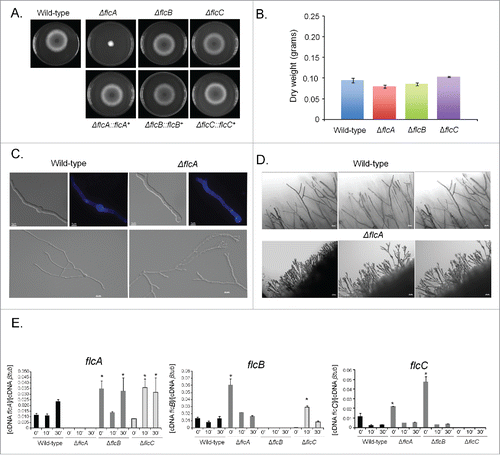
Figure 4. The ΔflcA mutant strain is more sensitive to calcium stress, cell wall damaging and oxidative stressing agents. (A) The wild type, the mutant, and the complemented strains were grown on YAG medium with increasing concentrations of EGTA, cyclosporin, calcofluor white (CFW), congo red (CR), Sodium Dodecyl Sulfate (SDS), t-butyl hydroxyperoxide, and paraquat for 48 h at 37 °C. (B) Radial growth of the wild-type and ΔflcA mutant strains grown in YAG medium for 120 hours at 37 °C in the absence and presence of increasing CaCl2 concentrations. The results are the average ± standard deviation of 3 repetitions. Statistical analysis was performed by using One-way Anova with post test Dunnett (*** p < 0.05).
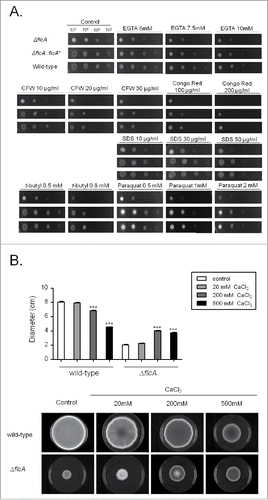
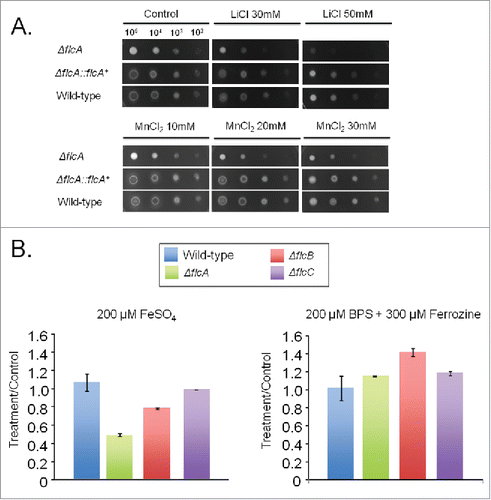
The ΔflcA mutant was more sensitive than the wild-type strain to the calcium chelating-agent ethylene glycol tetraacetic acid (EGTA), calcofluor white (CFW), congo red (CR), t-butyl hydroperoxide, and paraquat (). The increased sensitivity of ΔflcA to EGTA suggests that this mutant has a calcium shortage. Increasing CaCl2 concentrations in YAG medium improved significantly the ΔflcA growth and conidiation (), indicating that ΔflcA mutant has calcium insufficiency. The ΔflcA mutant was also more sensitive to metals, such as lithium, manganese and iron, but not to iron starvation ().
Figure 5. The A. fumigatus ΔflcA is more sensitive to metals. (A) The wild type, the mutant, and the complemented strains were grown on YAG medium with increasing concentrations of LiCl and MnCl2. (B) The wild type and the mutant strains were grown for 48 h at 37 °C in MM+200 µM FeSO4 or AMM+ferrozine+BPS. The data were normalized by dividing the dry weight of the treatments by the dry weight of the corresponding strains grown for 48 h at 37 °C in MM.
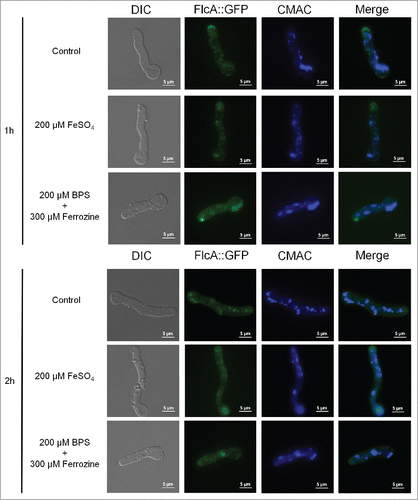
To verify FlcA-C cellular locatization, we generated FlcA-C::GFP strains which behaved identical to the wild-type strain (data not shown). Very low fluorescence was observed for FlcB::GFP and FlcC::GFP, not allowing us to determine its subcellular location (data not shown). In contrast, we were able to observe FlcA::GFP expressed as a single band of 103.6 kDa (Fig. S10) and when the FlcA::GFP strain was grown in minimal media for 16 hours at 30°C, a weak and diffuse fluorescent signal was distributed along the germlings in the cytosol and in some structures resembling vesicles, as confirmed by vacuolar staining with CMAC (in about 100 % of the germlings; Fig. S11). In addition, strong staining was visible in the apical tip (about 50 % of the germlings, ). The sub-cellular localization or intensity of the signal was not altered when the germlings were either exposed to iron excess or starvation for 1 or 2 hours () or paraquat (Fig. S11). Furthermore, we have not observed any difference in the FlcA::GFP localization in the presence of high calcium chloride concentrations (Fig. S11). The same results were observed at 37 °C (Fig. S11).
Figure 6. FlcA::GFP accumulates in the apical tip, cytoplasm and in vesicles. The FlcA::GFP strain was grown for 12 h at 30 °C in MM and transferred to either MM+FeSO4 or AMM+ferrozine+BPS for 1or 2 h. Bars, 5 μm.
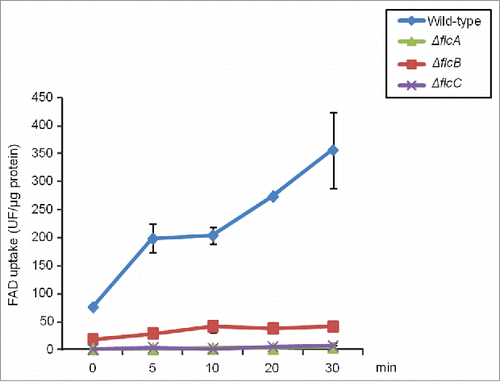
We also measured the retention of FAD into microsomes by using wild-type and ΔflcA-C protoplasts, and measured retained FAD by fluorescence spectrophotometry. We observed that wild-type protoplasts showed a vigorous accumulation of FAD while the ΔflcA-C protoplasts exhibited very low accumulation of FAD (). Taken together these results suggest that FlcA is important for morphogenetic development, and sensitivity to calcium, cell wall damaging agents, ions and oxidative stress. Additionally, all 3 genes are important for FAD accumulation.
The flcA-C are important for virulence in a neutropenic murine model of invasive pulmonary aspergillosis
We investigated the involvement of FlcA-C in A. fumigatus pathogenicity by using a neutropenic murine model of invasive pulmonary aspergillosis (). The infection with the wild-type strains produced 100 % mortality 12 to 15 d post-infection, but the infection with ΔflcA, ΔflcB, and ΔflcC yielded a significantly reduced mortality rate, at approximately 20 to 30% 15 d post-infection (, no statistical differences between the mutants and PBS negative control, p > 0 .1951 to 0.2024 Log−rank (Mantel−Cox) test and p > 0 .1967 to 0.2024 Gehan−Breslow−Wilcoxon test). The complemented strains (single ectopic re-integrations of the wild-type flcA-C loci) have not shown statistical difference with the wild−type strain (, p > 0 .2099 to 0.5886 and p > 0 .3029 to 0.7557 for the comparison between the wild-type and the complemented strains, Log−rank, Mantel−Cox and Gehan−Breslow−Wilcoxon tests, respectively).
Figure 8. A. fumigatus ΔflcA-C mutants are avirulent. (A) Comparative analysis of wild type, mutant, and complemented strains in a neutropenic murine model of pulmonary aspergillosis. Mice in groups of 10 per strain were infected intranasally with a 20 μl suspension of conidia at a dose of 105. Fungal burden was determined 48 h post-infection by real-time qPCR based on 18 S rRNA gene of A. fumigatus and an intronic region of the mouse GAPDH gene. Fungal and mouse DNA quantities were obtained from the Ct values from an appropriate standard curve. Fungal burden was determined through the ratio between ng of fungal DNA and mg of mouse DNA. The results are the means (± standard deviation) of 5 lungs for each treatment. Statistical analysis was performed by using t-test. (A) The ΔflcA mutant compared to the wild-type and ΔflcA::flcA+ strains. (B) Fungal burden for ΔflcA mutant, wild-type and ΔflcA::flcA+ strains. (C) The ΔflcB mutant compared to the wild type and ΔflcB::flcB+ strains. (D) Fungal burden for ΔflcB mutant, wild-type and ΔflcB::flcB+ strains. (E) The ΔflcC mutant compared to the wild type and ΔflcC::flcC+ strains. (F) Fungal burden for ΔflcC mutant, wild-type and ΔflcC::flcC+ strains. PBS = phosphate Buffer Saline.
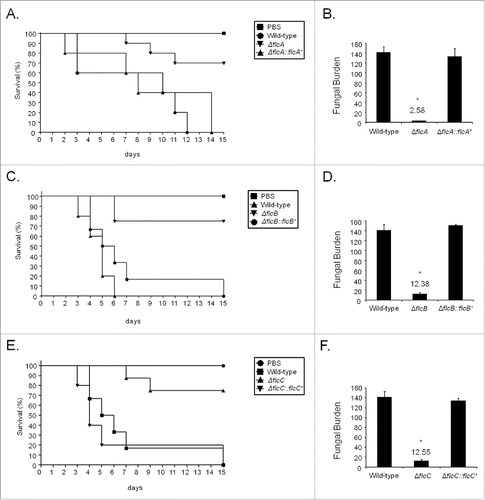
As an additional measurement of the fungal growth into the lungs, we have used fungal burden estimation by qPCR (). The ΔflcA-C strains were not able to grow within the lungs as well as the wild-type and the complemented strains (, p < 0 .0001 for the comparison between the wild-type and the deletion mutant, and p > 0 .05 between the wild-type and the complemented strain). Taken together, our results strongly indicate that FlcA-C are important for A. fumigatus virulence.
Discussion
In A. fumigatus FlcA belongs to a small protein family composed by 3 members, FlcA-C. In S. cerevisiae and C. albicans, the 3 homologues, FLC1-3, were required for the uptake of FAD into endoplasmic reticulum and heme iron uptake. These proteins were detected in the endoplasmic reticulum. Here, we constructed functional GFP fusions for all 3 A. fumigatus FlcA-C proteins, using their endogenous promoters. However, we were only able to observe FlcA::GFP localization, because the expression of the other fusions was very low (data not shown). Bioinformatic predictions of FlcA-C localization suggested they were resident endoplasmic reticulum proteins. However, FlcA::GFP localized to the apical tip and in vesicles that resemble the sorting multivesicular bodies (MVB) of the endocytic pathway.Citation41,42 We were also able to show that FlcA-C were important for FAD accumulation, which suggests that this function is conserved among different fungi.
All three genes flcA-C were important for A. fumigatus virulence. The flcB gene (but not flcA and flcC) is more expressed in vivo during initiation of murine infectionCitation43; however, we have not observed increased flcA, -B and -C mRNA accumulation induced either by human platelets in vitro or during airway epithelial cells interacting with A. fumigatus conidia.Citation44,45 This represents the first evidence that a FLC homolog to be involved in virulence, since the impact of the C. albicans FLC null mutants on virulence was not evaluated. It is surprising that ΔflcB and ΔflcC mutant strains are avirulent since the single phenotypes observed in vitro for these strains are: (i) a decrease in FAD accumulation and (ii) more specifically in the ΔflcC a transcriptional compensatory mechanism upon calcium exposure that increases the flcA and flcC mRNA accumulation. However, both phenotypes do not cause any in vitro growth and conidiation defects. It is tempting to speculate that flB and flcC genes are essential for in vivo growth but dispensable for in vitro growth. It is very intriguing how the dramatic reduction of FAD accumulation in these 2 strains that do not affect in vitro growth could impact in vivo growth. It is possible that FlcB and FlcC are collaborating during in vivo growth and this could help to explain the lack of virulence in the corresponding mutant strains. We were not able to construct a double ΔflcB ΔflcC mutant (data not shown), what could suggest FlcB and FlcC are interacting redundantly in vitro. However, all these possibilities remain to be investigated.
The A. fumigatus ΔflcA mutant had several phenotypes that phenocopy calcium deficiency, such as those observed in calcineurin and calcium transport or channel mutants.Citation46-48 For example, similar to the ΔcnaA (catalytic subunit of the calcineurin) mutant, we found that FlcA was required for A. fumigatus colony extension and influenced hyphal branching. The ΔflcA mutant was more sensitive to calcium and cyclosporine, excess of metals, and cell wall and oxidative stressing agents. Similar to ΔcnaA, the ΔflcA can also grow better in liquid than in solid medium, what could be due to the effect of defective apical branching of these strains on fungal growth on solid medium. In addition, the ΔflcA growth is improved in the presence of high calcium concentrations. All these different phenotypes suggest that FlcA was involved in calcium release and/or modulation. Actually, Rigamonti et al.Citation49 proposed that the S. cerevisiae hypertonic stress response, which was mediated by calcium release, involved FLC2. These authors performed extensive bioinformatics analysis and suggested that the 3 S. cerevisiae homologues, the Schizosaccharomyces pombe pkd2 and Neurospora crassa calcium-related spray protein are members of the fungal branch of TRP-like ion transporters.Citation49-51 Taken together, this evidence suggests that FlcA may also be involved in calcium transport.
In conclusion, this study identified a novel protein family related to calcium metabolism and virulence in A. fumigatus. FlcA was identified as regulated by CrzA upon calcium stress and it is important for FAD metabolism. It is important now to understand the connections among these pathways during the A. fumigatus pathogenicity. It remains to be determined the precise in vivo role played by FlcB and FlcC during A. fumigatus virulence and if these 2 putative transporters are interacting.
Materials and methods
Strains, media and culture methods
The A. fumigatus strains used in this study were CEA17 (pyrG+ and pyrG−), ΔcrzA, ΔflcA, ΔflcB, ΔflcC, ΔflcA::flcA+, ΔflcB::flcB+, and ΔflcC::flcC+.Citation52,53 All the comparisons with the deletion strains were performed with the CEA17 pyrG+. The media used were: complete medium composed for 2% w/v glucose, 0.5% w/v yeast extract, 2% w/v agar, trace elements (YAG) or YUU [YAG supplemented with 1.2 g (each) of uracil and uridine], and liquid YG or YG + UU medium with the same composition (but without agar). The minimal medium (MM) consist of 1% glucose, trace elements (22.0 g/l ZnSO4, 11 g/l boric acid, 5 g/l MnCl2, 5 g/l FeSO4, 1.6 g/l CoCl2, 1.6 g/l CuSO4, 1.1 g/l (NH4)2MoO4, 50 g/l ethylenediaminetetraacetic acid (EDTA)] and adjusted to pH 6.5 with NaOH ) and salt solution 20x, 2% agar, pH 6.5.Citation54 Strains were grown at 37°C or at 30 °C for microscopy experiments.
For the iron starvation experiments, the strains were grown in MM for 24 hours at 37 °C and transferred the mycelia to modified MM [consist of 1% glucose, trace elements without iron and salt solution 20x (NaNO3 272 g/l; KCl 10.4 g/l, KH2PO4 30.4 g/l, MgO4.7H2O 10.4 g/l, and 50 ml of this solution are added to 1 l of MM) plus BPS 200 µM [Bathophenanthrolinedisulfonic acid (4,7-diphenyl-1,10-phenanthrolinedisulfonic acid) and 3-(2-pyridyl)-5,6-bis(4-phenylsulfonic acid)-1,2,4-triazine (ferrozine)] 300 µM for 1 or 2 hours at 37 °C. For the iron excess experiments, the strains were grown in MM for 24 hours and then FeSO4.7H2O 200 µM or FeCl3 200 µM were added for 1 or 2 hours at 37 °C.
Construction of the A. fumigatus mutants
We have used the ‘in vivo” recombination method in S. cerevisiae as previously described by Colot et al.Citation55 for the construction of gene replacement cassettes. Thus, about 1.0 kb from the 5′-UTR and 3′-UTR flanking region of the targeted ORF regions were used for designing primers. The primers 5F and 3R also contains a short homolog sequence to the MCS of the plasmid pRS426. Both fragments, 5- and 3-UTR, were PCR-amplified from A. fumigatus genomic DNA (gDNA). The pyrG used in the A. fumigatus cassette for generating the mutant strains were used as marker for prototrophy and was amplified from pCDA21 plasmid.Citation56 The DNA fragments together with plasmid linearized pRS426 BamHI/EcoRI were transformed into S. cerevisiae strain SC9721 (FGSC) by the lithium acetate methodCitation57 and DNA of the transformants extracted as previously describedCitation58 TaKaRa Ex Taq™ DNA Polymerase (Clontech Takara Bio) was used for DNA amplification and Southern blot analyses demonstrated that the transformation cassettes had integrated homologously at the targeted A. fumigatus loci. A. fumigatus transformation was performed as described by de Castro et al.Citation48
The complementing strains were constructed by first isolating from the corresponding deletion strain, a pyrG− auxotroph sector resistant to 0.75 mg/ml of 5-FOA (5-Fluoroorotic acid, Sigma-Aldrich), a fluorinated derivative of the pyrimidine precursor orotic acid. This analog was used to select for the absence of a functional pyrG+ gene, which encodes the enzyme for the decarboxylation of 5-fluoroorotic acid to 5-fluorouracil, a toxic metabolite. The flcA-C gene deletions were confirmed in these strains and were complemented by co-transforming a DNA fragment (approximately 1 kb from each 5′ and 3′- flanking regions plus the ORF) together with the PCDA21 vector and selecting for the ability to grow in medium without uridine and uracil. Homologous recombination and gene replacement were confirmed by PCR or Southern blot analyses (Fig. S3).
To FlcA-C::GFP strains were constructed by cloning the flcA-C ORF in frame with the green fluorescent protein (GFP) gene. We linked GFP to the FlcA-C C terminus and separated them by 4 additional codons that, after translation, produce a 4-amino-acid linker (glycine-threonine-arginine-glycine).Citation59 The S. cerevisiae in vivo recombination system was used for production of the transformation cassette. First, the flcA-C ORF and approximately 500 pb its 5′-UTR flanking region were amplified from gDNA of the wild-type strain by the use of the primers FlcA-C pRS426 5 Fw and FlcA SPACER GFP Rv. The stop codon of the flcA-C gene was omitted in this construction. The GFP ORF was amplified from the pMCB17apx plasmid (provided by Vladimir P. Efimov) by the use of the primers Spacer GFP Fw and GFP VE3′ AF. The selective marker pyrG fragment was PCR amplified from the pCDA21 plasmid and the primers used for this PCR amplification were GFP pyrG Fw and pyrG Rv. The amplification of the 3′-UTR (approximately 600pb) was done with the Afu flcA-C 3 Fw and Afu FlcA-C 3 Rv primers. The PCR-amplified cassette was transformed into the A. fumigatus wild-type strain. The primers used are described in Table S1.
DNA manipulation
We have used Southern blot analysis to prove the cassettes had integrated homologously at the targeted A. fumigatus flcA-C loci. Genomic DNA was extracted as previously described.Citation60 Standard techniques for manipulation of DNA and Southern blot analyses were carried out as described.Citation61 Southern blot schemes are shown in Figure S3.
Microscopy
For microscopy, we have grown FlcA::GFP conidiospores on coverslips in 4 ml of MM media for 16 h at 30°C. After incubation, the coverslips with adherent germlings were left untreated or treated with iron starvation or excess. Subsequently, the coverslips were rinsed with phosphate-buffered saline (PBS; 140 mM NaCl, 2 mM KCl, 10 mM NaHPO4, 1.8 mM KH2PO4, pH7.4) and mounted for examination. Slides were visualized on an Observer Z1 fluorescence microscope using a 100x objective oil immersion lens for GFP, filter set 38-high efficiency [HE], excitation wavelength of 450 to 490 nm, and emission wavelength of 500 to 550 nm. DIC (differential interference contrast) images and fluorescent images were captured with an AxioCam camera (Carl Zeiss) and processed using AxioVision software (version 4.8).
RNA extraction and real-time PCR reactions
RNA extraction and real-time PCR experiments RNase free DNase I treatment were performed as previously described by Semighini et al.Citation62 All the PCR reactions were performed using an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, USA) and Taq-Man™ Universal PCR Master Mix kit (Applied Biosystems, USA). The reactions and calculations were performed according to Semighini et al.Citation62 The primers and fluorescent probes (TaqMan®, Applied Biosystems) used in this work are described in Table S1.
Murine model of pulmonary aspergillosis, lung histopathology and fungal burden
We have housed outbreed female mice (BALB/c strain; body weight, 20 to 22 g) in vented cages with 5 animals each. Mice immunosuppression was performed with cyclophosphamide (150 mg per kg of body weight), administered intraperitoneally on days −4, −1, and 2 before and after infection. Hydrocortisonacetate (200mg/ kg body weight) was injected subcutaneously on day −3. A. fumigatus strains were grown on YAG for 3 d prior to infection. Fresh conidia were harvested in PBS and filtered through a Miracloth (Calbiochem). Conidial suspensions were spun for 5 min at 3,000 × g, washed 3 times with PBS, counted using a hemocytometer, and resuspended at a concentration of 5.0 × 106 conidia/ ml. The viability of the administered inoculum was determined by incubating a serial dilution of the conidia on YAG medium, at 37°C. Mice were anesthetized by halothane inhalation and infected by intranasal instillation of 1.0 × 105 conidia in 20μl of PBS. As a negative control, a group of 5 mice received PBS only. Mice were weighed every 24 h from the day of infection and visually inspected twice daily. The statistical significance of the comparative survival values was calculated using log rank analysis and the Prism statistical analysis package.
Fungal burden was investigated in murine lungs, mice were immunosuppressed with cyclophosphamide (150 mg/ kg of body weight), which was administered intraperitoneally on days −4 and −1, while hydrocortisonacetate was injected subcutaneously (200 mg/ kg) on day-3. Five mice per group were intranasally inoculated with 1 × 106 conidia/ 20 μl of suspension. A higher inoculum, in comparison to the survival experiments, was used to increase fungal DNA detection. Animals were sacrificed 72 h post-infection, and the lungs were harvested and immediately frozen in liquid nitrogen. Samples were ground to a fine powder under liquid N2 and DNA was extracted via the phenol-chloroform method. DNA quantity and quality were assessed using a NanoDrop 2000 spectrophotometer (Thermo Scientific). At least 500 μg of total DNA from each sample was used for quantitative real-time PCRs. PCR reactions were performed using an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, USA) and SYBR Green detection. SYBR® Green PCR Master Mix (Applied Biosystems, USA) was used for reaction mixture preparation. The primer sets for the analyses were used to amplify the 18 S rRNA region of A. fumigatus (18 S rRNA Afu sybr FW, 5′-GACCTCGGCCCTTAAATAGC-3′ and 18 S rRNA Afu sybr Rv, 5′-CTCGGCCAAGGTGATGTACT-3′) and an intronic region of mouse GAPDH, encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH mouse sybr FW, 5′-GAGGGACTTGGAGGACACAG-3′ and GAPDH mouse sybr Rv, 5′-ACATCACCCCCATCACTCAT-3′). Six-point standard curves were calculated using serial dilutions of gDNA from all the A. fumigatus strains used and the uninfected mouse lung. Fungal and mouse DNA quantities were obtained from the threshold cycle (CT) values from an appropriate standard curve. Fungal burden was determined as the ratio between picograms of fungal and micrograms of mouse DNA. Virulence survival and fungal burden were repeated at least twice.
FAD transport assay
Protoplasts were produced for all the strains as previously described.Citation48 Protoplasts were resuspended in 1 ml of reaction buffer (20 mM HEPES pH 6.8, 150 mM potassium acetate, 250 mM sorbitol, 5 mM magnesium acetate). Protoplasts (1.25 × 106) were washed 3 times with reaction buffer at 4 °C and resuspended in the same buffer. Protoplasts were incubated with 1 mM of flavin adenine dinucleotide (Sigma, USA) in the absence of light for 0, 5, 10, 20, and 30 minutes at 30 °C. The reaction was stopped by adding 1 ml of reaction buffer at 4 °C, and washed 3 times with the same buffer. Finally, the protoplasts were lysed in 2 % Triton X-100, and the fluorescence of the supernatant was evaluated at 450 nm excitation and 530 nm emission.
Immunoblot analysis
To detect FlcA::GFP fusion, fresh harvested conidia (1×107) of the wild-type and mutant strains were inoculated in 50 ml liquid MM at 37°C for 20 h (160 rpm) and after this period, the mycelia were treated with CaCl2 (200mM) for 10 and 30 min. The mycelia was frozen and ground in liquid nitrogen. For protein extraction, 0.5 ml lysis bufferCitation63 containing 10% (v/v) glycerol, 50 mM Tris–HCl pH 7.5, 1% (v/v) Triton X-100, 150 mM NaCl, 0.1% (w/v) SDS, 5 mM EDTA, 5 mM sodium orthovanadate, 1 mM PMSF, and 1X Complete Mini-protease inhibitor (Roche Applied Science) was added to the ground mycelium. Extracts were centrifuged at 20,000 g for 40 minutes at 4°C. The supernatants were collected and the protein concentrations were determined using the Bradford methodCitation64 (BioRad). 50 µg of protein from each sample were resolved in a 12% (w/v) SDS–PAGE and transferred polyvinylidene difluoride (PVDF) membranes using the iBlot® 2 Dry Blotting System (Thermo Scientific™). The flc::GFP fusion was detected by the anti GFP antibody (Sigma G1544) at a 1:1,000 dilution in TBST containing. Incubation was performed at 4°C for 16 hours. The primary antibody was detected with an HRP-conjugated secondary antibody raised in rabbits (A0545; Sigma) in TBST buffer for an one-hour incubation, at room temperature. Chemoluminescent detection was performed by using an ECL Prime Western Blot detection kit (GE HealthCare). Images were generated by exposing the PVDF membranes to the ChemiDoc XRS gel imaging system (BioRad).
KVIR_S_1239010.zip
Download Zip (5.6 MB)Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) for providing financial support. We would like to thank Dr. Miguel Peñalva for his suggestions about the FlcA subcellular localization. We also would like to thank the 5 reviewers for their suggestions and comments.
References
- Thewes S. Calcineurin-Crz1 Signaling in Lower Eukaryotes. Eukaryot Cell 2014; 13:694-705; PMID:24681686; http://dx.doi.org/10.1128/EC.00038-14
- Juvvadi PR, Lee SC, Heitman J, Steinbach WJ. Calcineurin in fungal virulence and drug resistance: Prospects for harnessing targeted inhibition of calcineurin for an antifungal therapeutic approach. Virulence 2016; 20:1-12; http://dx.doi.org/10.1080/21505594.2016.1201250
- Cyert MS. Calcineurin signaling in Saccharomyces cereviside: how yeast go crazy in response to stress. Biochem Biophys Res Commun 2003; 311:1143-50; PMID:14623300; http://dx.doi.org/10.1016/S0006-291X(03)01552-3
- Steinbach WJ, Reedy JL, Cramer RA, Jr, Perfect JR, Heitman J. Harnessing calcineurin as a novel anti-infective agent against invasive fungal infections. Nat Rev Microbiol 2007; 5:418-30; PMID:17505522; http://dx.doi.org/10.1038/nrmicro1680
- Stie J, Fox D. Calcineurin regulation in fungi and beyond. Eukaryotic Cell 2008; 7:177-86; PMID:18065652; http://dx.doi.org/10.1128/EC.00326-07
- Carafoli E. Calcium signaling: a tale for all seasons. Proc Natl Acad Sci USA 2002; 99:1115-22; PMID:11830654; http://dx.doi.org/10.1073/pnas.032427999
- Brown NA, Goldman GH. The contribution of Aspergillus fumigatus stress responses to virulence and antifungal resistance. J Microbiol 2016; 54:243-53; PMID:26920884; http://dx.doi.org/10.1007/s12275-016-5510-4
- Rasmussen C, Garen C, Brining S, Kincaid RL, Means RL, Means AR. The calmodulin-dependent protein phosphatase catalytic subunit (calcineurin A) is an essential gene in Aspergillus nidulans. EMBO J 1994; 13:3917-24; PMID:8070419
- Juvvadi PR, Kuroki Y, Arioka M, Nakajima H, Kitamoto K. Functional analysis of the calcineurin-encoding gene cnaA from Aspergillus oryzae: evidence for its putative role in stress adaptation. Arch Microbiol 2003; 179:416-22; PMID:12709783
- Viaud MC, Balhadere PV, Talbot NJ. A Magnaporthe grisea cyclophilin acts as a virulence determinant during plant infection. Plant Cell 2002; 14:917-30; PMID:11971145; http://dx.doi.org/10.1105/tpc.010389
- Kothe GO, Free SJ. Calcineurin subunit B is required for normal vegetative growth in Neurospora crassa. Fungal Genet Biol 1998; 23:248-58; PMID:9680955; http://dx.doi.org/10.1006/fgbi.1998.1037
- Stathopoulos-Gerontides A, Guo JJ, Cyert MS. Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes and Development 1999; 13:798-803; PMID:10197980; http://dx.doi.org/10.1101/gad.13.7.798
- Stathopoulos AM, Cyert MS. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev 1997; 11:3432-44; PMID:9407035; http://dx.doi.org/10.1101/gad.11.24.3432
- Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med 2012; 4:165rv13; PMID:23253612; http://dx.doi.org/10.1126/scitranslmed.3004404
- Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med 2012; 4:165rv13; PMID:23253612; http://dx.doi.org/10.1126/scitranslmed.3004404
- Odom A, Muir S, Lim E, Toffaletti DL, Perfect J, Heitman J. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J 1997; 16:2576-89; PMID:9184205; http://dx.doi.org/10.1093/emboj/16.10.2576
- Chen Y-L, Lehman VN, Lewit Y, Averette AF, Heitman J. Calcineurin governs thermotolerance and virulence of Cryptococcus gattii. G3: Genes/Genomes/Genetics 2013; 3:527-39
- Chen YL, Brand A, Morrison EL, Silao FGS, Bigol UG, Malbas FF, Nett JE, Andes DR, Solis NV, Filler SG, Averette A, Heitman J. Calcineurin controls drug tolerance, hyphal growth, and virulence in Candida dubliniensis. Eukaryotic Cell 2011; 10:803-19; PMID:21531874; http://dx.doi.org/10.1128/EC.00310-10
- Blankenship JR, Wormley FL, Boyce MK, Schell WA, Filler SG, Perfect JR, Heitman J. Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryotic Cell 2003; 2:422-30; PMID:12796287; http://dx.doi.org/10.1128/EC.2.3.422-430.2003
- Bader T, Bodendorfer B, Schroppel K, Morschhauser J. Calcineurin is essential for virulence in Candida albicans. Infect Immun 2003; 71:5344-54; PMID:12933882; http://dx.doi.org/10.1128/IAI.71.9.5344-5354.2003
- Campos CB, Di Benedette JP, Morais FV, Ovalle R, Nobrega MP. Evidence for the role of calcineurin in morphogenesis and calcium homeostasis during mycelium-to-yeast dimorphism of Paracoccidioides brasiliensis. Eukaryot Cell. 2008; 7:1856-64; PMID:18776037; http://dx.doi.org/10.1128/EC.00110-08
- Steinbach WJ, Cramer RA, Perfect BZ, Asfaw YG, Sauer TC, Najvar LK, Kirkpatrick WR, Patterson TF, Benjamin DK, Jr, Heitman J, et al. Calcineurin controls growth, morphology, and pathogenicity in Aspergillus fumigatus. Eukaryotic Cell 2006; 5:1091-103; PMID:16835453; http://dx.doi.org/10.1128/EC.00139-06
- Ferreira MES, Heinekamp T, Hartl A, Brakhage AA, Semighini CP, Harris SD, Savoldi M, de Gouvea PF, de Souza Goldman MH, Goldman GH. Functional characterization of the Aspergillus fumigatus calcineurin. Fungal Genet Biol 2007; 44:219-30; PMID:16990036; http://dx.doi.org/10.1016/j.fgb.2006.08.004
- Harel A, Bercovich S, Yarden O. Calcineurin is required for sclerotial development and pathogenicity of Sclerotinia sclerotiorum in an oxalic acid-Independent manner. Mol Plant-Microbe Interact 2006; 19:682-93; PMID:16776301; http://dx.doi.org/10.1094/MPMI-19-0682
- Schumacher J, de Larrinoa IF, Tudzynski B. Calcineurin-responsive zinc finger transcription factor CRZ1 of Botrytis cinerea is required for growth, development, and full virulence on bean plants. Eukaryotic Cell 2008; 7:584-601; PMID:18263765; http://dx.doi.org/10.1128/EC.00426-07
- Choi JH, Kim Y, Lee YH. Functional analysis of MCNA, a gene encoding a catalytic subunit of calcineurin, in the rice blast fungus Magnaporthe oryzae. J Microbiol Biotechnol 2009; 19:11-6; PMID:19190403
- Egan JD, Garcıa-Pedrajas MD, Andrews DL, Gold SE. Calcineurin is an antagonist to PKA protein phosphorylation required for postmating filamentation and virulence, while PP2A is required for viability in Ustilago maydis. Mol Plant-Microbe Interact 2009; 22:1293-301; PMID:19737102; http://dx.doi.org/10.1094/MPMI-22-10-1293
- Cervantes-Chavez JA, Ali S, Bakkeren G. Response to environmental stresses, cell-wall integrity, and virulence are orchestrated through the calcineurin pathway in Ustilago hordei. Mol Plant-Microbe Interact 2010; 24:219-32; http://dx.doi.org/10.1094/MPMI-09-10-0202
- Juvvadi PR, Lamoth F, Steinbach WJ. Calcineurin as a multifunctional regulator: unraveling novel functions in fungal stress responses, hyphal growth, drug resistance, and pathogenesis. Fungal Biol Rev 2014; 28:56-69; PMID:25383089; http://dx.doi.org/10.1016/j.fbr.2014.02.004
- Ueno K, Namiki Y, Mitani H, Yamaguchi M, Chibana H. Differential cell wall remodeling of two chitin synthase deletants Dchs3A and Dchs3B in the pathogenic yeast Candida glabrata. FEMS Yeast Res 2011; 11:398-407; PMID:21453325; http://dx.doi.org/10.1111/j.1567-1364.2011.00728.x
- Fortwendel JR, Juvvadi PR, Perfect BZ, Rogg LE, Perfect JR, Steinbach WJ. Transcriptional regulation of chitin synthases by calcineurin controls paradoxical growth of Aspergillus fumigatus in response to caspofungin. Antimicrob Agents Chemother 2010; 54:1555-63; PMID:20124000; http://dx.doi.org/10.1128/AAC.00854-09
- Katiyar SK, Alastruey-Izquierdo A, Healey KR, Johnson ME, Perlin DS, Edlind TD. Fks1 and Fks2 are functionally redundant but differentially regulated in Candida glabrata: implications for echinocandin resistance. Antimicrob Agents Chemother 2012; 56:6304-9; PMID:23027185; http://dx.doi.org/10.1128/AAC.00813-12
- Mazur P, Morin N, Baginsky W, el-Sherbeini M, Clemas JA, Nielsen JB, Foor F. Differential expression and function of two homologous subunits of yeast 1,3-β-D-glucan synthase. Mol Cell Biol 1995; 15:5671-81; PMID:7565718; http://dx.doi.org/10.1128/MCB.15.10.5671
- Greenberger PA. Allergic bronchopulmonary aspergillosis. J Allergy Clin Immunol 2002; 110:685-92; PMID:12417875; http://dx.doi.org/10.1067/mai.2002.130179
- Dagenais TR, Keller NP. Pathogenesis of Aspergillus fumigatus in Invasive Aspergillosis. Clin Microbiol Rev 2009; 22:447-65; PMID:19597008; http://dx.doi.org/10.1128/CMR.00055-08
- Brown GD, Denning DW, Levitz SM. Tackling human fungal infections. Science 2012; 336:647; PMID:22582229; http://dx.doi.org/10.1126/science.1222236
- Lackner M, Lass-Flörl C. Up-date on diagnostic strategies of invasive aspergillosis. Curr Pharm Des 2013; 19:3595-614; PMID:23278540; http://dx.doi.org/10.2174/1381612811319-9990323
- Brakhage AA. Systemic fungal infections caused by Aspergillus species: Epidemiology, infection process and virulence determinants. The Journal of Current Drug Targets 2005; 6:875-86; http://dx.doi.org/10.2174/138945005774912717
- de Castro PA, Chen C, de Almeida RS, Freitas FZ, Bertolini MC, Morais ER, Brown NA, Ramalho LN, Hagiwara D, Mitchell TK, et al. ChIP-seq reveals a role for CrzA in the Aspergillus fumigatus high-osmolarity glycerol response (HOG) signalling pathway. Mol Microbiol 2014; 94:655-74; http://dx.doi.org/10.1111/mmi.12785
- Protchenko O, Rodriguez-Suarez R, Androphy R, Bussey H, Philpott CC. A screen for genes of heme uptake identifies the FLC family required for import of FAD into the endoplasmic reticulum. J Biol Chem 2006; 281:21445-57; PMID:167170-99; http://dx.doi.org/10.1074/jbc.M512812200
- Peñalva MA. Endocytosis in filamentous fungi: Cinderella gets her reward. Curr Opin Microbiol 2010; 13:684-92; http://dx.doi.org/10.1016/j.mib.2010.09.005
- Peñalva MA, Galindo A, Abenza JF, Pinar M, Calcagno-Pizarelli AM, Arst HN, Pantazopoulou A. Searching for gold beyond mitosis: Mining intracellular membrane traffic in Aspergillus nidulans. Cell Logist 2012; 2:2-14; http://dx.doi.org/10.4161/cl.19304
- McDonagh A, Fedorova ND, Crabtree J, Yu Y, Kim S, Chen D, Loss O, Cairns T, Goldman G, Armstrong-James D, et al. Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog 2008; 4:e1000154; PMID:18787699; http://dx.doi.org/10.1371/journal.ppat.1000154
- Perkhofer S, Zenzmaier C, Frealle E, Blatzer M, Hackl H, Sartori B, Lass-Flörl C. Differential gene expression in Aspergillus fumigatus induced by human platelets in vitro. Int J Med Microbiol 2015; 305:327-38; PMID:25661519; http://dx.doi.org/10.1016/j.ijmm.2015.01.002
- Oosthuizen JL, Gomez P, Ruan J, Hackett TL, Moore MM, Knight DA, Tebbutt SJ. Dual organism transcriptomics of airway epithelial cells interacting with conidia of Aspergillus fumigatus. PLoS One 2011; 6:e20527; PMID:21655222; http://dx.doi.org/10.1371/journal.pone.0020527
- da Silva Ferreira ME, Heinekamp T, Härtl A, Brakhage AA, Semighini CP, Harris SD, Savoldi M, de Gouvêa PF, de Souza Goldman MH, Goldman GH. Functional characterization of the Aspergillus fumigatus calcineurin. Fungal Genet Biol 2007; 44:219-30; PMID:16990036; http://dx.doi.org/10.1016/j.fgb.2006.08.004
- Dinamarco TM, Freitas FZ, Almeida RS, Brown NA, dos Reis TF, Ramalho LN, Savoldi M, Goldman MH, Bertolini MC, Goldman GH. Functional characterization of an Aspergillus fumigatus calcium transporter (PmcA) that is essential for fungal infection. PLoS One 2012; 7:e37591; PMID:22649543; http://dx.doi.org/10.1371/journal.pone.0037591
- de Castro PA, Chiaratto J, Winkelströter LK, Bom VL, Ramalho LN, Goldman MH, Brown NA, Goldman GH. The involvement of the Mid1/Cch1/Yvc1 calcium channels in Aspergillus fumigatus virulence. PLoS One 2014; 9:e103957; PMID:25083783; http://dx.doi.org/10.1371/journal.pone.0103957
- Rigamonti M, Groppi S, Belotti F, Ambrosini R, Filippi G, Martegani E, Tisi R. Hypotonic stress-induced calcium signaling in Saccharomyces cerevisiae involves TRP-like transporters on the endoplasmic reticulum membrane. Cell Calcium 2015; 57:57-68; PMID:25573187; http://dx.doi.org/10.1016/j.ceca.2014.12.003
- Palmer CP, Aydar E, Djamgoz MB. A microbial TRP-like polycystic-kidney-disease-related ion channel gene. Biochem J 2005; 387:211-19; PMID:15537393; http://dx.doi.org/10.1042/BJ20041710
- Bok JW, Sone T, Silverman-Gavrila LB, Lew RR, Bowring FJ. Catcheside DE, Griffiths AJ. Structure and function analysis of the calcium-related gene spray in Neurospora crassa. Fungal Genet Biol 2001; 32:145-58; PMID:11343401; http://dx.doi.org/10.1006/fgbi.2000.1259
- da Silva Ferreira ME, Kress MR, Savoldi M, Goldman MH, Hartl A, Heinekamp T, Brakhage AA, Goldman GH. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryotic Cell 2006; 5:207-11; PMID:16400184; http://dx.doi.org/10.1128/EC.5.1.207-211.2006
- Soriani FM, Malavazi I, da Silva Ferreira ME, Savoldi M, Von Zeska Kress MR, de Souza Goldman MH, Loss O, Bignell E, Goldman GH. Functional characterization of the Aspergillus fumigatus CRZ1 homologue, CrzA. Mol Microbiol 2008; 67:1274-91; PMID:18298443; http://dx.doi.org/10.1111/j.1365-2958.2008.06122.x
- Kafer E. Meiotic and mitotic recombination in Aspergilllus and its chromosomal aberrations. Adv Genet 1977; 19:33-131; PMID:327767
- Colot HH, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. A highthroughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA 2006; 103:10352-7; PMID:16801547; http://dx.doi.org/10.1073/pnas.0601456103
- Chaveroche MK, Ghigo JM, d'Enfert C. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res 2000; 28:E97; PMID:11071951; http://dx.doi.org/10.1093/nar/28.22.e97
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 1989; 16:339-46; PMID:2692852; http://dx.doi.org/10.1007/BF00340712
- Goldman GH, dos Reis Marques E, Duarte Ribeiro DC, de Souza Bernardes LA, Quiapin AC, Vitorelli PM, Savoldi M, Semighini CP, de Oliveira RC, Nunes LR, et al. Expressed sequence tag analysis of the human pathogen Paracoccidioides brasiliensis yeast phase: identification of putative homologues of Candida albicans virulence and pathogenicity genes. Eukaryot Cell 2003; 2:34-48; PMID:12582121; http://dx.doi.org/10.1128/EC.2.1.34-48.2003
- Teepe AG, Loprete DM, He Z, Hoggard TA, Hill TW. The protein kinase C orthologue PkcA plays a role in cell wall integrity and polarized growth in Aspergillus nidulans. Fungal Genet Biol 2007; 44:554-62; PMID:17118679; http://dx.doi.org/10.1016/j.fgb.2006.10.001
- Malavazi I, Goldman GH. Gene disruption in Aspergillus fumigatus using a PCR-based strategy and in vivo recombination in yeast. Methods Mol Biol 2012; 845:99-118; PMID:22328370; http://dx.doi.org/10.1007/978-1-61779-539-8_7
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual 2001; 3rd ed. London: CSHL Press
- Semighini CP, Marins M, Goldman MHS, Goldman GH. Quantitative analysis of the relative transcript levels of ABC transporter Atr genes in Aspergillus nidulans by real-time reverse transcription-PCR assay. Appl Environ Microbiol 2002; 68:1351-7; PMID:11872487; http://dx.doi.org/10.1128/AEM.68.3.1351-1357.2002
- Valiante V, Jain R, Heinekamp T, Brakhage AA. The MpkA MAP kinase module regulates cell wall integrity signaling and pyomelanin formation in Aspergillus fumigatus. Fungal Genet Biol 2009; 46:909-18; PMID:19715768; http://dx.doi.org/10.1016/j.fgb.2009.08.005
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976; 72:248-54; PMID:942051; http://dx.doi.org/10.1016/0003-2697(76)90527-3