ABSTRACT
The emergence of multiresistant Gram-negative bacteria requires new therapies for combating bacterial infections. Targeting the biogenesis of virulence factors could be an alternative strategy instead of killing bacteria with antibiotics. The outer membrane (OM) of Gram-negative bacteria acts as a physical barrier. At the same time it facilitates the exchange of molecules and harbors a multitude of proteins associated with virulence. In order to insert proteins into the OM, an essential oligomeric membrane-associated protein complex, the ß-barrel assembly machinery (BAM) is required. Being essential for the biogenesis of outer membrane proteins (OMPs) the BAM and also periplasmic chaperones may serve as attractive targets to develop novel antiinfective agents. Herein, we aimed to elucidate which proteins belonging to the OMP biogenesis machinery have the most important function in granting bacterial fitness, OM barrier function, facilitating biogenesis of dedicated virulence factors and determination of overall virulence. To this end we used the enteropathogen Yersinia enterocolitica as a model system. We individually knocked out all non-essential components of the BAM (BamB, C and E) as well as the periplasmic chaperones DegP, SurA and Skp. In summary, we found that the most profound phenotypes were produced by the loss of BamB or SurA with both knockouts resulting in significant attenuation or even avirulence of Ye in a mouse infection model. Thus, we assume that both BamB and SurA are promising targets for the development of new antiinfective drugs in the future.
Introduction
The use of traditional antibiotics that target essential processes in bacteria (e.g. DNA or RNA synthesis, protein synthesis, cell division) has the great disadvantage to select for resistant subpopulations and thereby promotes the emergence of drug resistant bacteria.Citation1,2 The spread of these resistances profoundly limits the treatment options for many bacterial infections and has become a serious global health problem. More recently, particularly Gram-negative bacteria, e.g. carbapenem-resistant or enhanced spectrum β-lactamase producing Enterobacteriaceae (ESBLs) have become a great problem especially in hospital settings.Citation3,4,5 We urgently need to develop alternative strategies to combat Gram-negative bacterial infections. One attractive approach is to control pathogens without killing them in first place while preserving the endogenous microbiota that is usually severely affected by antibiotic treatment.Citation1,6,7 This could be achieved by drugs that interfere with the pathogens virulence mechanisms but do not kill them directly (e.g. by blocking adhesion to host cells or by rendering bacteria more susceptible to killing by the host immune system). The development of drugs directed against Gram-negative bacteria is a great challenge due to the OM, which prevents substances from entering the cell because of its barrier function.Citation2 One approach is to define targets that are in close proximity to the OM or reside in the periplasm. Since a lot of functions of the OM of Gram-negative bacteria are related to OMPs and particularly virulence associated proteins are often OMPsCitation8,9 it seems worthwile to investigate if non-essential factors of the OMP biogenesis pathway might provide potential targets for the development of new antiinfective drugs. Additionally, drugs that are able to induce global rearrangements of the OM that lead to a disruption of the OM barrier function could be exploited as sensitizers administered along with classical antibiotics.
The OM of Gram-negative bacteria is an asymmetrical lipid bilayer. The inner leaflet consists of phospholipids whereas the outer leaflet consists mainly of lipopolysaccharides (LPS).Citation10 OMPs are synthesized in the cytoplasm with an N-terminal signal sequence. These precursors are then translocated across the inner membrane via the Sec-machinery having their signal sequence cleaved off to reach the periplasm.Citation11 There, periplasmic chaperones like SurA or Skp bind to the nascent OMPs to keep them in a protected, unfolded state and guide them to the OM.Citation12 Another chaperone, DegP, functions primarily as a protease to degrade misfolded and aggregated OMPs.Citation13 There exist two pathways of chaperoning OMPs across the periplasm: (I) the SurA pathway and (II) the Skp/DegP pathway.Citation12 Although most OMPs seem to prefer the SurA pathway, they can use Skp/DegP as a rescue pathway under stress conditions.Citation12,14 Actually, it has been shown that a distinct subset of proteins that are strongly intertwined with the pathogenic potential of bacteria strictly relies on the interaction with SurA in order to be inserted properly into the OM.Citation15,16,17 For folding and insertion of ß-barrel proteins into the OM the BAM complex is essential.Citation18 In E. coli this multiprotein complex is composed of five proteins: the two essential components BamA and BamD and the three non-essential components BamB, BamC and BamE.Citation18,19 BamA is the central component of the BAM complex.Citation18 The N-terminal periplasmic part of BamA consists of five polypeptide transport-associated (POTRA) domains.Citation20 The POTRA-domains enable BamA to interact directly or indirectly with the other components of the BAM complex, its substrates (i.e. nascent OMPs) and periplasmic chaperones like SurA.Citation12,21,22 BamB has been suggested to increase the efficiency of OMP biogenesis by providing a scaffold during ß-barrel folding of nascent OMPs at the BAM complex.Citation23 The BamC/D/E subcomplex has been proposed to regulate the function of the BAM complex by driving conformational changes of BamA which enable the insertion of ß-barrel proteins into the OM.Citation24,25 Even though the general pathway for OMP biogenesis from the bacterial cytoplasm to the BAM complex has already been addressed,Citation26 it is still unknown how OMPs are inserted into the OM exactly and how they adopt their final conformation. Nevertheless, the recently described crystal structure of the entire BAM complex of E. coliCitation24 will allow us to experimentally address concrete hypotheses that may explain the mechanisms underlying OMP biogenesis in the future.
Yersinia enterocolitica (Ye) is an enteropathogenic Gram-negative bacterium that belongs to the family of Enterobacteriaceae. This human pathogenic bacterium can be transmitted on oral-fecal route, usually by the uptake of water and food contaminated with excrements of infected pigs or other animals.Citation27 In the small bowel, Ye adheres to and invades epithelial cellsCitation28 leading to mostly self-limiting enterocolitis with diarrhea, fever and occasionally to mesenterial lymphadenitis and secondary diseases like erythema nodosum and arthritis. In some cases, the infection can lead to life-threatening diseases like septicemia.Citation27,29 In the established infection model of murine yersiniosis it has been observed that Ye causes similar diseases compared to humans.Citation30 Thus this model is perfectly suited to address all questions concerning the contribution of distinct factors of the OMP biogenesis machinery to virulence. The virulence of Ye is determined by several virulence factors, the most important ones being the Yersinia adhesin A (YadA),Citation31,32 the adhesin Invasin (Inv)Citation33 and a type III secretion system.Citation34,35 YadA and Inv mediate binding to host cells via extracellular matrix proteins and/or ß1-integrins.Citation36,37,38 Besides facilitating attachment to host cells, YadA also mediates serum resistance, an important virulence trait. Serum resistance is achieved by the interaction with various negative regulators of the complement cascade, namely factor H, C3b and iC3bCitation39,40,41,42 and vitronectin.Citation43
Our research aims at the identification of new targets worthwhile to be addressed in the development of new antiinfective drugs directed against Gram-negative pathogens. We therefore addressed the role of the non-essential components of the OMP biogenesis machinery (BamB, C, E, DegP, Skp, SurA) for virulence associated phenotypes and overall virulence. To this end we used the model organism Ye, generated single gene knockouts, phenotypically characterized the resulting strains and finally tested their virulence in a mouse model of infection.
Materials and methods
Bacterial strains and growth conditions
Bacteria used in this study are listed in Table S1. Ye strains were routinely grown at 27°C in lysogeny broth (LB) containing suitable antibiotics overnight with shaking. Antibiotics were added at the following concentrations: nalidixic acid 10 µg/ml, kanamycin 25 µg/ml, spectinomycin (Sp) 100 µg/ml, chloramphenicol (Cm) 25 µg/ ml and tetracyclin (Tet) 6 µg/ml. Overnight cultures were diluted into fresh medium to an OD600 of 0,1. To generate growth curves, bacteria were grown in 50 ml LB medium with shaking and growth was recorded by measuring the OD at 600 nm every 30 min over 9 h at 37°C, respectively. Expression of bamB from pASK-IBA4C was induced using anhydrotetracyclin (AHTC) in a final concentration of 200 ng/ml in overnight culture and subculture.
Generation of knockout strains
The knockout strains, that are listed in Table S1 were generated using the suicide plasmid pSB890Y (a derivative of pSB890Citation46 where internal PstI restriction sites were removed). The sequences of primers used are listed in Table S2. First, pSB890Y was linearized by PCR using the primers gib_uni_890_r2 and gib_uni_890_f2. Also the flanking regions ∼1000 bp up- and downstream of the target genes were amplified by PCR using the primer pairs listed in the category “Generation of deletion mutants” in Table S2. The vector pSB890Y and the flanking regions were then assembled using a selfmade Gibson Assembly Master MixCitation47 containing T5 Exonuclease (10U µl, Epicentre), Phusion Polymerase (2U µl, New England Biolabs) and Taq-DNA-Ligase (40U µl, New England Biolabs). Next, the strain E. coli 118λpir was transformed with the resulting plasmid. TetR clones (indicative for the presence of pSB890Y) were selected and the presence of the inserts comprising the up- and downstream regions of the target genes was verified by PCR with primers pSB890_seq_f and pSB890_seq_r. The plasmid was then isolated using the peqGold plasmid miniprep kitI (VWR) and transformed into the diaminopimelic acid auxotroph strain E. coli ß2163. Finally, the plasmid was transferred into the Ye wildtype strain via mating, which resulted in TetR merodiploid Ye. After growth at 27°C without antibiotics for 24 h, Ye were counterselected on sucrose for loss of pSB890Y. pSB890Y encodes for a Levansucrase (sacB) that produces a toxic metabolite if grown on sucrose. Only if the plasmid was lost due to a second recombination event, Ye could grow on sucrose-containing media. The knockout of target genes was verified by PCR using the primer pairs listed in the category “Verification of deletions (genomic DNA)” in Table S2.
Generation of complementation constructs
For complementation of the bamB, surA and skp mutant, the coding sequences were amplified by PCR from genomic DNA of Ye wildtype strain and were assembled with the vector pACYC or pASK-IBA4C for inducible expression (bamB complementation) using Gibson Assembly Master Mix. Primer pairs used for the generation of the different pACYC constructs are listed under the category “Cloning of expression vectors for complementation of deletions” in Table S2.
Antibiotic sensitivity
Sensitivity of the different mutant strains to rifampicin, erythromycin, vancomycin and bacitracin was measured by a disk diffusion assay using 6 mm filter paper-disks (BD) after growth at 27°C for 20 h, as described previously.Citation48 Three independent experiments were carried out for each strain.
Sensitivity against bile salts/ SDS
Sensitivity against bile salts or SDS was assessed by the ability to grow on Mac Conkey agar (1.5 mg/ml bile salts) or LB agar containing 0.0125 % SDS by plating a dilution series and determining the presence of bacterial growth after 24 h at 27°C.
Preparation of OM fractions
For preparation of an OM protein fraction 50 ml of bacterial culture grown at 37°C for 4 h with matched numbers of bacterial cells were used. Cells were harvested and resuspended in 500 µl resuspension buffer (0.2 M Tris, 1 M sucrose, 1 mM EDTA, pH 8). Then 500 µg lysozyme (20 units µg; MSB) and 3.2 ml water were added and the samples were incubated for 20 min at room temperature. After the addition of lysis buffer (2% Triton X-100, 50 mM Tris, 10 mM MgCl2, pH 8) protoplasts were lysed. Released DNA was digested with 50 µg DNaseI (10 mg/ml, Roche Applied Science). OMs were pelleted by centrifugation at 77.100 × g for 45 min at 4°C. Resulting pellet was washed in buffer containing 50 mM Tris-HCl pH8, 2% (w/v) Triton-X-100 and 10 mM MgCl2, centrifuged at 85.000 × g for 15 min at 4°C. After three washing steps with water, the membranes were resuspended in SDS sample buffer. The OM proteins in the samples were visualized by staining SDS gels with Coomassie Brilliant Blue (Bio-Rad) for 1 h followed by destaining. For recording of images the Odyssey imaging system (Li-Cor Biosciences) was used.
Western blot analysis
For analysis of whole cell lysates, samples were taken after subculturing for 4 h at 27°C (analysis of Inv expression) or 37°C (analysis of YadA expression). Pellets were resuspended in H2O and Laemmli buffer (Bio-Rad) to obtain 5×106 bacteria/ml. Samples were boiled for 10 min at 95°C before loading on the gel. After SDS-PAGE proteins were transferred onto nitrocellulose membranes. The membranes were blocked overnight with 5% milk powder in PBS at 4°C. Membranes were probed with antisera against Inv (rabbit anti-Inv, 1:2000), YadA (rabbit anti-YadA, 1:1000), OmpA (rabbit anti-OmpA, 1:1000), OmpF (rabbit anti-OmpF, 1:1000), BamB (rabbit anti-BamB, 1:1000) and RNA-Polymerase β subunit (mouse anti-RNA-Polymerase, Neoclone, 1:1000) and secondary antibodies goat anti-rabbit IgG DyLight 680 conjugate or goat anti-mouse IgG DyLight 800 conjugate (Thermo Fisher scientific). For detection of bands, the Odyssey imaging system (Li-Cor Biosciences) was used.
Quantification of Inv and YadA surface display by flow cytometry
After growth for 4 h at 27°C (analysis of Inv expression) or 37°C (analysis of YadA expression), 5×107 bacteria were harvested, washed with PBS, fixed with 4% paraformaldehyde and blocked with 1% BSA in PBS. Cells were incubated with rabbit anti-Inv or rabbit anti-YadA (1:200) antibodies overnight at 4°C followed by incubation with secondary antibody donkey anti rabbit-APC (1:200, Jackson ImmunoResearch) for 1 h at room temperature. Surface display of Inv or YadA was measured by flow cytometry using an LSRFortessa cell analyzer (BD Biosciences). Data were analyzed with WinMDI (J. Trotter) software.
RNA extraction and quantitative real-time PCR
Relative mRNA levels of target genes were determined using quantitative real-time PCR (RT-PCR). Bacteria were grown for 4 h at 27°C (for verification of deletions at mRNA level) or at 37°C (for quantification of mRNA levels in relation to the MS data which were obtained at 37°C) in LB medium. RNA of bacteria was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer protocol. Genomic DNA was digested with DNAseI (recombinant, RNAse free 10 U µl; Roche). Primer pairs used for RT-PCR are listed in the category “Verification of deletions (qRT-PCR)” and “Quantification of expression levels of selected OMPs” in Table S2. For quantitative RT-PCR the Brilliant II SYBR© Green 1-Step Master Mix (Agilent) was used. Relative quantifications were performed with a LightCycler480II instrument (Roche). For determination of transcriptional levels of target genes with the expression of gyrB as a reference the advanced relative quantification tool of the LightCycler480 software 1.5 was used. Experiments were performed three times independently.
NanoLC-MS/MS analysis
OM protein fractions were subjected to 1D SDS-PAGE and stained with Coomassie Brilliant blue (Bio-Rad) for 1 h followed by destaining. Lanes were cut into three slices of comparable size and digested in gel with trypsin as described previously.Citation49 Peptide mixtures were separated on the EasyLC nano-HPLC (Proxeon Biosystems) coupled to an LTQ Orbitrap XL (Thermo Fisher Scientific). Binding and chromatographic separation of the peptides was performed on a 15 cm fused silica emitter with an inner diameter of 75 µm (Proxeon Biosystems), in-house packed with reversed-phase ReproSil-Pur C18-AQ 3 µm resin (Dr. Maisch GmbH). The peptide mixtures were injected in HPLC solvent A (0.5% acetic acid) at a flow rate of 500 nl/min and subsequently eluted with an 127 minute segmented gradient of 5–33–50–90% of HPLC solvent B (80% acetonitrile in 0.5% acetic acid) at a flow rate of 200 nl/min. The mass spectrometer was operated in the data-dependent mode to automatically switch between MS and MS/MS acquisition. Precursor ions were acquired in the mass range from m/z 300 to 2000 in the Orbitrap mass analyzer at a resolution of 60.000. Accumulation target value of 106 charges was set and the lock mass option was used for internal calibration.Citation50 The 10 most intense ions were sequentially isolated and fragmented in the linear ion trap using collision-induced dissociation (CID) at the ion accumulation target value of 5000 and default CID settings. The ions already selected for MS/MS were dynamically excluded for 90 s. The resulting peptide fragment ions were recorded in the linear ion trap.
MS data processing and analysis
Acquired MS spectra were processed with MaxQuant software package version 1.5.2.8Citation51 with integrated Andromeda search engine.Citation52 Database search was performed against a target-decoy Ye serotype O:8 specific database obtained from Uniprot, containing 4172 protein entries, and 245 commonly observed contaminants. This database did not include an entry for TolC. Therefore, data were also analysed with a Ye Uniprot database containing 23,675 protein entries. Endoprotease trypsin was defined as the protease with a maximum missed cleavage of two. Oxidation of methionines and N-terminal acetylation were specified as variable modifications, whereas carbamidomethylation on cysteines was defined as a fixed modification. Initial maximum allowed mass tolerance was set to 4.5 ppm (for the survey scan) and 0.5 Da for CID fragment ions. A false discovery rate of 1% was applied at the peptide and protein level. The label-free algorithm was enabled, as was the “match between runs” option.Citation53 MaxQuant output table was filtered for OM proteins (lipoproteins and ß-barrel proteins). Fourty-seven OM proteins could be identified and were kept for further analysis. Perseus software (version 1.5.0.15), a module from the MaxQuant suite,Citation51 was used for calculation of the significance B (psigB) for each protein ratio (calculated based on averaged LFQ intensities) with respect to the distance of the median of the distribution of all protein ratios as well as its LFQ intensity. All proteins with psigB < 0.05 in a pairwise comparison were considered to be differentially expressed. The mass spectrometry proteomics data have been deposited to the ProteomeXchangeCitation54 Consortium via the PRIDECitation55 partner repository with the dataset identifier PXD004429 (http://proteomecentral.proteomexchange.org/cgi/GetDataset Username: [email protected] Password: XBxYXb4E) and in Table S4. Hydropathy indices of proteins were calculated using the tool http://www.gravy-calculator.de/.
LPS analysis
LPS analysis was performed as described previouslyCitation56,57 using 4% stacking and 12% separating deoxycholate–polyacrylamide gel electrophoresis (DOC-PAGE). After the run the material was visualized by silver staining.Citation58
Quantification of O-antigen surface display by flow cytometry
After growth for 4 h at 27°C, 1×108 bacteria were harvested, washed with PBS, fixed with 4% paraformaldehyde and blocked with 1% BSA in PBS. Cells were incubated with Ye O:8-antigen-specific mouse Fu26–1F1–1 (1:20) monoclonal antibodyCitation59 overnight at 4°C followed by incubation with secondary antibody goat anti mouse IgG Cy2 (1:200,Dianova) for 1 h at room temperature. Surface display of O-antigen was measured by flow cytometry using an LSRFortessa cell analyzer (BD Biosciences). Data were analyzed with WinMDI (J. Trotter) software.
Microscopy
For preparation of electron microscopy pictures bacteria were grown at 37°C for 5 h, fixed in Karnovsky's fixative, embedded in agarose, coagulated, cut in small blocks and fixed again in Karnovsky's solution. After post-fixation and embedding in glycid ether blocks were cut using an ultramicrotome. Sections (30 nm) were mounted on copper grids and analyzed using a Zeiss LIBRA 120 transmission electron microscope.
Mouse infection experiments
Mouse infection experiments were performed according to German law with permission of the Regierungspräsidium Tübingen (permission number H1/14). Groups of five female C57/Bl6 mice (Harlan) of similar age (6–8 weeks) and weight (∼17 g), were injected intraveniously with 1×105 Ye wildtype or mutant bacteria in PBS that had been grown for 4 h at 37°C. Mice were sacrificed after one or three days post infection and the colony forming units (CFU) in the spleen were determined. For oral infection groups of seven BALB/c mice (Harlan) of similar age (6–8 weeks) and weight (∼17 g) were fed 1×108 Ye wildtype or mutant bacteria in PBS that had been grown for 4 h at 27°C. At one, three and five days post infection the CFU in feces were determined. At day five post infection mice were sacrificed and the CFU in spleen, peyers patches and mesenteric lymph nodes were determined. Organs were homogenized in 4 ml (spleen), 2 ml (peyers patches) or 1 ml (mesenteric lymph nodes and feces) sterile PBS (Life Technologies). The number of bacteria and the detection limit were determined as described previously.Citation60 The detection limit of CFU per gram was 500 (log10 500 = 2.69; spleen), 1000 (log10 1111 = 3.05; peyers patches), 333 (log10 333 = 2.47; mesenteric lymph nodes) and 654 (log10 654 = 2.8; feces).
Serum killing assay
Normal human serum (NHS) was collected from at least 4 healthy volunteers and pooled. The IgG and IgM antibody titer against Yersinia in serum was tested by ELISA. Only IgG and IgM negative pooled serum was used throughout the experiments. Aliquots were stored at −80°C and thawed only once. Heat inactivated serum was generated by incubating the serum at 56°C for 30 min immediately before use. To analyze the susceptibility of Ye against complement mediated killing, 5×106 bacteria were incubated in 5% HIS or NHS for 15 min at 37°C. After that the samples were placed on ice for 5 min and an equal volume of brain heart infusion medium was added to stop complement activity. Serial dilutions of the samples were prepared and plated on selective agar plates. After incubation at 27°C for 24 h CFU were determined by counting.
Statistics
Data are means ± SD. Statistical analysis was carried out using GraphPad Prism 5 (GraphPad Software, La Jolla, CA) applying one-way ANOVA analysis with either Dunett´s multiple comparison test or with a Kruskal Wallis test and Dunn´s multiple comparisons test as described in the figure legends. Differences were considered significant if *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Results
Generation of knockout strains and impact of knockouts on growth of Ye
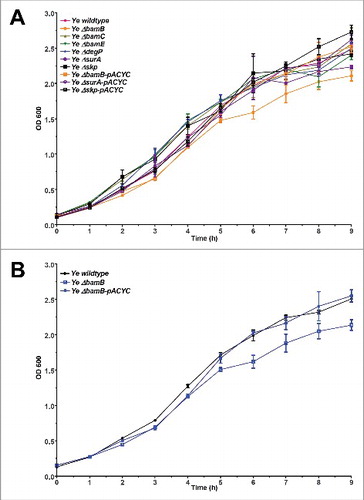
In order to address the importance of BamB, BamC, BamE, DegP, SurA and Skp for fitness and virulence of Ye, we generated single gene knockouts by a mating and homologous recombination strategy using a suicide plasmid (see Materials and Methods section for details). To this end we used the strain Ye WA-314.Citation61 The resulting single gene knockout strains were verified by PCR using genomic DNA as template (). PCR products obtained with the knockout strains had the size expected for successful deletion. Additionally, the presence of mRNA transcripts was analysed by realtime RT-PCR (). mRNA transcripts for the targeted genes were not detectable in the knockout strains. The presence of protein was tested by Western blots using antibodies raised against the corresponding E. coli proteins. However, only the BamB and BamC-specific antibodies did cross-react with the Yersinia proteins (). Though, PCR and RT-PCR clearly demonstrate that our single gene deletions were successful for all targeted genes. To test if the individual knockout of BamB, BamC, BamE, SurA, DegP, or Skp had a general impact on Ye growth at host temperature, we performed growth curves at 37°C (). Compared to the wildtype strain only the BamB deficient strain displayed slightly reduced growth. This turned obvious at 4 h after inoculation and sustained throughout recording of the growth curve (). The growth phenotype of the ΔbamB strain could be rescued by ectopic constitutive expression of BamB from a plasmid (, Ye ΔbamB-pACYC).
Figure 1. Verification of knockout strains. (A) Genomic DNA of indicated strains was isolated and the deletion of target genes was verified by PCR. The expected product sizes are indicated in the table on the right-hand side. (B) Total RNA was extracted from Ye wildtype and the different mutant strains. The deletion of target genes was verified by qRT-PCR using gyrA as reference gene. (C) Ye whole cell lysates were probed with antibodies raised against E. coli BamB or BamC that exhibited cross-reactivity with the respective Ye proteins (all other available antibodies did not cross-react with the Ye homologs). The calculated molecular weight of the tested proteins from E. coli and Ye homologs are indicated in the table on the right-hand side.
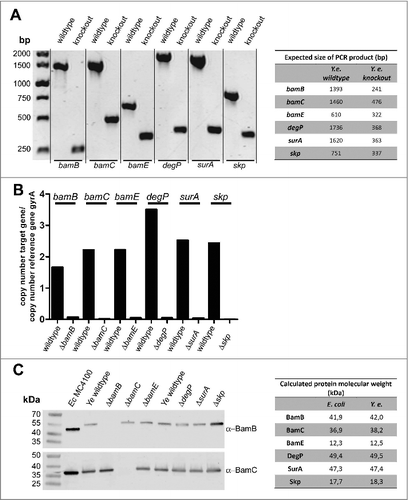
Figure 2. Growth of single-gene knockout strains. Bacteria were grown at 37°C for 9 h in LB. Samples were taken every 30 min and the OD of appropriate dilutions was determined at 600 nm. (A) Comparison of growth of the parent strain Ye wildtype, all mutant strains Ye ΔbamB, ΔbamC, ΔbamE, ΔdegP, ΔsurA and Δskp and the complemented mutants Ye ΔbamB-pACYC, ΔsurA-pACYC and Δskp-pACYC. (B) Growth curves of Ye wildtype strain, ΔbamB strain and ΔbamB strain carrying wildtype BamB on a plasmid (ΔbamB-pACYC).
Assessment of OM integrity
There are different means to assess the integrity and permeability barrier function of the Gram-negative OM. Usually the OM of Gram-negatives quite effectively excludes several antibiotics from entry into the cell. It was reported earlier that the loss of BamB in E. coli leads to an increase in the susceptibility against various antibiotics as a result of increased permeability of the OM.Citation48,62 We therefore investigated the susceptibility of the different Ye knockout strains to the antibiotics rifampicin, erythromycin, vancomycin and bacitracin using a disk diffusion assay (). We found that the sensitivity of the ΔbamC and the ΔdegP mutant strains to all antibiotics used was comparable to that of the wildtype strain, whereas the ΔbamE mutant showed increased susceptibility to erythromycin. Also the Δskp mutant was more sensitive to erythromycin compared to the wildtype strain. Additionally, the inhibition zone around the vancomycin disk was hazy, which could mean that the concentration of vancomycin was subinhibitory for this mutant. More striking phenotypes were observed with the ΔbamB mutant that displayed increased sensitivity to rifampicin, erythromycin, vancomycin but not to bacitracin. The greatest impact on antibiotic sensitivity was observed in the ΔsurA mutant strain that displayed increased sensitivity to all four antibiotics tested as compared to the wildtype strain. By introducing plasmids harbouring wildtype bamB, surA or skp into the corresponding deletion strains the wildtype phenotype could be restored at least partially. These results demonstrate that in Ye the loss of either BamB or SurA significantly enhances the efficacy of selected antibiotics. The observed effect is most likely the result of impaired OM integrity. Skp and BamE seem to play only minor or redundant roles in keeping up OM integrity. The loss of BamC or DegP did not affect the OM permeability in our hands.
Table 1. Sensitivity of knockout strains against antibiotics, bile salts and SDS.
Another possibility to assess the integrity and function of the OM is to challenge the bacteria with detergents like bile salts or SDS. As for antibiotics, the OM usually is an efficient barrier for these substances.Citation62,63,64 We therefore tested growth of the different knockout strains on MacConkey agar, which contains bile salts, or agar containing SDS. We found that the sensitivity of the ΔbamC, ΔbamE, ΔdegP and Δskp mutants to bile salts and SDS was comparable to wildtype (). In contrast, the ΔbamB and the ΔsurA mutants displayed increased sensitivity to both bile salts and SDS compared to the wildtype strain. Again, these phenotypes were restored by introducing wildtypic bamB or wildtypic surA into the corresponding mutants. Overall these results corroborate our findings obtained by the disk diffusion assays and demonstrate that BamB and SurA are essential for maintaining envelope integrity in Ye. Moreover, BamB and SurA seem to fulfill non-redundant functions, whereas BamC, BamE, DegP and Skp appear to play only a minor role.
OM protein composition of knockout strains
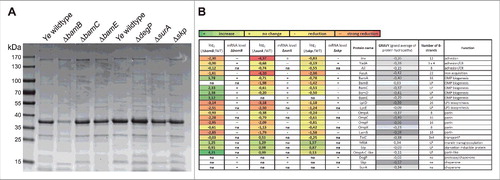
OM integrity is significantly determined by OMP abundance and composition.Citation18,62,65 As we had observed that the OM integrity is disturbed in some of the mutant strains we wanted to get a more in-depth view into their OMP profile. To do so, we prepared OMP fractions of equal bacterial numbers as described elsewhereCitation66 and assessed gross OMP composition by SDS-PAGE and Coomassie blue staining. We found that the pattern and the intensity of numerous bands in all knock-out strains differed to variable extent compared to the wildtype strain (). The most noticeable effect was observed in the ΔbamB mutant strain where the levels of almost all OMPs were drastically reduced. To get a more comprehensive insight into which set of OMPs is affected by the absence of BamB, SurA or Skp (i.e. the strains which had shown some phenotype in the experiments carried out before) we performed proteome analysis of OM fractions and compared it to the wildtype strain (). Semiquantitative proteomic analysis was performed using tryptic in-gel digestion and LC-MS/MS analysis. We could identify 47 OMPs (lipoproteins and ß-barrel proteins; for sake of clarity only some of these are included in ; for the full list please refer to http://proteomecentral.proteomexchange.org/cgi/GetDataset using the Dataset Identifier PXD004429 or Table S4), that were kept for further analysis. Compared to Ye wildtype six OMPs were identified as having significantly higher or lower signal intensities by applying a stringent selection (p < 0.05). In the ΔbamB mutant strain BamE (Protein ID A1JKI3: log2 ΔbamB/WT = 3.12) and a OmpA-C like protein (Protein ID A1JT57: log2 ΔbamB/WT = 4.21) were significantly more abundant, in the Δskp mutant strain we found a membrane-bound lytic murein transglycosylase A precursor (Protein ID A1JPB3: log2 Δskp/WT = 1.57) and a starvation-inducible outer membrane lipoprotein (Protein ID A1JRD0: log2 Δskp/WT = 0.87) at significantly higher intensities. In both the ΔsurA and Δskp mutant strains, FyuA (Protein ID A1JTG3: log2 ΔsurA/WT = −4.10; log2 Δskp/WT = −2.36) was significantly less abundant. Additionally, we observed a significant reduction of Inv (Protein ID A1JT35: log2 ΔsurA/WT = −4.37) in the ΔsurA mutant strain. Being especially interested in the effects of the knockouts on the abundance of virulence factors, the BAM itself and the most abundant OMPs we generated a summary about the effects on protein levels shown as the log2 (mutant/Ye wildtype)-fold change of label-free quantification (LFQ) intensities (). To decipher if the observed changes in protein abundance might be attributed to downregulation of gene expression, we assessed the relative mRNA levels of representative candidates in the bamB, surA or skp deletion mutants by quantitative RT-PCR (; changes in mRNA levels are indicated by +, =, -- and –; for precise numbers please refer to Table S3). As expected, a reduction of mRNA was associated with reduced abundance of protein for all of the highly abundant major OMPs we tested (OmpA, OmpF, LamB). Interestingly, we found that in some cases the strong reduction of protein abundance could not be explained by massive downregulation of mRNA expression. This holds true for Inv and FyuA in the ΔsurA and LptD in the ΔbamB, ΔsurA and Δskp mutant strain. Inv, FyuA and LptD might thus be defined as “true” substrates of SurA, strictly relying on its presence in order to be inserted efficiently into the bacterial OM. We also found that in the ΔbamB mutant all BAM-complex components were more abundant in the OM whereas the corresponding mRNA levels were at wildtype levels. A possible explanation for this is that upon the downregulation of the highly abundant porins (OmpA, C, F, X and LamB) the spectrum of BAM substrates changes profoundly. Consequently, other BAM substrates whose mRNA expression remains unchanged - such as BamA - are inserted into the OM at a higher rate. This might result in the assembly of more BAM complexes in the OM of the ΔbamB mutant strain compared to the wildtype strain. Taken together, we identified the Ye virulence factor Inv as a new true substrate of SurA and figured out that also other important OMPs that contribute either to virulence or bacterial fitness (i.e. FyuA as the receptor for Yersiniabactin that is decisive for efficient iron uptake;Citation67 and LptD, an essential protein in E. coli that is involved in LPS transportCitation68 are less abundant in the mutant strains. Their loss might consequently influence the virulence especially of the Ye ΔbamB, ΔsurA and possibly also of the Δskp mutant strain.
Figure 3. Changes in OM protein composition upon loss of BamB, BamC, BamE, DegP, SurA or Skp. (A) OM proteins were separated on an SDS-PAGE and stained with Coomassie brilliant blue. Gross changes in OMP pattern and intensity of individual bands can be observed. (B) Semiquantitative changes in the abundance of 19 OMPs of Ye ΔbamB, ΔsurA or Δskp as assessed by mass spectrometry. The log2 (mutant/Ye wildtype)-fold change of LFQ intensities is shown, comparing Ye ΔbamB, ΔsurA or Δskp with Ye wildtype. Data are means of three independent experiments. *TolC was not present in the Ye serotype O:8-specific database that was initially used for analysis of the mass spectrometry data. Therefore, data had to be reanalysed using a different Ye (not serotype-specific) database that included also TolC. Corresponding mRNA levels were determined using gyrB as a reference gene (relative quantification) in three independent experiments. Ratios were then calculated as log2 (mutant/Ye wildtype) using relative mRNA concentrations that were calculated based on Cp values and a standard curve comparing Ye ΔbamB, ΔsurA or Δskp with Ye wildtype. Increased mRNA levels are indicated as + (increase; 0.335 to 1.27). Unchanged mRNA levels are indicated as = (no change; −0.6 to 0.335). Decreased mRNA levels are indicated as -- (strong reduction; −2.47 to −1.535) or - (reduction; −1.535 to −0.6). LP = lipoprotein, CR = complement resistance, na = not assessed. The number of β-strands composing the OMP β-barrels is indicated as well as the GRAVY (grand average of protein hydropathy) and the function that has been assigned to the individual proteins.
Expression and surface presentation of Inv and YadA in knockout strains
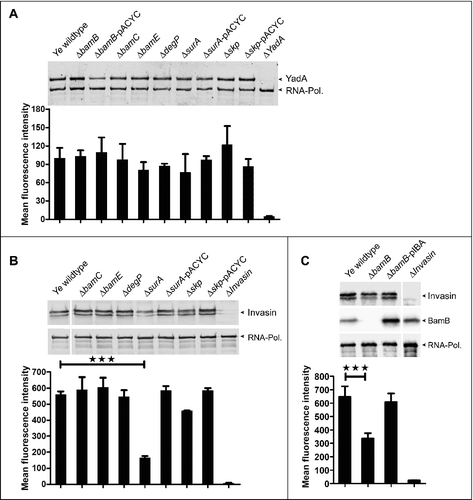
One interesting finding of our MS analyses was that YadA and Inv were affected to different extent by the knockout of bamB, surA or skp. Both YadA and Inv belong to the same membrane protein family that consists of numerous virulence factors and is termed the type V secretion system,Citation8,9,69 also called autotransporters (AT). Whereas YadA belongs to the type Vc AT and forms trimers, Inv is a monomeric, inverted AT belonging to the type Ve AT. Both YadA and Inv are known to be inserted into the OM BAM-dependentlyCitation70,71 and both are anchored to the OM by a 12-stranded β-barrel (consisting of 3 monomers contributing 4 β-strands each in the case of YadA). Thus, we wondered why Inv biogenesis was affected more severely by the absence of BamB or SurA than that of YadA. To this end, we first wanted to verify our MS data by quantifiying the OM abundance of YadA and Inv by other means. We prepared whole cell lysates and performed Western blots and additionally carried out flow cytometry analysis with intact bacteria using antibodies directed against the passenger domains of YadA or Inv, respectively (). Whereas YadA protein levels were hardly affected by any of the knockouts (), we observed a clear reduction of the Inv total protein amount in the Ye ΔbamB () and ΔsurA () strains and consequently also of passenger domain surface display as assessed by flow cytometry ( and ). This reduction could be restored by the ectopic expression of SurA from a plasmid (samples labelled - pACYC for complementation). However, in the case of BamB it seemed that the expression level achieved with this plasmid was not adequate to complement for the absence of BamB in the bacterial chromosome. Therefore, we generated another plasmid for the complementation of the ΔbamB mutant strain that allowed inducible expression and therefore titration of the amount of BamB produced. Using this plasmid the reduction of Inv could be restored in the ΔbamB mutant strain. Taken together, it seems that the BAM complex can efficiently catalyze YadA β-barrel assembly and OM insertion even in the absence of BamB or SurA. In the case of YadA, SurA and Skp thus seem to fulfill redundant roles. To verify this finding we attempted to generate a surA-skp double knockout mutant, but the deletion of both chaperones seems to be synthetically lethal in Ye. This observation has been described before for E. coli.Citation72 In contrast, the efficient folding and insertion of Inv is significantly impaired if BamB or SurA are absent. Possible reasons for this will be discussed later.
Figure 4. Differential impact of single gene knockouts on YadA and Inv protein levels and surface presentation. (A upper panel) Ye wildtype and the different mutant strains were grown in LB. Whole cell lysates were separated by SDS-PAGE, transferred to a nitrocellulose membrane and probed with antibodies raised against YadA. As a loading control, all samples were probed with antibodies directed against bacterial RNA-Polymerase I (RNA-Pol.). (A lower panel) Flow cytometry analysis of YadA cell surface exposure was investigated using antibodies directed against YadA. Bar charts show values of the mean fluorescence intensities +/− SD of 3 independent bacterial cultures. (B upper panel) Western blot analysis of Ye whole cell lysates as described above, but probed with anti-Inv antibodies. (B lower panel) Flow cytometry analysis of Inv cell surface exposure was carried out using antibodies directed against Inv. (C upper panel) Western Blot analysis of Ye whole cell lysates as described above for Ye wildtype, ΔbamB, ΔbamB-pIBA and ΔInvasin strains, using antibodies directed against Inv, BamB and RNA-Pol. (C lower panel) Flow cytometry analysis of Inv cell surface exposure in Ye wildtype, ΔbamB, ΔbamB-pIBA and ΔInvasin strains was carried out as described above. Bar charts show values of the mean fluorescence intensities +/− SD of 3 independent bacterial cultures. The main p value was determined by one-way ANOVA (B, flow cytometry: p < 0.0001). Multiple comparisons were performed by one-way ANOVA with a Dunnett's multiple comparisons test and the p values are indicated with asterisks. *p ≤ 0.05; **p ≤ 0.01, ***p ≤ 0.001.
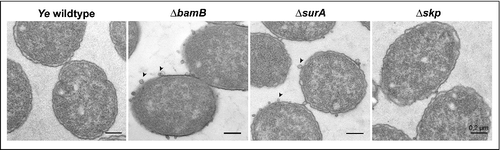
Morphology of selected knockout strains
So far, our experiments demonstrated that the deletion of BamB, SurA and, to a lesser extent, of Skp lead to changes in the OM integrity and abundance of selected OMPs. As the OM composition is intrinsically tied to shaping of the bacterial morphology, we asked whether our mutant strains displayed visible morphological changes. To address this question we prepared electron microscopy pictures of the Ye wildtype and the ΔbamB, ΔsurA and Δskp mutant strain. In the Ye ΔbamB, the ΔsurA and the Δskp mutant strains we observed the formation of outer membrane vesicles (OMVs; ). The phenotype seemed to be somehow less pronounced in the Δskp mutant. The release of OMVs is a common mechanism of all Gram-negative bacteria to secrete OM and periplasmic materialCitation73 and serves as an indicator for cell envelope stress.Citation74 Thus, our results indicate that the loss of BamB, SurA or Skp induces cell envelope stress and the accumulation of envelope components in Ye, which may then be released from the cell surface contained in OMV.
Impact of knockouts on Ye virulence in mouse infection
All our data obtained point at a profound change in OM integrity and OMP composition upon the loss of BamB, SurA and Skp. Therefore, we wanted to know how these changes translate into overall virulence of Ye. In order to assess the effect of the individual deletions of BamB, BamC, BamE, DegP, SurA or Skp during systemic infection, we injected female C57BL/6 mice intravenously with Ye. The bacterial burden (CFU) in the spleen was assessed one and three days post infection (). One day post infection the Ye wildtype strain resulted in a bacterial burden of 7.33 ± 0.42 log10 per gram spleen whereas bacterial counts were below detection limit in the spleens of mice infected with the ΔbamB mutant (p < 0.001). The ΔbamC (7.32 ± 0.12 log10 CFU), the ΔbamE (7.01 ± 0.34 log10 CFU) and the ΔdegP (7.61 ± 0.14 log10 CFU) mutant strains exhibited CFU comparable to the wildtype. The ΔsurA (3.97 ± 0.33 log10 CFU; p < 0.01) strain was significantly attenuated, whereas the Δskp (6.97 ± 0.33 log10 CFU) mutant was only slightly attenuated. To figure out the further development of the CFU of the ΔsurA and the Δskp mutant strains we assessed the bacterial burden in the spleen also at three days post infection (). We were not able to detect Ye ΔsurA (p < 0.01) in the spleens of mice at this later timepoint. The CFU of the Δskp mutant strain (5.38 ± 3.23 log10 CFU; p ≤ 0.01) was still below wildtype levels (9.77 ± 0.11 log10 CFU). This indicates that both the Δskp and the ΔsurA mutant are not able to replicate at wildtype levels in the host and are cleared more efficiently by the host.
Figure 6. Impact of single gene knockouts on virulence of Ye. (A left panel) C57BL/6 wildtype mice were infected intravenously with Ye wildtype and the different mutant strains. Bacterial burden in the spleen was determined one day post infection. (A right panel) C57BL/6 wt mice were infected intravenously with Ye wildtype, ΔsurA and Δskp mutant strains. Bacterial burden in the spleen was determined three days post infection. (B) Balb/C mice were infected orally with Ye wildtype, ΔbamB, ΔsurA and Δskp mutant strains. Bacterial burden in feces was determined one, three and five days post infection. (C) At five days post infection the bacterial burden in peyer´s patches and mesenteric lymph nodes was also determined. The p value for the comparison of wildtype and mutant strains was determined by one-way ANOVA (p < 0.0001). Multiple comparisons were performed by one-way ANOVA with a Kruskal Wallis test and Dunn´s multiple comparisons test and the p value is indicated with asterisks. The horizontal lines denote the mean, the error bars the SD. * p≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, n = 5–17.
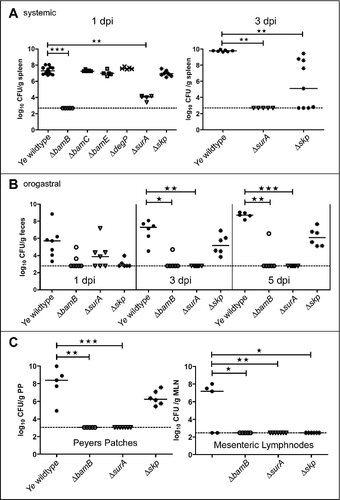
The systemic mouse infection model is very helpful to determine the overall pathogenic potential of a Ye strain. However, the natural route of Ye infection is orogastric. In contrast to systemic infection where the bacteria are immediately confronted with host immune cells and also the complement system, stressors are rather different in the oral infection model. Here, the bacteria have to cope with the passage through the stomach and gut and to compete with the endogenous microbiota. To test how our mutant strains behave under these circumstances, we infected mice orally with the Ye wildtype strain or the ΔbamB, ΔsurA or Δskp mutant strains (). The bacterial burden in feces was then determined at one, three and five days post infection. In animals infected with the wildtype strain we measured a bacterial burden of 5.52 ± 1.79 log10 CFU per gram feces at day one post infection. At day three and five the bacterial numbers had increased (6.86 ± 1.30 log10 CFU at day 3; 8.71 ± 0.35 log10 CFU at day 5). In contrast, we found that the bacterial numbers were slightly lower in animals infected with ΔbamB (3.23 ± 0.83 log10 CFU), ΔsurA (4.02 ± 1.56 log10 CFU) and Δskp (3.04 ± 0.47 log10 CFU) mutant strains at day one post infection. After three days the CFU of the ΔbamB (p ≤ 0.05) and the ΔsurA (p ≤ 0.001) mutant strains dropped below our detection limit. The Δskp mutant was still detectable at day three and five but at slightly lower CFU compared to the wildtype strain (5.27 ± 1.16 log10 CFU at day 3; 6.15 ± 1.04 log10 CFU at day 5). These results demonstrate that the ΔbamB and ΔsurA mutant are able to reach the gut, but are then cleared more efficiently. To analyze the ability of the mutant strains to disseminate from the gut into peyer´s patches and subsequently into the mesenteric lymph nodes and spleen, we also determined the CFU in these organs after five days of infection (). In the peyer´s patches, the ΔbamB (p ≤ 0.01) and the ΔsurA (p ≤ 0.001) mutant strains were not detectable, whereas Ye Δskp (6.39 ± 0.80 log10 CFU/ g tissue) was detectable, but with slightly lower CFU compared to the wildtype (7.98 ± 1.89 log10 CFU/ g tissue). In the mesenteric lymph nodes we were able to detect wildtype only (4.54 ± 4.15 log10 CFU). The results of our animal infection models demonstrate that ΔbamB and ΔsurA are unable to disseminate, whereas the Δskp mutant is able to enter the peyer´s patches but cannot disseminate to the mesenteric lymph nodes. We could not detect any bacteria in the spleen five days after oral infection. Overall these data show that the loss of BamB or SurA and to a lesser degree Skp leads to attenuation of Ye in both systemic and oral infection.
Serum killing assay
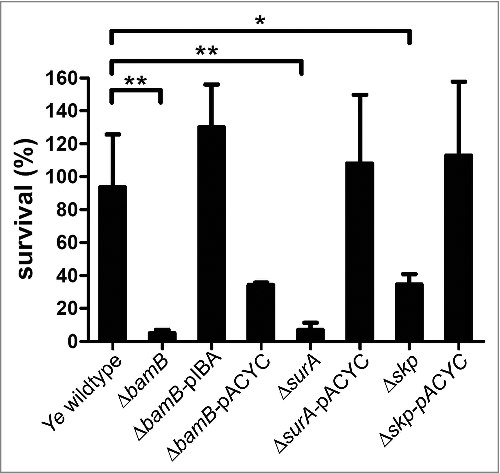
One important virulence trait of Ye is serum resistance. Ye has evolved a multitude of mechanisms to evade the action of the serum complement and can therefore withstand complement-mediated lysis quite efficiently.Citation39,40,41,42 Therefore, we wondered if this aspect of Ye virulence is influenced in the strains that were significantly attenuated in systemic infection and might thus explain the findings of our animal infection experiments. To compare the serum resistance of wildtype and the ΔbamB, the ΔsurA and the Δskp mutant strains we carried out serum killing assays (). Actually, we found that the ΔbamB, the ΔsurA and the Δskp mutant (although to a lesser extent) are highly susceptible to serum killing. We could restore this serum sensitivity phenotype by ectopic expression of BamB, SurA or Skp from plasmids. For complementation of the ΔsurA and the Δskp strain we used a plasmid (pACYC) that leads to constitutive expression of SurA or Skp respectively. For BamB we used an inducible plasmid, since the constitutive expression of BamB from a plasmid did not restore the phenotype. Serum sensitivity could be a decisive factor explaining the quick elimination of the ΔbamB, ΔsurA and the Δskp mutant strains in the systemic infection model either due to enhanced direct lysis by the terminal complement complex or due to more efficient opsonophagocytosis.
Figure 7. Ye ΔsurA, ΔbamB and Δskp are more susceptible to serum killing. Serum killing assay of Ye wildtype, ΔsurA, ΔbamB and Δskp mutant and complemented strains thereof. As a full rescue of the absence of bamB could not be achieved by pACYC, we additionally included a rescue with an inducible plasmid (pIBA). Bacteria were incubated in HIS or NHS. After stopping the reaction the samples were plated on selective agar plates and the CFU was determined. The bar chart depicts the percentage of survival in comparison to samples incubated in HIS in parallel (100% survival). Data are means ± SD of at least three individual experiments. The main p value was determined by one-way ANOVA (p < 0.001). Multiple comparisons were performed by one-way ANOVA with a Dunnett's multiple comparisons test and the p value is indicated with asterisks. *p≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, n = 4.
Discussion
In our study we wanted to investigate if single BAM-complex components and periplasmic chaperones could serve as targets to develop antivirulence drugs. We could show that SurA and BamB are key factors for virulence of Ye and might therefore serve as good candidates. Their loss led to significant attenuation in a mouse infection model and significantly enhanced killing by NHS. BamB and SurA are not only key factors determining virulence of Ye, but also in a number of other important Gram-negative pathogens. The importance of BamB has been described for E. coli and Salmonella.Citation62,75 A key role of SurA for virulence was reported also for Salmonella enterica,Citation76,77 Shigella flexneri,Citation78 uropathogenic E. coli,Citation79,80 Yersinia pseudotuberculosisCitation15,81 and Yersinia pestis.Citation82 Thus the spectrum of pathogens that could be treated by new drugs that aim at reduction of virulence and impairment of OM barrier could be extended or adapted to a large range of Gram-negatives. In fact, the sequence homology of BamB and SurA in a variety of enteropathogens is quite high, particularly in the family of Enterobacteriaceae comprising e.g. Ye, E. coli, Salmonella enterica or Klebsiella pneumoniae (70% protein sequence identity and higher). Therefore, it is possible that a drug that targets BamB or SurA of Enterobacteriaceae can hit several pathogens. Besides targeting OM virulence factors of pathogens by interfering with their biogenesis a drug inhibiting SurA or BamB may also be very helpful to sensitize pathogens to treatment with available antibiotic substances due to the general membrane defect that is induced. By lowering minimal inhibitory concentrations we additionally could reduce the consumption of antibiotics. This would be an important contribution to the avoidance of the emergence of antibiotic resistance and spread.
Our main interest here was to decipher the impact of the knockouts of non-essential BAM proteins and periplasmic chaperones on Ye virulence. Among the best-studied virulence factors of Ye are its adhesins Inv and YadA. We know that a deletion of Inv does not lead to reduced virulence, however, the course of Ye infection is somehow delayed in such mutant strains.Citation33 In contrast, YadA deficient mutants are avirulent in mouse infection models.Citation31,60,83 Even a reduction of YadA molecules in the OM may lead to a significant decrease in virulence.Citation60 However, we think that the reduced virulence of the ΔbamB and ΔsurA mutant strains in the mouse infection is confounded by several aspects of Ye virulence. One candidate protein that has been shown to be required for full virulence of Ye in mice is the iron uptake receptor FyuA.Citation67,84 FyuA abundance was strongly affected in the ΔsurA strain, also in the Δskp and to a lesser extent in the ΔbamB mutant strain and might thus contribute to reduced virulence. Another important virulence trait of Ye is serum resistance. The main factor determining serum resistance is YadA,Citation39,40,41,42 but also LPS and Ail have been shown to contribute.Citation39,56 However, since both YadA and Ail protein levels and surface display were hardly affected in the ΔbamB and ΔsurA mutant strains it is unlikely that changes in YadA or Ail abundance are the reason for the rapid and efficient serum killing we have observed. Therefore, we suspected that changes in LPS could contribute to this phenotype. However, although we had observed profound changes in the abundance of the LPS transport proteins LptD and LptE, the amount of surface exposed LPS (as revealed by O-antigen surface stainings; Fig. S1) of the ΔsurA and Δskp mutant strain was comparable to wildtype. Thus, enhanced serum killing does not seem to be an effect of impaired LPS transport onto the bacterial surface at least in the ΔsurA and Δskp mutant strain.
Actually, we found that in the ΔbamB mutant the LPS phenotype indeed was altered, i.e., it lacked O-antigen, whereas in the ΔsurA and Δskp mutant strains the O-antigen structure was not significantly different compared to the wildtype bacteria (Fig. S1).
The O-antigen does not contribute directly to serum resistance,Citation40 however, it has been shown earlier that Ye lacking LPS O-antigen are highly susceptible to serum killing.Citation39,56 Strikingly, YadA expression levels were comparable to wildtype in these strains.Citation56 But even so, YadA-mediated binding to extracellular matrix proteins (collagen I and laminin) was reduced. It was hypothesized that this effect might be induced by conformational changes in the YadA tertiary structure caused by the absence of O-antigen. Such alterations have been shown to influence the interaction of YadA with collagen I.Citation85 It is also conceivable that changes in the YadA tertiary structure might directly influence the serum resistance by changing the efficiency or the pattern of interactions with negative regulators of complement (factor H, C4BP, C3b, iC3b; reviewed in.Citation86). This is intriguing, as we have observed some time ago that a strain of Ye that expresses a YadA version that carries a single point mutation within its membrane anchor domain was present on the bacterial surface at wildtype levels, but also displayed reduced serum resistance.Citation60 Also here, slight changes in tertiary structure that might alter the interaction with complement regulatory factors were one possible reason for explanation. Finally, an altered function of YadA might also be explained this way: the presence of O-antigen masks other factors on the surface of Ye that efficiently compete with YadA for the binding of ECM or serum proteins and thereby induce reduced serum resistance.
We claim that also the general reduction of OMP abundance might facilitate enhanced binding of terminal complement complex factors, simply due to better accessibility and possibly also due to changes in membrane charge. Taken together, we provide evidence that impaired serum resistance might contribute to the enhanced clearance of both Ye ΔbamB and ΔsurA in the systemic mouse infection model. The more pronounced phenotype of Ye ΔbamB regarding CFU in spleen is in good agreement with the greater importance of BamB also in maintenance of OM barrier integrity as assessed by sensitivity towards antibiotics, SDS and bile salts. Moreover, the lack of O-antigen in the ΔbamB mutant (Fig. S1) might also lead to further reduction of virulence. Indeed, it was shown previously that the O-antigen is a virulence factor in Ye and that a rough-mutant is attenuated in mouse infection.Citation87 In the orogastric infection model, the disrupted OM barrier function of Ye ΔbamB and ΔsurA and the absence of O-antigen of Ye ΔbamB might preclude the survival of Ye during the gastrointestinal passage as both strains have increased susceptibility to detergents and are unable to grow at pH 4.5 (own observation).
Besides deciphering the effect on virulence we wanted to investigate which effects the loss of single BAM-complex components and periplasmic chaperones has on the molecular level in Ye. We therefore analyzed the composition of OMPs and virulence factors. Data obtained by mass spectrometry analysis allowed us some insight into substrate preferences regarding BAM and periplasmic chaperones. We found that the absence of SurA or BamB led to significantly reduced levels of Inv in the OM but not of YadA, suggesting different contributions of these factors to the biogenesis of Inv and YadA. The loss of SurA or BamB significantly reduced the surface display of Inv, but did not alter the OM abundance of YadA. Thus, we assume that Inv is a true substrate of SurA, since its mRNA levels remained unchanged in the absence of SurA, whereas the OM insertion was reduced. Skp obviously is not able to take over the role of SurA for Inv transit and delivery to the BAM. In contrast, the decisive Ye virulence factor YadA can use both the chaperones SurA and Skp for periplasmic transit and BamB is dispensable for its assembly and OM insertion. YadA and Inv are both autotransporter adhesins with Inv being a monomer and YadA being a trimer made of three identical subunits that together form a single ß-barrel. The actual mechanism of how these autotransporters are inserted and assembled into the OM via the BAM complex is still not fully understood, but our results support the hypothesis that the structural differences of Inv and YadA result in different requirements for efficient assembly and insertion. YadA as a trimer is inserted independently of BamB. This is in good agreement with findings for another trimeric OMP of E. coli, namely TolC. TolC is inserted independently of non-essential BAM proteins and periplasmic chaperonesCitation65,88 but is strongly affected by limitation of the essential BAM proteins BamA and BamD.Citation89 TolC has recently been suggested to confound a new class of BAM substrates. This class of atypical multimeric proteins is characterized as being assembled independently of non-essential BAM proteins and periplasmic chaperones, but being strongly affected by a limitation of the number of functional BAM complexes. Like the ß-barrel of TolC,Citation90 the ß-barrel of YadA is made of three identical subunits, each contributing four ß-strands to build a 12-stranded ß-barrelCitation91 and its multimeric ß-barrel is structurally related to TolC. Moreover, YadA is hardly affected by knockouts of the non-essential Bam proteins and the periplasmic chaperones but its assembly is strongly affected even at very early time points of BamA depletion.Citation70 These criteria support the idea that YadA might belong to the class of TolC like BAM substrates. Consequently, we would like to support the recent suggestion that the susceptibility of these multimeric proteins specifically to BAM limitations is caused by the requirement of several BAM complexes to insert a multimer into the OM.Citation89
We also observed that not only the level of Inv (12 ß-strands) but also the levels of a variety of other OMPs, FyuA (22 ß-strands), LptD (26 ß-strands), OmpC (16 ß-strands), OmpF (16 ß-strands) and LamB (18 ß-strands), were reduced when BamB was absent. In the case of OmpF and LamB this can at least partly be attributed to a downregulation at mRNA level as a result of a σE stress response.Citation92,93,94 Interestingly, we did not observe a markedly reduction of OmpA and OmpX in the ΔbamB mutant strain. The ß-barrels of these OMPs are made of 8 ß-strands, whereas the ß-barrels of OMPs that show high dependency on BamB for proper insertion, are made of 12 to 26 ß-strands. It was hypothesized previously, that BamB is more important for the biogenesis of OMPs that have large ß-barrels by providing an increased ß-strand binding capacity to the BAM.Citation95 BamB forms an eight bladed ß-propeller and is suggested to enhance OMP biogenesis efficiency by interacting with ß-strands from the nascent OMPs through ß-augmentation before delivering them to BamA.Citation23,95,96 Our results are in line with observations suggesting that BamB is important for assembly and insertion of OMPs consisting of monomeric ß-barrels with a large number of ß-strands.Citation95 Interestingly, we found that the OMPs that showed a high dependency on BamB for proper insertion (Inv, FyuA, LptD) appeared to be also more dependent on SurA, suggesting that OMPs delivered by SurA preferentially made use of BamB for assembly. Palomino and coworkers observed comparable results for the fimbrial usher FimD, an OMP from E. coli.Citation95 Interestingly, BamA was kind of an exception from this rule. Although its β-barrel is composed of 16 β-strands, its abundance was not reduced in the BamB deficient strain. Given the central function of BamA for bacterial viability, we think that it makes sense that it owns some unique features that render it more independent in how it is being inserted into the OM. Indeed, it has been shown previously that in contrast to other OMPs bearing many ß-strands, BamA is inserted efficiently by a BamACDE complex independently of BamB.Citation97 Thus, BamA might have structural features which facilitate its own assembly and render it less reliant on preassembled Bam-complexes compared to other OMPS. Additionally, we have calculated the grand averages of protein hydropathy (GRAVY) of all proteins (). Actually, there is quite a good correlation for the impact of the absence of BamB on the number of ß-strands and the hydropathy. The lower the hydropathy, the more striking the reduction of OM protein levels. Exceptions from this rule are FyuA which has hydropathy of −0,42 albeit consisting of 22 strands and BamA which is rather unaffected by the absence of BamB and SurA although having 16 ß-strands and a GRAVY index of −0,40.
Finally, we would like to state that both BamB and SurA seem to be targets worthwhile to be addressed in future studies, possibly leading to the development of antivirulence or sensitizer drugs that will provide then new options to treat infections caused by Gram-negative bacteria.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KVIR_S_1278333.zip
Download Zip (399 KB)Acknowledgements
We thank Tanja Späth for excellent technical assistance and Birgit Fehrenbacher for preparation of EM pictures. We also thank Silke Wahl and Irina Droste-Borel for technical assistance in MS sample preparation and Philipp Oberhettinger and Samuel Wagner for helpful discussions.
Funding
This work was funded by grants from the German Research Council (DFG) within the SFB 766 to M.S., the German Center for Infection Research (DZIF) to M.S. and by the gender equality program E.05.00390 of the University Clinics Tübingen to M.S.
References
- Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov 2010; 9:117-28; PMID: 20081869; http://dx.doi.org/10.1038/nrd3013
- Gill EE, Franco OL, Hancock RE. Antibiotic adjuvants: Diverse strategies for controlling drug-resistant pathogens. Chem Biol Drug Des 2015; 85:56-78; PMID: 25393203; http://dx.doi.org/10.1111/cbdd.12478
- Brolund A, Edquist PJ, Makitalo B, Olsson-Liljequist B, Soderblom T, Wisell KT, Giske CG. Epidemiology of extended-spectrum beta-lactamase-producing Escherichia coli in Sweden 2007–2011. Clin Microbiol Infect 2014; 20:O344-52; PMID: 24118431; http://dx.doi.org/10.1111/1469-0691.12413
- Colpan A, Johnston B, Porter S, Clabots C, Anway R, Thao L, Kuskowski MA, Tchesnokova V, Sokurenko EV, Johnson JR, et al. Escherichia coli sequence type 131 (ST131) subclone H30 as an emergent multidrug-resistant pathogen among US veterans. Clin Infect Dis 2013; 57:1256-65; PMID: 23926176; http://dx.doi.org/10.1093/cid/cit503
- Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol 2008; 29:1099-106; PMID: 18973455; http://dx.doi.org/10.1086/592412
- Alksne LE. Virulence as a target for antimicrobial chemotherapy. Expert Opin Investig Drugs 2002; 11:1149-59; PMID: 12150708; http://dx.doi.org/10.1517/13543784.11.8.1149
- Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 2013; 14:685-90; PMID: 23778796; http://dx.doi.org/10.1038/ni.2608
- Henderson IR, Navarro-Garcia F, Nataro JP. The great escape: Structure and function of the autotransporter proteins. Trends Microbiol 1998; 6:370-8; PMID: 9778731; http://dx.doi.org/10.1016/S0966-842X(98)01318-3
- Leo JC, Grin I, Linke D. Type V secretion: Mechanism(s) of autotransport through the bacterial outer membrane. Philos Trans R Soc Lond B Biol Sci 2012; 367:1088-101; PMID: 22411980; http://dx.doi.org/10.1098/rstb.2011.0208
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 2003; 67:593-656; PMID: 14665678; http://dx.doi.org/10.1128/MMBR.67.4.593-656.2003
- Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem 2008; 77:643-67; PMID: 18078384; http://dx.doi.org/10.1146/annurev.biochem.77.061606.160747
- Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev 2007; 21:2473-84; PMID: 17908933; http://dx.doi.org/10.1101/gad.1581007
- Ge X, Wang R, Ma J, Liu Y, Ezemaduka AN, Chen PR, Fu X, Chang Z. DegP primarily functions as a protease for the biogenesis of beta-barrel outer membrane proteins in the Gram-negative bacterium Escherichia coli. FEBS J 2014; 281:1226-40; PMID: 24373465; http://dx.doi.org/10.1111/febs.12701
- Denoncin K, Schwalm J, Vertommen D, Silhavy TJ, Collet JF. Dissecting the Escherichia coli periplasmic chaperone network using differential proteomics. Proteomics 2012; 12:1391-401; PMID: 22589188; http://dx.doi.org/10.1002/pmic.201100633
- Obi IR, Francis MS. Demarcating SurA activities required for outer membrane targeting of Yersinia pseudotuberculosis adhesins. Infect Immun 2013; 81:2296-308; PMID: 23589578; http://dx.doi.org/10.1128/IAI.01208-12
- Ruiz-Perez F, Henderson IR, Leyton DL, Rossiter AE, Zhang Y, Nataro JP. Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of Enterobacteriaceae. J Bacteriol 2009; 191:6571-83; PMID: 19734313; http://dx.doi.org/10.1128/JB.00754-09
- Vertommen D, Ruiz N, Leverrier P, Silhavy TJ, Collet JF. Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. Proteomics 2009; 9:2432-43; PMID: 19343722; http://dx.doi.org/10.1002/pmic.200800794
- Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 2005; 121:235-45; PMID: 15851030; http://dx.doi.org/10.1016/j.cell.2005.02.015
- Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A 2007; 104:6400-5; PMID: 17404237; http://dx.doi.org/10.1073/pnas.0701579104
- Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science 2007; 317:961-4; PMID: 17702946; http://dx.doi.org/10.1126/science.1143993
- Bennion D, Charlson ES, Coon E, Misra R. Dissection of beta-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol Microbiol 2010; 77:1153-71; PMID: 20598079; http://dx.doi.org/10.1111/j.1365-2958.2010.07280.x
- Vuong P, Bennion D, Mantei J, Frost D, Misra R. Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. J Bacteriol 2008; 190:1507-17; PMID: 18165306; http://dx.doi.org/10.1128/JB.01477-07
- Heuck A, Schleiffer A, Clausen T. Augmenting beta-augmentation: structural basis of how BamB binds BamA and may support folding of outer membrane proteins. J Mol Biol 2011; 406:659-66; PMID: 21236263; http://dx.doi.org/10.1016/j.jmb.2011.01.002
- Bakelar J, Buchanan SK, Noinaj N. The structure of the beta-barrel assembly machinery complex. Science 2016; 351:180-6; PMID: 26744406; http://dx.doi.org/10.1126/science.aad3460
- Noinaj N, Kuszak AJ, Balusek C, Gumbart JC, Buchanan SK. Lateral Opening and Exit Pore Formation Are Required for BamA Function. Structure 2014; 22:1055-62 PMID: 24980798; PMID: 24980798; http://dx.doi.org/10.1016/j.str.2014.05.008
- Hagan CL, Silhavy TJ, Kahne D. beta-Barrel membrane protein assembly by the Bam complex. Annu Rev Biochem 2011; 80:189-210; PMID: 21370981; http://dx.doi.org/10.1146/annurev-biochem-061408-144611
- Bottone EJ. Yersinia enterocolitica: The charisma continues. Clin Microbiol Rev 1997; 10:257-76; PMID: 9105754
- Autenrieth IB, Firsching R. Penetration of M cells and destruction of Peyer's patches by Yersinia enterocolitica: An ultrastructural and histological study. J Med Microbiol 1996; 44:285-94; PMID: 8606357; http://dx.doi.org/10.1099/00222615-44-4-285
- Bottone EJ. Yersinia enterocolitica: A panoramic view of a charismatic microorganism. CRC Crit Rev Microbiol 1977; 5:211-41; PMID: 844324; http://dx.doi.org/10.3109/10408417709102312
- Carter PB. Pathogenicity of Yersinia enterocolitica for mice. Infect Immun 1975; 11:164-70; PMID: 1116874
- Roggenkamp A, Neuberger HR, Flügel A, Schmoll T, Heesemann J. Substitution of two histidine residues in YadA protein of Yersinia enterocolitica abrogates collagen binding, cell adherence and mouse virulence. Mol Microbiol 1995; 16:1207-19; PMID: 8577254; http://dx.doi.org/10.1111/j.1365-2958.1995.tb02343.x
- Tamm A, Tarkkanen AM, Korhonen TK, Kuusela P, Toivanen P, Skurnik M. Hydrophobic domains affect the collagen-binding specificity and surface polymerization as well as the virulence potential of the YadA protein of Yersinia enterocolitica. Mol Microbiol 1993; 10:995-1011; PMID: 7934875; http://dx.doi.org/10.1111/j.1365-2958.1993.tb00971.x
- Pepe JC, Miller VL. The biological role of invasin during a Yersinia enterocolitica infection. Infect Agents Dis 1993; 2:236-41; PMID: 8173802
- Bliska JB, Wang X, Viboud GI, Brodsky IE. Modulation of innate immune responses by Yersinia type III secretion system translocators and effectors. Cell Microbiol 2013; 15:1622-31; PMID: 23834311
- Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol 2006; 4:811-25; PMID: 17041629; http://dx.doi.org/10.1038/nrmicro1526
- Hamburger ZA, Brown MS, Isberg RR, Bjorkman PJ. Crystal structure of invasin: A bacterial integrin-binding protein. Science 1999; 286:291-5; PMID: 10514372; http://dx.doi.org/10.1126/science.286.5438.291
- Isberg RR, Leong JM. Multiple beta 1 chain integrins are receptors for invasin, a protein that promotes bacterial penetration into mammalian cells. Cell 1990; 60:861-71; PMID: 2311122; http://dx.doi.org/10.1016/0092-8674(90)90099-Z
- Leong JM, Morrissey PE, Isberg RR. A 76-amino acid disulfide loop in the Yersinia pseudotuberculosis invasin protein is required for integrin receptor recognition. J Biol Chem 1993; 268:20524-32; PMID: 8376409
- Biedzka-Sarek M, Venho R, Skurnik M. Role of YadA, Ail, and Lipopolysaccharide in Serum Resistance of Yersinia enterocolitica Serotype O:3. Infect Immun 2005; 73:2232-44; PMID: 15784567; http://dx.doi.org/10.1128/IAI.73.4.2232-2244.2005
- Kirjavainen V, Jarva H, Biedzka-Sarek M, Blom AM, Skurnik M, Meri S. Yersinia enterocolitica serum resistance proteins YadA and ail bind the complement regulator C4b-binding protein. PLoS Pathog 2008; 4:e1000140; PMID: 18769718; http://dx.doi.org/10.1371/journal.ppat.1000140
- Pilz D, Vocke T, Heesemann J, Brade V. Mechanism of YadA-mediated serum resistance of Yersinia enterocolitica serotype O3. Infect Immun 1992; 60:189-95; PMID: 1729182
- Schindler MK, Schütz MS, Mühlenkamp MC, Rooijakkers SH, Hallström T, Zipfel PF, Autenrieth IB. Yersinia enterocolitica YadA mediates complement evasion by recruitment and inactivation of C3 products. J Immunol 2012; 189:4900-8; PMID: 23071281; http://dx.doi.org/10.4049/jimmunol.1201383
- Mühlenkamp MC, Hallström T, Autenrieth IB, Bohn E, Linke D, Rinker J, Riesbeck K, Singh B, Leo JC, Hammerschmidt S, et al. Vitronectin binds to a specific stretch within the head region of yersinia adhesin a and thereby modulates yersinia enterocolitica host interaction. J Innate Immun 2017; 9: 33-35; PMID: 27798934; http://dx.doi.org/10.1159/000449200
- Peters JE, Thate TE, Craig NL. Definition of the Escherichia coli MC4100 genome by use of a DNA array. J Bacteriol 2003; 185:2017-21; PMID: 12618467; http://dx.doi.org/10.1128/JB.185.6.2017-2021.2003
- Portnoy DA, Moseley SL, Falkow S. Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect Immun 1981; 31:775-82; PMID: 7216474
- Hapfelmeier S, Ehrbar K, Stecher B, Barthel M, Kremer M, Hardt WD. Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun 2004; 72:795-809; PMID: 14742523; http://dx.doi.org/10.1128/IAI.72.2.795-809.2004
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 2009; 6:343-5; PMID: 19363495; http://dx.doi.org/10.1038/nmeth.1318
- Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: A genetic strategy to probe organelle assembly. Cell 2005; 121:307-17; PMID: 15851036; http://dx.doi.org/10.1016/j.cell.2005.02.014
- Borchert N, Dieterich C, Krug K, Schütz W, Jung S, Nordheim A, Sommer RJ, Macek B. Proteogenomics of Pristionchus pacificus reveals distinct proteome structure of nematode models. Genome Res 2010; 20:837-46; PMID: 20237107; http://dx.doi.org/10.1101/gr.103119.109
- Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics 2005; 4:2010-21; PMID: 16249172; http://dx.doi.org/10.1074/mcp.T500030-MCP200
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008; 26:1367-72; PMID: 19029910; http://dx.doi.org/10.1038/nbt.1511
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res 2011; 10:1794-805; PMID: 21254760; http://dx.doi.org/10.1021/pr101065j
- Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J, Akira S, Wiegand M, Hochrein H, O'Keeffe M, et al. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 2010; 32:279-89; PMID: 20171123; http://dx.doi.org/10.1016/j.immuni.2010.01.013
- Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, Dianes JA, Sun Z, Farrah T, Bandeira N, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 2014; 32:223-6; PMID: 24727771; http://dx.doi.org/10.1038/nbt.2839
- Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 2016; 44:D447-56; http://dx.doi.org/10.1093/nar/gkv1145
- Bengoechea JA, Najdenski H, Skurnik M. Lipopolysaccharide O antigen status of Yersinia enterocolitica O:8 is essential for virulence and absence of O antigen affects the expression of other Yersinia virulence factors. Mol Microbiol 2004; 52:451-69; PMID: 15066033; http://dx.doi.org/10.1111/j.1365-2958.2004.03987.x
- Krauss JH, Weckesser J, Mayer H. Electrophoretic analysis of lipopolysaccharides of purple nonsulfur bacteria. International Journal of Systematic Bacteriology 1988; 38:157-163; http://dx.doi.org/10.1099/00207713-38-2-157
- al-Hendy A, Toivanen P, Skurnik M. Rapid method for isolation and staining of bacterial lipopolysaccharide. Microbiol Immunol 1991; 35:331-3; PMID: 1719357; http://dx.doi.org/10.1111/j.1348-0421.1991.tb01562.x
- Gripenberg-Lerche C, Skurnik M, Zhang L, Soderstrom KO, Toivanen P. Role of YadA in arthritogenicity of Yersinia enterocolitica serotype O:8: Experimental studies with rats. Infect Immun 1994; 62:5568-75; PMID: 7525487
- Schütz M, Weiss EM, Schindler M, Hallström T, Zipfel PF, Linke D, Autenrieth IB. Trimer stability of yada is critical for virulence of Yersinia enterocolitica. Infect Immun 2010; 78:2677-90; PMID: 20308293; http://dx.doi.org/10.1128/IAI.01350-09
- Heesemann J, Laufs R. Construction of a mobilizable Yersinia enterocolitica virulence plasmid. J Bacteriol 1983; 155:761-7; PMID: 6874644
- Fardini Y, Trotereau J, Bottreau E, Souchard C, Velge P, Virlogeux-Payant I. Investigation of the role of the BAM complex and SurA chaperone in outer-membrane protein biogenesis and type III secretion system expression in Salmonella. Microbiology 2009; 155:1613-22; PMID: 19372159; http://dx.doi.org/10.1099/mic.0.025155-0
- Ruiz N, Wu T, Kahne D, Silhavy TJ. Probing the barrier function of the outer membrane with chemical conditionality. ACS Chem Biol 2006; 1:385-95; PMID: 17163776; http://dx.doi.org/10.1021/cb600128v
- Urfer M, Bogdanovic J, Lo Monte F, Moehle K, Zerbe K, Omasits U, Ahrens CH, Pessi G, Eberl L, Robinson JA. A peptidomimetic antibiotic targets outer membrane proteins and disrupts selectively the outer membrane in escherichia coli. J Biol Chem 2016; 291:1921-32; PMID: 26627837; http://dx.doi.org/10.1074/jbc.M115.691725
- Charlson ES, Werner JN, Misra R. Differential effects of yfgL mutation on Escherichia coli outer membrane proteins and lipopolysaccharide. J Bacteriol 2006; 188:7186-94; PMID: 17015657; http://dx.doi.org/10.1128/JB.00571-06
- Thein M, Sauer G, Paramasivam N, Grin I, Linke D. Efficient subfractionation of gram-negative bacteria for proteomics studies. J Proteome Res 2010; 9:6135-47; PMID: 20932056; http://dx.doi.org/10.1021/pr1002438
- Heesemann J, Hantke K, Vocke T, Saken E, Rakin A, Stojiljkovic I, Berner R. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65,000 Da and pesticin sensitivity. Mol Microbiol 1993; 8:397-408 PMID: 8316088; PMID: 8316088; http://dx.doi.org/10.1111/j.1365-2958.1993.tb01583.x
- Braun M, Silhavy TJ. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol Microbiol 2002; 45:1289-302; PMID: 12207697; http://dx.doi.org/10.1046/j.1365-2958.2002.03091.x
- Salacha R, Kovacic F, Brochier-Armanet C, Wilhelm S, Tommassen J, Filloux A, Voulhoux R, Bleves S. The Pseudomonas aeruginosa patatin-like protein PlpD is the archetype of a novel Type V secretion system. Environ Microbiol 2010; 12:1498-512; PMID: 20192961
- Lehr U, Schütz M, Oberhettinger P, Ruiz-Perez F, Donald JW, Palmer T, Linke D, Henderson IR, Autenrieth IB. C-terminal amino acid residues of the trimeric autotransporter adhesin YadA of Yersinia enterocolitica are decisive for its recognition and assembly by BamA. Mol Microbiol 2010; 78:932-46; PMID: 20815824; http://dx.doi.org/10.1111/j.1365-2958.2010.07377.x
- Oberhettinger P, Schütz M, Leo JC, Heinz N, Berger J, Autenrieth IB, Linke D. Intimin and invasin export their C-terminus to the bacterial cell surface using an inverse mechanism compared to classical autotransport. PLoS One 2012; 7:e47069; PMID: 23056583; http://dx.doi.org/10.1371/journal.pone.0047069
- Rizzitello AE, Harper JR, Silhavy TJ. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J Bacteriol 2001; 183:6794-800; PMID: 11698367; http://dx.doi.org/10.1128/JB.183.23.6794-6800.2001
- Kulp A, Kühn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 2010; 64:163-84; PMID: 20825345; http://dx.doi.org/10.1146/annurev.micro.091208.073413
- McBroom AJ, Kühn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol 2007; 63:545-58 PMID: 17163978; PMID: 17163978; http://dx.doi.org/10.1111/j.1365-2958.2006.05522.x
- Rolhion N, Barnich N, Claret L, Darfeuille-Michaud A. Strong decrease in invasive ability and outer membrane vesicle release in Crohn's disease-associated adherent-invasive Escherichia coli strain LF82 with the yfgL gene deleted. J Bacteriol 2005; 187:2286-96; PMID: 15774871; http://dx.doi.org/10.1128/JB.187.7.2286-2296.2005
- Sydenham M, Douce G, Bowe F, Ahmed S, Chatfield S, Dougan G. Salmonella enterica serovar typhimurium surA mutants are attenuated and effective live oral vaccines. Infect Immun 2000; 68:1109-15; PMID: 10678914; http://dx.doi.org/10.1128/IAI.68.3.1109-1115.2000
- Tamayo R, Ryan SS, McCoy AJ, Gunn JS. Identification and genetic characterization of PmrA-regulated genes and genes involved in polymyxin B resistance in Salmonella enterica serovar typhimurium. Infect Immun 2002; 70:6770-8; PMID: 12438352; http://dx.doi.org/10.1128/IAI.70.12.6770-6778.2002
- Purdy GE, Fisher CR, Payne SM. IcsA surface presentation in Shigella flexneri requires the periplasmic chaperones DegP, Skp, and SurA. J Bacteriol 2007; 189:5566-73; PMID: 17526712; http://dx.doi.org/10.1128/JB.00483-07
- Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, Hultgren SJ. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 2003; 301:105-7; PMID: 12843396; http://dx.doi.org/10.1126/science.1084550
- Justice SS, Lauer SR, Hultgren SJ, Hunstad DA. Maturation of intracellular Escherichia coli communities requires SurA. Infect Immun 2006; 74:4793-800; PMID: 16861667; http://dx.doi.org/10.1128/IAI.00355-06
- Obi IR, Nordfelth R, Francis MS. Varying dependency of periplasmic peptidylprolyl cis-trans isomerases in promoting Yersinia pseudotuberculosis stress tolerance and pathogenicity. Biochem J 2011; 439:321-32; PMID: 21726196; http://dx.doi.org/10.1042/BJ20110767
- Southern SJ, Scott AE, Jenner DC, Ireland PM, Norville IH, Sarkar-Tyson M. Survival protein A is essential for virulence in Yersinia pestis. Microb Pathog 2016; 92:50-3; PMID: 26724738; http://dx.doi.org/10.1016/j.micpath.2015.12.013
- Pepe JC, Wachtel MR, Wagar E, Miller VL. Pathogenesis of defined invasion mutants of Yersinia enterocolitica in a BALB/c mouse model of infection. Infect Immun 1995; 63:4837-48; PMID: 7591144
- Rakin A, Saken E, Harmsen D, Heesemann J. The pesticin receptor of Yersinia enterocolitica: A novel virulence factor with dual function. Mol Microbiol 1994; 13:253-63; PMID: 7984105; http://dx.doi.org/10.1111/j.1365-2958.1994.tb00420.x
- Tahir YE, Kuusela P, Skurnik M. Functional mapping of the Yersinia enterocolitica adhesin YadA. Identification Of eight NSVAIG - S motifs in the amino-terminal half of the protein involved in collagen binding. Mol Microbiol 2000; 37:192-206; PMID: 10931316; http://dx.doi.org/10.1046/j.1365-2958.2000.01992.x
- Mühlenkamp M, Oberhettinger P, Leo JC, Linke D, Schütz MS. Yersinia adhesin A (YadA)–beauty & beast. Int J Med Microbiol 2015; 305:252-8; PMID: 25604505; http://dx.doi.org/10.1016/j.ijmm.2014.12.008
- Zhang L, Radziejewska-Lebrecht J, Krajewska-Pietrasik D, Toivanen P, Skurnik M. Molecular and chemical characterization of the lipopolysaccharide O-antigen and its role in the virulence of Yersinia enterocolitica serotype O:8. Mol Microbiol 1997; 23:63-76; PMID: 9004221; http://dx.doi.org/10.1046/j.1365-2958.1997.1871558.x
- Werner J, Augustus AM, Misra R. Assembly of TolC, a structurally unique and multifunctional outer membrane protein of Escherichia coli K-12. J Bacteriol 2003; 185:6540-7; PMID: 14594826; http://dx.doi.org/10.1128/JB.185.22.6540-6547.2003
- Mahoney TF, Ricci DP, Silhavy TJ. Classifying beta-barrel assembly substrates by manipulating essential bam complex members. J Bacteriol 2016; 198(14):1984-92; PMID: 27161117
- Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 2000; 405:914-9; PMID: 10879525; http://dx.doi.org/10.1038/35016007
- Shahid SA, Bardiaux B, Franks WT, Krabben L, Habeck M, van Rossum BJ, Linke D. Membrane-protein structure determination by solid-state NMR spectroscopy of microcrystals. Nature methods 2012; 9:1212-7; PMID: 23142870; http://dx.doi.org/10.1038/nmeth.2248
- Dartigalongue C, Missiakas D, Raina S. Characterization of the Escherichia coli sigma E regulon. J Biol Chem 2001; 276:20866-75; PMID: 11274153; http://dx.doi.org/10.1074/jbc.M100464200
- Onufryk C, Crouch ML, Fang FC, Gross CA. Characterization of six lipoproteins in the sigmaE regulon. J Bacteriol 2005; 187:4552-61; PMID: 15968066; http://dx.doi.org/10.1128/JB.187.13.4552-4561.2005
- Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol 2006; 4:e2; PMID: 16336047; http://dx.doi.org/10.1371/journal.pbio.0040002
- Palomino C, Marin E, Fernandez LA. The fimbrial usher FimD follows the SurA-BamB pathway for its assembly in the outer membrane of Escherichia coli. J Bacteriol 2011; 193:5222-30; PMID: 21784935; http://dx.doi.org/10.1128/JB.05585-11
- Noinaj N, Fairman JW, Buchanan SK. The crystal structure of BamB suggests interactions with BamA and its role within the BAM complex. J Mol Biol 2011; 407:248-60; PMID: 21277859; http://dx.doi.org/10.1016/j.jmb.2011.01.042
- Hagan CL, Westwood DB, Kahne D. Bam lipoproteins assemble bama in vitro. Biochemistry 2013; 52:6108-13; PMID: 23919461; http://dx.doi.org/10.1021/bi400865z