HTLV-1 infects approximately 5–10 millions of individuals in the world and the majority of them remain lifelong asymptomatic carriers (AC). However, up to 5% of HTLV-1 patients may present inflammatory neurological or proliferative disorders as HTLV-1- associated myelopathy / tropical spastic paraparesis (HAM/TSP) and adult T-cell leukemia (ATL), respectively. In HAM/TSP, the spinal cord inflammation, leads to a chronic disabling disturbance. In ATL, HTLV induces T-cell proliferation and transformation, with a lethal consequence to the host.Citation1 High proviral load is associated with disease progression. However, factors that may influence viral replication are unclear. Some viral encoded proteins, such as p12, induces infected T-cell proliferation and stimulates immune evasion.Citation2-15 Previous studies demonstrated that mutations in p12 may influence the HTLV-1 infection outcome.Citation3,5,8,16,17 Here, we identified, at the first time, a HTLV-1 strain with a premature stop codon in p12 infecting an individual from Brazil, presenting a very low proviral load. We hypothesize if this mutation would have a protective role against HTLV-1 replication.
Genomic DNA was extracted from peripheral blood mononuclear cells (PBMCs) of an asymptomatic individual that is followed in the Neuroinfection outpatient clinic (HUGG/UNIRIO). The patient was a Brazilian Caucasian, 53 years old, female, married, with no family history of HTLV-1 infection, blood transfusion or drug abuse. The HTLV-1 infection was detected by ELISA and Western Blot fifteen years ago, when the patient was tested for blood donation. The HTLV-1 proviral load was 0.03 copies of Tax/100 PBMCs (TaqMan Real Time PCR).Citation18 For DNA extraction QIAamp DNA mini kit (Quiagen, CA) was used. Nested polymerase chain reaction (n-PCR) was performed in order to amplify p12Citation18 and LTR. The PCR product was purified using Illustra GFX PCR DNA and Gel band purification kit (GE HealthCare) and directly sequenced using ABI model 373A automated DNA sequencer and the manufacturer's Dye Terminator FS Kit instructions. The electropherogram was analyzed by Chromas Lite 2.1 (Thechnelysium) (). The p12 sequence was aligned and translated on BioEdit editor (North Carolina State University). The analyzed sequence (HUGGRJ25) presented a mutation () that resulted in a premature stop codon at position 82 (Genbank accession number: KR337634) (). The phylogeny on LTR (634bp) showed that the sequence belongs to HTLV-1aA subtype (GenBank accession number KR337613).
Figure 1. Electropherogram of DNA sequence obtained from peripheral blood of a Brazilian asymptomatic individual. The point mutation (G245A) that resulted in the premature stop codon is highlighted and indicated with an arrow. It is localized at position 286 in the present electropherogram.
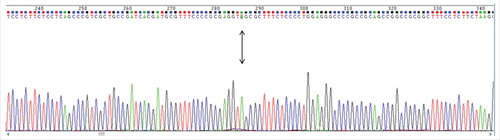
Figure 2. HTLV-1 p12 alignment. Predict aminoacid sequence of p12 of HTLV-1 asymptomatic carrier from Rio de Janeiro, Brazil (HUGG25), HTLV-1aA Prototype (BOI: L36905) and other previously described p12 truncated sequences that are available on GenBank.*: represents stop codon; HAM: HAM/TSP individuals; AC: HTLV-1 asymptomatic carriers; WA: West Africa; CA: Central Africa. The cleavage sites (yellow) and the ubiquitylation (88) positions (red) are highlighted. SH3-binding motifs are underlined. Calcineurin-binding motif is shown in bracket.
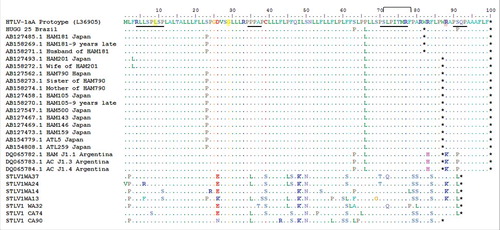
Truncated p12 proteins had already been identified in AC, HAM/TSP and ATL individuals from Argentina, Japan as also as in Simian T Lymphotropic virus (STLV).Citation19 In this set of sequences two truncated p12 proteins were observed: the major group presented 87 residues and the other 81 residues. Our truncated p12 sequence belongs to the later type. One patient from Japan with a truncated protein (with the stop codon at position 82) was a HAM/TSP and presented a high proviral load.Citation20 This is the first report of a truncated protein in a HTLV-1aA from Brazil. In fact, we evaluated a large set of p12 sequences available in GenBank from 76 new HTLV-1 complete genome sequences from BrazilCitation21 and none presented a premature stop codon at p12 protein. Moreover, Igñez et al (2005) analyzed 26 ORF-I sequences from Brazil (20 from AC, and 6 from HAM/TSP) and also did not find any individual carrying a provirus with a truncated p12.Citation22 In a very interesting study, ORF-I sequences of 160 HTLV-1 infected individuals from different areas of the world (including Brazil) were cloned and none of 1600 obtained sequences had a premature stop codon either.Citation3 In Japan, the prevalence of truncated p12 protein was 4% (n = 10) among HTLV-1 infected individuals.Citation20
An in vitro study demonstrated that strains producing a truncated p12 protein (without the amino acids 87–99) presented a dramatic reduction of NFAT activation.Citation23 As NFAT activation is associated with the upregulation of IL-2, this deletion may cause a decrease on T-cell proliferation.Citation23 The p12 protein presents different mechanisms to induce T-cell activation and proliferation. One mechanism is associated with NFAT activation. It is know that p12 localizes at the endoplasmatic reticulum (ER), where it can interacts with different cellular proteins. It can bind to calreticulin/calnexin, increasing the release of calcium from ER. The increase in the cytosolic calcium, activate the calmodulin, which stimulates calcineurin (calmodulin phosphatase dependent). Calcineurin dephosphorilates NFAT, activating it. Then, NFAT is translocated into the cell nucleus where it increases the IL-2 transcription.Citation6 In the other hand, p12 can bind to calcineurin at the ER, competing with NFAT. This induces a negative modulation of NFAT activation.Citation6,24 It is know that there is a calcineurin/calnexin binding motif in p12 protein, localized in position 70–75.Citation24 It was also demonstrated that there are four different SH3 binding motifs in p12. While SH3–2 and SH3–4 presents a positive regulation on NFAT, SH3–1 and SH3–3 presents a negative impact of NFAT activation.Citation23 Therefore, NFAT regulation by p12 is complex. The truncated form still present the calcineurin binding motif, two inhibitory SH3 motifs and only one SH3 with a positive regulation of NFAT. The same picture was observed in the truncated protein produced by Ding et al (2003), which presented a marked reduced NFAT activation in vitro.Citation23
Moreover, the p12 protein undergoes to complex post-translational modifications. It can be cleaved at two positions (at position 9 and 29). When p12 is cleaved at position 9 it lost the ER retention signal. When it is cleaved at position 29 it produces the p8 protein. While p12 remains at the ER, p8 traffic to cell surface where it plays an important role. At the ER, p12 can bind to different molecules: 1) IL-2R (increasing the Jack/STAT signaling), 2) MHC-I heavy chain (impairing the expression of this molecule), 3) Calnexin/calmodulin (increasing cytosolic calcium, activating NFAT). In contrast; p8 localizes at immunological synapses (IS) and interacts with TCR, causing a down modulation to the TCR signaling. Moreover, p8 increases cell adhesion and viral transmission via cell-cell synapses. These mechanisms are important to maintain a persistent viral infection, as it interferes in both immune response and viral transmission. The exact cellular pathway that p8 utilize for achieve this objectives are not elucidated. By hypothesis, the C-terminus truncated protein can influence in both p12 and p8 function.
Others post translational modifications identified in p12 includes ubiquitylation, palmitoylation and dimerization.Citation25-28 Some mutations, such as C39A abrogated dimerization and palmitoylation of both proteins, without interfering neither in the localization of p8 nor in the increase in the cellular adhesion. As C39 is maintained in our truncated p12, the palmitoylation may occur in this protein.Citation27,28 In vitro studies showed that ubiquitylation occur at a Lysine localized at position 88, and it influences the protein stability.Citation25 p12 is ubiquitylated and subsequently degraded in the proteasomes. For example, a mutation to an Arginine in position 88 (K88R), inhibits the ubiquitylation and increases p12 stability.Citation25 Therefore, as the premature stop codon is on position 82, it may influence the protein stability.
The protein localization is important for it function. In this context, a mutant lacking the C-terminal amino acid residues (1–86) maintained the wild type p12 protein localization.Citation7
Different p12 alleles present different profiles of p12 and p8 expression. Interestingly, all described truncated proteins presented alleles that were previously associated with an increased expression of p12 (F3L, S23P, F34L/F61L, S63P).Citation3 A transfection study, as performed by Pise-Masison et al. (2014) to evaluate the expression profile of both proteins would be interesting.
It is possible that, in situations with limited concentration of IL-2 (as usually occurs in vivo) p12 will be important in the initial phase of infection (causing a T cell activation and proliferation and immune evasion).Citation29 At a later stage, p8 becomes more important causing infected T cell anergy and increasing cell-cell adhesion to transfer viral particles through the virological synapses. The post-translational mechanisms could control these opposing effects by changing the production profile of p8 and p12, according to the stage of infection (mechanism similar to what occurs with HIV Nef). Finally, in situations where the individual presents high levels of IL-2, the importance of p12 for HTLV-1 replication would be limited. This may occur in the presence of some co-infections, such as with Hepatitis C virus (HCV).Citation30 This hypothesis may explain the differences observed between this current case and previous described patients.Citation20
This is the first report of a truncated p12 protein in a patient from Brazil presenting a very low proviral load.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The PDTIS/FIOCRUZ sequencing platform.
Funding
This work was supported by Fundação de Amparo a Pesquisa do Estado do Rio de Janeiro (FAPERJ) and by Post-Graduation on Infectious and Parasitic Diseases (UFRJ), a PhD fellowship to C.R. from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and a PhD fellowship to M.J.C.C. and L.Z. from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
References
- Gessain A, Cassar O. Epidemiological Aspects and World Distribution of HTLV-1 Infection. Front Microbiol 2012; 3:388; PMID:23162541; http://dx.doi.org/10.3389/fmicb.2012.00388
- Collins ND, Newbound GC, Albrecht B, Beard JL, Ratner L, Lairmore MD. Selective ablation of human T-cell lymphotropic virus type 1 p12I reduces viral infectivity in vivo. Blood 1998; 91:4701-7; PMID:9616168
- Pise-Masison CA, de Castro-Amarante MF, Enose-Akahata Y, Buchmann RC, Fenizia C, Washington Parks R, Edwards D, Fiocchi M, Alcantara LC, Bialuk I, et al. Co-dependence of HTLV-1 p12 and p8 functions in virus persistence. PLoS Pathog 2014; 10:e1004454; PMID:25375128; http://dx.doi.org/10.1371/journal.ppat.1004454
- Ding W, Kim S-J, Nair AM, Michael B, Boris-Lawrie K, Tripp A, Feuer G, Lairmore MD. Human T-cell lymphotropic virus type 1 p12I enhances interleukin-2 production during T-cell activation. J Virol 2003; 77:11027-39; PMID:14512551; http://dx.doi.org/10.1128/JVI.77.20.11027-11039.2003
- Van Prooyen N, Andresen V, Gold H, Bialuk I, Pise-Masison C, Franchini G. Hijacking the T-cell communication network by the human T-cell leukemia/lymphoma virus type 1 (HTLV-1) p12 and p8 proteins. Mol Aspects Med 2010; 31:333-43; PMID:20673780; http://dx.doi.org/10.1016/j.mam.2010.07.001
- Bai XT, Nicot C. Overview on HTLV-1 p12, p8, p30, p13: accomplices in persistent infection and viral pathogenesis. Front Microbiol 2012; 3:400; PMID:23248621; http://dx.doi.org/10.3389/fmicb.2012.00400
- Ding W, Albrecht B, Luo R, Zhang W, Stanley JR, Newbound GC, Lairmore MD. Endoplasmic reticulum and cis-Golgi localization of human T-lymphotropic virus type 1 p12(I): association with calreticulin and calnexin. J Virol 2001; 75:7672-82; PMID:11462039; http://dx.doi.org/10.1128/JVI.75.16.7672-7682.2001
- Martins ML, Soares BC, Ribas JG, Thorun GW, Johnson J, Kroon EG, Carneiro-Prioetti AB, Bonjardim CA. Frequency of p12K and p12R alleles of HTLV Type 1 in HAM/TSP patients and in asymptomatic HTLV type 1 carriers. AIDS Res Hum Retroviruses 2002; 18:899-902; PMID:12230932; http://dx.doi.org/10.1089/088922202760265560
- Ding W, Albrecht B, Kelley RE, Muthusamy N, Kim S-J, Altschuld RA, Lairmore MD. Human T-cell lymphotropic virus type 1 p12(I) expression increases cytoplasmic calcium to enhance the activation of nuclear factor of activated T cells. J Virol 2002; 76:10374-82; PMID:12239314; http://dx.doi.org/10.1128/JVI.76.20.10374-10382.2002
- Kim S-J, Nair AM, Fernandez S, Mathes L, Lairmore MD. Enhancement of LFA-1-mediated T cell adhesion by human T lymphotropic virus type 1 p12I1. J Immunol 2006; 176:5463-70; PMID:16622014; http://dx.doi.org/10.4049/jimmunol.176.9.5463
- Nicot C. HTLV-1 p12I protein enhances STAT5 activation and decreases the interleukin-2 requirement for proliferation of primary human peripheral blood mononuclear cells. Blood 2001; 98:823-9; PMID:11468184; http://dx.doi.org/10.1182/blood.V98.3.823
- Edwards D, Fenizia C, Gold H, de Castro-Amarante MF, Buchmann C, Pise-Masison CA, Franchini G. Orf-I and orf-II-encoded proteins in HTLV-1 infection and persistence. Viruses 2011; 3:861-85; PMID:21994758; http://dx.doi.org/10.3390/v3060861
- Johnson JM, Mulloy JC, Ciminale V, Fullen J, Nicot C, Franchini G. The MHC class I heavy chain is a common target of the small proteins encoded by the 3’ end of HTLV type 1 and HTLV type 2. AIDS Res Hum Retroviruses 2000; 16:1777-81; PMID:11080826; http://dx.doi.org/10.1089/08892220050193308
- Koralnik IJ, Gessain A, Klotman ME, Lo Monico A, Berneman ZN, Franchini G. Protein isoforms encoded by the pX region of human T-cell leukemia/lymphotropic virus type I. Proc Natl Acad Sci U S A 1992; 89:8813-7; PMID:1528897; http://dx.doi.org/10.1073/pnas.89.18.8813
- Rosadas C, Puccioni-sohler M. HTLV-1 ORF-I Encoded Proteins and the Regulation of Host Immune Response: Viral induced dysregulation of intracellular signaling. J Immunol Res 2015; 2015:498054
- Albrecht B, Collins ND, Burniston MT, Nisbet JW, Ratner L, Green PL, Lairmore MD. Human T-lymphotropic virus type 1 open reading frame I p12(I) is required for efficient viral infectivity in primary lymphocytes. J Virol 2000; 74:9828-35; PMID:11024109; http://dx.doi.org/10.1128/JVI.74.21.9828-9835.2000
- Barreto FK, Khouri R, de Almeida Rego FF, Santos LA, de Castro-Amarante MF, Bialuk I, Pise-Masison CA, Galvão-Castro B, Gessain A, Jacobson S, et al. Analyses of HTLV-1 sequences suggest interaction between ORF-I mutations and HAM/TSP outcome. Infect Genet Evol 2016; 45:420-425; Available from: http://www.ncbi.nlm.nih.gov/pubmed/27553711; PMID:27553711
- Rosadas C, Cabral-Castro MJ, Vicente ACP, Peralta JM, Puccioni-Sohler M. Validation of a quantitative real-time PCR assay for HTLV-1 proviral load in peripheral blood mononuclear cells. J Virol Methods 2013; 193:536-41; PMID:23911967; http://dx.doi.org/10.1016/j.jviromet.2013.07.040
- Iñiguez AM, Gastaldello R, Gallego S, Otsuki K, Vicente ACP. HTLV-1 p12I protein sequences from South America: truncated proteins and common genetic signatures. AIDS Res Hum Retroviruses 2006; 22:466-9; http://dx.doi.org/10.1089/aid.2006.22.466
- Furukawa Y, Usuku K, Izumo S, Osame M. Human T cell lymphotropic virus type I (HTLV-I) p12I is dispensable for HTLV-I transmission and maintenance of infection in vivo. AIDS Res Hum Retroviruses 2004; 20:1092-9; PMID:15585100; http://dx.doi.org/10.1089/aid.2004.20.1092
- Pessôa R, Watanabe JT, Nukui Y, Pereira J, Kasseb J, de Oliveira ACP, Segurado AC, Sanabani SS. Molecular characterization of human T-cell lymphotropic virus type 1 full and partial genomes by Illumina massively parallel sequencing technology. PLoS One 2014; 9:e93374; http://dx.doi.org/10.1371/journal.pone.0093374
- Iñiguez AM, Otsuki K, Magalhães GP, Silva EA, Vicente ACP. Genetic markers on the HTLV-1 p12I protein sequences from Brazilian HAM/TSP patients and asymptomatic HTLV-1 carrier isolates. AIDS Res Hum Retroviruses 2005; 21:580-2; http://dx.doi.org/10.1089/aid.2005.21.580
- Ding W, Kim S-J, Nair AM, Michael B, Boris-Lawrie K, Tripp A, Feuer G, Lairmore MD. Human T-cell lymphotropic virus type 1 p12I enhances interleukin-2 production during T-cell activation. J Virol 2003; 77:11027-39; PMID:14512551; http://dx.doi.org/10.1128/JVI.77.20.11027-11039.2003
- Kim S, Ding W, Albrecht B, Green PL, Lairmore MD. A conserved calcineurin-binding motif in human T lymphotropic virus type 1 p12I functions to modulate nuclear factor of activated T cell activation. J Biol Chem 2003; 278:15550-7; PMID:12601010; http://dx.doi.org/10.1074/jbc.M210210200
- Trovato R, Mulloy JC, Johnson JM, Takemoto S, de Oliveira MP, Franchini G. A lysine-to-arginine change found in natural alleles of the human T-cell lymphotropic/leukemia virus type 1 p12(I) protein greatly influences its stability. J Virol 1999; 73:6460-7; PMID:10400740
- Bidoia C. Human T-lymphotropic virus proteins and post-translational modification pathways. World J Virol 2012; 1:115; PMID:24175216; http://dx.doi.org/10.5501/wjv.v1.i4.115
- Edwards D, Fukumoto R, de Castro-Amarante MF, Alcantara LC, Galvão-Castro B, Washington Parks R, Pise-Masison C, Franchini G. Palmitoylation and p8-mediated human T-cell leukemia virus type 1 transmission. J Virol 2014; 88:2319-22; PMID:24284316; http://dx.doi.org/10.1128/JVI.03444-13
- Edwards D, Fukumoto R, Prooyen N, Van, Gold H, Castro-amarante MF De. Role of dimerization and palmitoylation on the function of HTLV-1 p12 and p8. Retrovirology 2011; 8:A124.
- Collins ND, D'Souza C, Albrecht B, Robek MD, Ratner L, Ding W, Green PL, Lairmore MD. Proliferation response to interleukin-2 and Jak/Stat activation of T cells immortalized by human T-cell lymphotropic virus type 1 is independent of open reading frame I expression. J Virol 1999; 73:9642-9; PMID:10516077
- Sofian M, Aghakhani A, Farazi AA, Banifazl M, Eslamifar A, Rashidi N, Khadem Sadegh A, Ramezani A. Serum profile of T helper 1 and T helper 2 cytokines in hepatitis C virus infected patients. Hepat Mon 2012; 12:e6156; PMID:23423691; http://dx.doi.org/10.5812/hepatmon.6156
