ABSTRACT
Enterococcus faecalis and Enterococcus faecium are common inhabitants of the human gastrointestinal tract, as well as frequent opportunistic pathogens. Enterococci cause a range of infections including, most frequently, infections of the urinary tract, catheterized urinary tract, bloodstream, wounds and surgical sites, and heart valves in endocarditis. Enterococcal infections are often biofilm-associated, polymicrobial in nature, and resistant to antibiotics of last resort. Understanding Enterococcal mechanisms of colonization and pathogenesis are important for identifying new ways to manage and intervene with these infections. We review vertebrate and invertebrate model systems applied to study the most common E. faecalis and E. faecium infections, with emphasis on recent findings examining Enterococcal-host interactions using these models. We discuss strengths and shortcomings of each model, propose future animal models not yet applied to study mono- and polymicrobial infections involving E. faecalis and E. faecium, and comment on the significance of anti-virulence strategies derived from a fundamental understanding of host-pathogen interactions in model systems.
Epidemiology of enterococcal infections
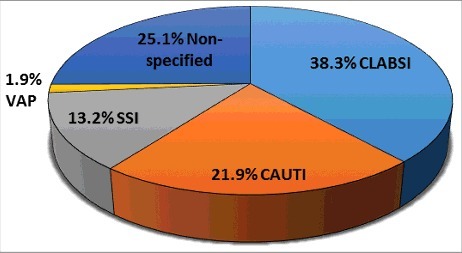
Enterococcus species are ubiquitous organisms present in dairy and fermented food products, natural environments (i.e. plants, soil and water bodies), and the gastrointestinal (GI) tract of humans, other mammals, reptiles and insects.Citation1 This broad distribution is likely due to its ability to survive and persist in a broad range of environments, such as pH, temperature, hyper- and hypotonic conditions.Citation1 In susceptible hosts, Enterococci can cause opportunistic infections. Enterococci are the second most common nosocomial pathogen causing up to 14% of all hospital-acquired infections (HAIs) in the US between 2011–2014 ().Citation2 Between 2006–2007 in the US, Enterococci caused 40% of device-associated infections in the medical intensive care unit (ICU), including central-line associated bloodstream infection (CLABSI), catheter-associated urinary tract infection (CAUTI), surgical site infection (SSI), and ventilator-associated pneumonia (VAP) (). These infections often lead to other clinical manifestations such as infective endocarditis (IE), urinary tract infection (UTI), bacteremia, peritonitis, prosthetic joint infection (PJI), and endophthalmitis; all of which can be serious and life-threatening if left untreated.Citation3,4 Furthermore, infection-associated Enterococci are often antibiotic resistant, making it more complicated to treat.
Figure 1. Eight common pathogens account for 83% of the reported HAIs in the United States. Data adapted from the summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014.Citation2
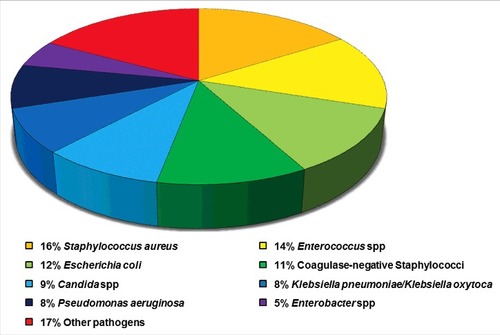
Figure 2. Prevalence of E. faecalis and E. faecium in device-associated HAIs. Data adapted from the summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, January 2006-October 2007.Citation6
Among Enterococcus species, E. faecalis and E. faecium are the 2 most commonly identified species in the human GI tract, and E. faecalis is responsible for 80–90% of Enterococcal-associated nosocomial infections, followed by E. faecium (10–15%).Citation5 This over-representation of E. faecalis among clinical isolates may be related to its natural abundance in the GI tract, where E. faecalis is approximately 100 times more prevalent than E. faecium. However, during the last 20 years, a major epidemiological shift has been noted in the incidence of E. faecium in both US and European hospitals where E. faecium has become increasingly prevalent. The reason for the ecological replacement of E. faecalis with E. faecium is unknown but it has been speculated to be due to the extensive use of antibiotics in hospitals. Currently 90% and 80% of E. faecium from HAIs are resistant to ampicillin and to vancomycin, respectively, while E. faecalis is still largely susceptible to both of these antibiotics.Citation6 The reasons for the difference in antibiotic susceptibility between these 2 Enterococcal species are not well understood.
Many infections are polymicrobial, in which bacteria exist within mixed-species biofilms on host tissues or on medical devices and are more tolerant to antibiotic treatment or environmental stresses. Clustering of microorganisms within biofilms can facilitate and enhance horizontal gene transfer (HGT) of determinants that may increase the capacity of the organisms to colonize, infect, and persist in patients in the clinical setting.Citation7,8 Emerging strains of multidrug resistant Enterococci are a major medical problem, as its resistance profile has extended to include vancomycin and daptomycin, leaving limited options for treatment.
Enterococcal-associated polymicrobial infections
Polymicrobial infections involving several multidrug-resistant pathogens are implicated with increased mortality, hospitalization care, healthcare, and treatment costs.Citation9 Enterococci can cause opportunistic, polymicrobial disease in immunocompromised hosts or in those with underlying health conditions.Citation1,10,11 Since the 1980s, polymicrobial infections of the urinary tract, catheterized urinary tract, wounds, diabetic soft tissues, heart valves, bloodstream, and intra-abdominal and pelvic sites have been reported to be Enterococci-associated.Citation9,11-19 Bacterial species that are frequently, but not always co-isolated with Enterococci in these infections include Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, Klebsiella spp. and Proteus spp.Citation9,15-21 Epidemiological reports describe the presence of Enterococci in polymicrobial infections, but there is limited literature defining the role of Enterococcus spp at infection sites, prompting the need for in vitro and in vivo studies to identify Enterococcal virulence factors, and their mechanism and contribution during interspecies interactions.
Multispecies biofilms rely strongly on interspecies interaction to successfully colonize a niche, either to cause disease (e.g. CAUTI) or to establish colonization resistance (e.g., in the gut).Citation22,23 In vitro models can serve as a preliminary platform to recapitulate human infections for screening antimicrobial agents and anti-biofilm therapeutics, or simulate conditions to discover synergistic or antagonistic effects of interspecies interactions.Citation23-26 In a recent study, Galván and colleagues found that E. faecalis attachment during biofilm formation in vitro can be partially inhibited by uropathogenic E. coli (UPEC) but biofilm formation by K. pneumoniae or UPEC are not affected by E. faecalis.Citation23 Similarly, E. faecalis can promote E. coli biofilm biomass accumulation.Citation27,28 Moreover, co-culture of an E. faecium probiotic strain with enteropathogenic E. coli increased the antibiotic susceptibility of E. coli to aminoglycosides, β-lactams and quinolones.Citation24 In vitro models have also proved significant in identifying virulence mechanisms such as Fusobacterium nucleatum Adherence Inducing Determinant 1 (aid1) mediating co-aggregation with E. faecalis and oral Streptococci but not Lactobacillus casei or Staphylococcus epidermidis,Citation26 and the growth promotion of S. aureus menaquinone biosynthesis-deficient variants in the presence of E. faecalis via menaquinone exchange.Citation25 Recently, Zackular and colleagues showed enhanced Enterococcal dissemination from the GI tract to the liver of Clostridium difficile-infected mice fed with a high-zinc diet, suggesting a possible synergistic relationship between the organisms during C. difficile infection.Citation29
Introduction of invertebrates (i.e., Caenorhabditis elegans) to model polymicrobial infections are an improvement to in vitro models as host survival can be assessed to measure lethality of co-culture combinations and can be useful in screening assays. The C. elegans worm model is a popular invertebrate to study pathogenesis (see section on C. elegans below). Lavigne and colleagues reported increased lethality of warms co-infected with E. faecalis and E. coli, associating enhanced mortality with polymicrobial infections.Citation30 On the other hand, E. faecalis is able to attenuate Candida albicans killing in the C. elegans model by antagonizing hyphal morphogenesis via the Fsr quorum sensing (QS) system.Citation31 However, in vivo mammalian models are the most ideal in simulating human infections due to the presence of a more similar host immune response. In particular, mice offer the ready possibility of genetically modified animals to assess infections in immune deficient backgrounds, while rats are amendable to surgical procedures that make them suitable for modeling post-surgical infections (see and later sections).
Virulence factors of Enterococcus faecalis and Enterococcus faecium
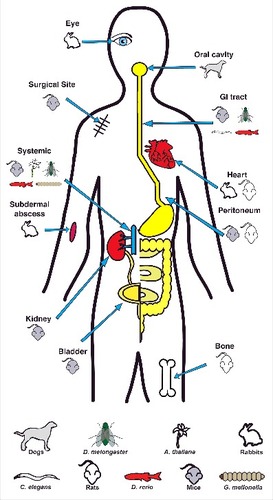
The pathogenesis of Enterococci in opportunistic infections is achieved, at least in part, by their production of virulence factors and their resistance to antimicrobials. These virulence factors are involved in the attachment to host cells or extracellular matrix (ECM) proteins, as well as in immune evasion. Absence of these virulence factors attenuate infection. Most Enterococcal virulence factors have been described in E. faecalis, whereas only a limited number of genes in E. faecium have been experimentally demonstrated to be involved in virulence. Many virulence factors of E. faecalis and E. faecium have been comprehensively reviewed by Garsin and colleagues;Citation4 in this review we will highlight virulence factors of both E. faecalis and E. faecium that have been characterized using various vertebrate and non-vertebrate model systems since 2000 ().
Table 1. Advantages and disadvantages of animal models used for Enterococcal colonization and infection
Table 2. E. faecalis and E. faecium virulence factors studied in various host model systems
Clinical manifestation and animal models for Enterococcus faecalis and Enterococcus faecium Infections
Animal models of infection that can mimic human diseases are crucial tools in establishing the microbial etiology for infectious diseases. Enterococci can colonize and infect the host at a variety of sites (). There is no single comprehensive model system for all Enterococcal infections. Models to study Enterococcal infections have been recently reviewed;Citation4 here we focus on significant features and findings in Enterococcal pathogenesis derived from these model systems in recent years.
Vertebrates
Infective endocarditis
Endocarditis is a heart condition characterized by damage of the vascular endothelium due to high-pressure blood flow through narrowed openings into low-pressure regions of the heart causing platelet-fibrin (vegetation) formation; often prevalent in individuals with pre-existing cardiac conditions.Citation32 In addition to formation of nonbacterial thrombotic vegetation, other endocarditis risk factors include those with medically implanted devices (e.g., artificial heart valves, pacemakers, or implantable defibrillators) that predisposes the individual to bacterial infection from the bloodstream, resulting in infective endocarditis (IE).Citation33 Vegetations serve as a substratum for adhesion and biofilm formation by the infecting bacteria, which limit clearance by host defenses necessitating intensive antibiotic therapy or surgical intervention for treatment.Citation33 IE can be categorized as nosocomial or community acquired IE, with coagulase-negative Staphylococci and Enterococci predominantly causing nosocomial IE.Citation34 Enterococci are the third leading cause of IE in North America, and fourth worldwide.Citation35 Enterococcal endocarditis accounts for 5–15% of all IE cases in the US.Citation36 As endocarditis rarely occurs in healthy hearts, catheters are implanted to induce aortic valve damage in laboratory animals, followed by bacterial infection to mimic IE in humans.Citation32
An experimental model of endocarditis was first established in rabbits, before the development of a rat model in 1978 by Santoro and Levision.Citation37 Rabbits are the most commonly used experimental model of IE, although at times rats are selected for the advantage of housing more animals at a lower expense. After anesthesia, aortic damage is caused by inserting a polyethylene catheter through the right carotid artery into the left ventricle by surgical incision. Catheters can either be removed following intravenous (IV) administration of bacteria or left in place for the duration of the study.Citation38-41
Aggregation substance (AS) is a highly conserved family of surface-anchored polypeptides with adherence properties that enable E. faecalis to interact with host cells and ECM proteins. AS mediates E. faecalis intercellular clumping and is one of the best studied endocarditis virulence factors, although Enterococcal endocarditis is not dependent on the presence of AS.Citation42 AS encoded on the E. faecalis pCF10 sex pheromone plasmid contributes to larger cardiac vegetations in rabbits, as well as complications such as liver necrosis and enlarged spleens.Citation41 Plasmid pCF10 carriage confers a selective advantage to E. faecalis during Enterococcal endocarditis, as seen in mixed infections using varying donor (harboring pCF10)/recipient (no plasmid) ratios.Citation41 Different chromosomal antibiotic resistance markers for differentiation showed consistent shifts in favor of cells harboring pCF10 at 3 d post infection (dpi).Citation41 Immunohistochemistry positively correlates AS expression with virulence in vivo using this model.Citation39 AS is a large 137 kDa protein that harbors an N-terminal signal sequence, lipoteichoic acid (LTA) binding aggregation domain, central aggregation domain (with 2 RGD motifs), and a C-terminal cell wall anchor.Citation43 AS is encoded by the prgB gene of the conjugative plasmid pCF10. Chuang and colleagues observed attenuated virulence in E. faecalis endocarditis upon substitution of glycine to alanine residues in the 2 RGD motifs of AS.Citation43 Active immunization using a purified AS44–331 fragment is ineffective in protecting rabbits from IE due to formation of platelet/fibrin-rich structures formed in endocardial vegetations shielding infecting E. faecalis from host antibodies and possibly immune cells of the humoral immune system.Citation39,44 A combination of studies showed that when challenged with AS-expressing (AS+) E. faecalis, neither active immunization against AS+ E. faecalis cells nor the N-terminal domain of the AS protein protects rabbits against IE; instead the disease is aggravated suggesting that IgG-mediated aggregation of AS+ E. faecalis promotes disease.Citation39,42 Rabbits infected with AS+ E. faecalis following active immunization with AS− E. faecalis showed lower mortality than rabbits actively immunized with AS+ E. faecalis.Citation42 Likewise, Fab fragments of IgG from rabbit antibodies raised against purified AS partially protected AS+ E. faecalis-challenged rabbits (immunized just before infection) by reducing endocardial lesion microbial counts.Citation42
Several other E. faecalis factors have been demonstrated as virulence factors in the rabbit IE model. AtlA is a major autolysin of E. faecalis that cleaves the β−1, 4 links between N-acetylglucosamine and N-acetylmuramic acid within the peptidoglycan. AtlA is a 3-domain enzyme composed of an N-terminal threonine- and glutamic acid (T/E) rich domain of unknown function, a central putative catalytic domain, and a C-terminal cell wall binding domain consisting of 6 LysM modules.Citation45 E. faecalis ΔatlA displayed reduced susceptibility to the bactericidal effects of amoxicillin compared with wildtype (WT) at 48 hours post infection (hpi) in rabbits, suggesting a role for AtlA during IE.Citation46
Recombinase-based in vivo expression technology (RIVET) during E. faecalis surgical site infection identified 2 genes highly expressed in rabbit subdermal abscesses: proB and eep.Citation47 proB encodes for glutamate 5-kinase, involved in proline metabolism, and proline production has been shown to contribute to the survival and pathogenicity of other Gram-positive bacteria in selected animal models under an osmolyte-depleted environment.Citation48 eep encodes an intramembrane metalloprotease that is a member of the site 2 protease family, a conserved class of enzymes that performs regulated intramembrane proteolysis.Citation49 Characterization of these deletion mutants and their role in IE showed stronger attenuation of the eep mutant compared with ΔproB.Citation47 An ortholog of the ArgR family transcription factor, AhrC, was identified in a transposon screen for in vitro biofilm mutants, and subsequently shown to also be strongly attenuated in IE.Citation38 However, not all biofilm-impaired mutants are attenuated in IE, suggesting that biofilms grown on abiotic surfaces in vitro and biotic surfaces in vivo have different properties.Citation38 Consistent with this observation, Leuck and colleagues reported inconsistent adherence phenotypes by E. faecalis clinical strains comparing polystyrene dish biofilm assays and an ex vivo heart valve assay.Citation50 Data from both studies suggest that biofilm formation in vitro and endocarditis development may not necessarily be linked.Citation38,47
Bacterial QS relies on 2-component systems involving a sensor transducer and response regulator for sensing cell density and regulates expression of virulence factors.Citation51 The S. aureus agr QS locus mediates S. aureus dissemination during animal models of infection.Citation52 E. faecalis expresses Agr-like proteins (FsrA, FsrB and FsrC) that are necessary for positive regulation of the virulence-associated proteases Gelatinase E (GelE) and serine protease (SprE).Citation51,53 FsrC is a histidine kinase that senses extracellular accumulation of a peptide lactone encoded at the C-terminus of the FsrB protein. FsrC sensing leads to activation of the response regulator and transcription factor FsrA. GelE, encoded by gelE, aids in the subversion of host immune responses to E. faecalis during IE.Citation40 Even though the fsr system and its products are important for virulence in all the infection models discussed in this review, the absence of GelE alone can limit dissemination from the primary vegetation and results in lower bacterial burdens in IE.Citation40 In contrast, the absence of SprE does not impact virulence in IE. Since gelatinase hydrolyzes fibrin to facilitate bacterial dissemination, aortic vegetations from rabbits infected with E. faecalis lacking GelE display more fibrin-rich matrices than WT-infected rabbits.Citation40 GelE also cleaves chemoattractants such as complement C5a resulting in decreased neutrophil migration in vitro.Citation40 In IE, GelE modulates the host immune response by reducing neutrophil-like cell migration to infected tissues.Citation40 Consistent with the rabbit model of IE and other infection models, rat cardiac colonization is attenuated in E. faecalis lacking gelE and sprE;Citation40,51,53-55 requiring a higher ID50 than WT to colonize rat heart valves and display reduced endocarditis induction rate.Citation56
E. faecalis and E. faecium virulence are also well studied using a rat model of IE. This model has been used to test the efficacy of new therapeutics and prophylactic countermeasures against E. faecalis and E. faecium.Citation56-67 IE is initiated in the same manner as in rabbits, and disease severity can be assessed by mortality rates, embolism, colonization efficiency, 50% infective doses (ID50), and mixed competitive infections.Citation57
E. faecalis glycosyltransferases, encoded by bgsA and bgsB, play important roles in synthesising cell wall glycolipid; mutants in an individual cell wall glycolipid display fewer endocarditic lesions and reduced bacterial colony-forming units (CFU) in vegetations.Citation58 Strains mutated in the general stress gene gls24 require a higher ID50 in both the mouse peritonitis and rat endocarditis model.Citation57,68 gls24 encodes a general stress protein involved in bile-salt resistance and its transcription was induced under glucose starvation and other stress conditions, including the presence of bile salts and cadmium chloride.Citation69 Two other E. faecalis virulence factors, Ace (collagen adhesion) and EfbA (Enterococcal fibronectin-binding protein) were studied by Singh and colleagues in the rat IE model. ace encodes a collagen-binding protein whose transcription is positively regulated by a 2-component system regulatory system, GrvRS (global regulator of virulence).Citation70 Another transcription regulator, Ers (PrfA-like regulator), can act as a repressor of Ace.Citation71 Ace is an adhesin that binds to collagen (type I and IV) and laminin and belongs to the microbial surface components recognizing adhesive matrix molecules (MSCRAMM) protein family, sharing sequence homology to the ligand-binding region with S. aureus Cna.Citation72 EfbA is encoded by the Enterococcal ortholog of Streptococcus pneumoniae pavA that binds with strong affinity to immobilized fibronectin, collagen I and collagen V.Citation73 Independent abrogation of either ace or efbA resulted in significant attenuation in a mixed infection.Citation63,64 Finally, rats pre-immunized with either Ace or EfbA were less susceptible to IE, highlighting the potential of Ace and EfbA as immunotherapeutic antigenic targets.Citation63,64 Another prophylactic treatment effective in protecting the host against IE includes passive protection of rats by injecting anti-EbpC (endocarditis and biofilm-associated pilus subunit protein C) monoclonal antibody (Mab), compared with control IgG, intravenously through the tail vein 24 h post-catheterization (hpc) and 1 h before bacterial inoculation. The endocarditis- and biofilm-associated pilus (Ebp) is encoded by a polycistronic gene locus which consists of 3 structural pilin genes, ebpA, ebpB and ebpC, and an adjacent downstream gene, srtC that is independently expressed from a second promoter.Citation74 Sortase C (SrtC) is a pilin-specific sortase that polymerizes the Ebp/Emp pili by a transpeptidation reaction before their attachment to the cell wall by Sortase A (SrtA). SrtA is a housekeeping enzyme that recognizes the LPXTG motif at the C-terminus of most cell wall protein precursors and is required for the attachment of these cell surface-anchored virulence factors on the cell wall. Rats passively immunized with anti-EbpC were less susceptible to IE by E. faecalis.Citation62 Laverde and colleagues performed similar passive immunization experiments by introducing polyclonal antibodies against LTA fragments into rats over 3 doses: (i) at the time of catheterization, (ii) 24 hpc, and (iii) 4 hpi by E. faecalis. When compared with controls, passively immunized rats had fewer cardiac vegetations and greater bacterial clearance.Citation59
A number of E. faecium virulence factors have been examined in the rat IE model, including the cell wall-anchored adhesion for collagen from E. faecium (acm),Citation60 Enterococcal surface protein (esp),Citation67 global transcriptional regulator (ccpA),Citation65 fibronectin-binding protein (fnm),Citation66 novel cell wall binding proteins containing a WxL motif,Citation75 biofilm and endocarditis-associated permease A (bepA)Citation61 and EmpA (previously ebpAfm).Citation76 Like Ace in E. faecalis, Acm is also a homolog of Cna, a collagen adhesin shown to be important for S. aureus endocarditis.Citation77 Acm is important for early adherence as well as vegetation formation.Citation60 Enterococcal surface protein Esp is encoded on a pathogenicity island in both E. faecalis and E. faecium. It contains multiple repeat motifs, a characteristic found in many bacteria surface protein adhesins involved in host-ligand binding.Citation78 esp transcription is regulated by Enterococcal biofilm regulator B (ebrB) located upstream of esp in E. faecium.Citation79 Esp contributes to colonization of heart valves at 24 hpi but not at 3 hpi and is therefore not likely to play a role in initial colonization.Citation67 EmpA, the tip subunit of the Emp pili, is essential for vegetation colonization at 48 hpi.Citation76 ccpA that encodes CcpA, a global transcriptional regulator of carbon catabolite repression in Gram-positive bacteria, affects the growth of E. faecium in a rat IE model.Citation65 In competitive infections, E. faecium ΔccpA is outcompeted in aortic valve colonization compared with WT; however, this could be due to reduced fitness of the ΔccpA strain as it has a growth defect when cultivated in brain heart infusion (BHI) broth.Citation65 The Fnm protein of E. faecium, is a homolog of S. pneumoniae PavA. Fnm binds to immobilized fibronectin in a concentration dependent manner, and can also bind to collagen type V and laminin.Citation66 Deletion of fnm resulted in reduced binding to fibronectin and this strongly attenuates cardiac colonization.Citation66 Cell wall binding proteins harboring a WxL sequence are involved in IE pathogenesis, since a strain defective in all 3 WxL (ΔwxlABC) genes was outcompeted in vegetations by WT at 48 hpi.Citation75 E. faecium bepA, encoding a carbohydrate phosphotransferase system (PTS) permease, is enriched in E. faecium hospital outbreak isolates; bepA mutants are also outcompeted by the WT strain in IE.Citation61 A common observation from these competitive mixed infections is that genetic mutants usually fail to colonize as efficiently as their WT counterpart, suggesting that the competing WT strain is unable to compensate for loss of gene function in the mutant.
Peritonitis
Peritonitis is an infection of the abdominal lining and can progress in 3 stages: (i) primary peritonitis - arises via hematogenous spread of bacteria which often affects immunocompromised individuals; (ii) secondary infection - arises via damage to a visceral organ, which may be postoperative (for example perforation, trauma, postoperative complication, etc.); (iii) tertiary peritonitis - defined as persistent or recurrent intra-abdominal infection. Peritonitis may be mono- or polymicrobial and can lead to bloodstream infection, organ failure, or in more serious cases, death.Citation11
Rodents are preferred over other animals to study peritonitis owing to their small size and low cost.Citation80 Typically, rodents are challenged with an intraperitoneal (IP) injection of 0.2–1 ml bacterial inoculum mixed with sterile rat fecal extract.Citation81-84 Modifications to rat infections may include making an abdominal incision before implantation of gelatin capsules harboring bacteria.Citation85 A combination of RIVET screens performed under different in vivo conditions (i.e., mouse bacteremia or mouse peritonitis) identified E. faecalis ef_0377 transcriptional activation during peritonitis.Citation86 Other genes identified in the RIVET screen have yet been characterized in vivo. Additionally, genes induced during E. faecalis-mediated mouse peritonitis include cytolysin (cylLL, cylLS, cylM), endocarditis-specific antigen (efaCBA), gelE, QS 2-component system (fsrABCD), ace adhesin, AS (prgB), a predicted adhesin (ef0149) and genes involved in glycerol metabolism (gldA and glpK).Citation87 Cytolysin is made up of 2 subunits; the large subunit encoded by cylLL and the small subunit encoded by cylLs, Both subunits are post-transcriptionally modified by the cylM gene product. Once modified, the cytolysin subunits are secreted from the cell by an ABC transporter encoded by cylB. Activation and subsequent maturation of the cytolysin subunits is performed by a protease encoded by cylA.Citation88 The endocarditis-specific antigen (efaCBA) is a 3 gene operon, predicted to encode components of an ABC-type transporter, with EfaA as its putative substrate-binding lipoprotein component.Citation89 Virulence determinants cytolysin,Citation90 Efa,Citation91 GelE,Citation51 FsrABCCitation51 and ASCitation92 were previously characterized and contribute to mouse peritonitis. Double deletion of gldA (glycerol dehydrogenase) and glpK (glycerol kinase) resulted in attenuated bacteremia in mice while single mutants of gldA and glpK were not attenuated.Citation87 Overall, screening methods have been highly informative for obtaining a global view of bacterial responses in a defined environment, and especially useful for identifying novel factors that are relevant to specific diseases. However, validation including reverse-transcription quantitative PCR (RT-qPCR), gene characterization, and mutational analyses must be performed to fully understand gene functions during infection.
Individual deletions of fsrA, fsrB and sprE delay mortality in mice suffering peritoneal infection while fsrB complementation restores killing.Citation51,53 A homology search for putative 2-component systems led to the identification of an Enterococcal 2-component system a (eta), with the etaR regulator mutant requiring a higher LD50 (50% lethal dose) compared with WT in the mouse peritonitis model.Citation93 Pathogenicity islands (PAIs) are large genomic regions containing a collection of virulence-related genes acquired via HGT. PAI-encoded virulence factors contain regulators that control virulence gene expression. A putative AraC/XylS-type transcriptional regulator, perA, encoded on a PAI of E. faecalis mediates persistence in spleens up to 72 hpi during mouse peritonitis.Citation94 PerA expression is variable and depends on the insertion of IS1191 in the perA promoter sequence; in strains where IS1191 is absent, PerA expression is not disrupted.Citation94 Low bacterial burden in liver and spleens at 24 and 48 hpi of perA mutants may be a result of sensitivity to oxidative stress, although dissemination was not hampered since both mutant and WT CFU were comparable in the blood in the mouse model of peritonitis.Citation94 Although attenuated in the peritonitis model, inactivating perA resulted in stronger biofilm formation in vitro.Citation94 PerA may negatively regulate expression of biofilm factors. The MafR (Mga/AtxA-like faecalis regulator) regulator of E. faecalis influences transcription of genes related to carbon source utilization and positively influences the transcription of genes during the growth of E. faecalis in blood and urine. Although a mafR deletion did not result in attenuated colonization of the peritoneal cavity, a reduction of IL-6 and neutrophil infiltration in the peritoneal fluid implied that MafR modulates the host inflammatory response during this infection.Citation95
It is worth highlighting that some studies have used the mouse model of peritonitis to study E. faecalis virulence using an in vivo-in vitro infection model. Infections are performed as described in the in vivo model but peritoneal macrophages are harvested from peritoneal lavage fluid 4 h following infection.Citation51,96 Giard and colleagues showed that the E. faecalis Ers (Enterococcal regulator of survival) mutant strain did not survive within murine peritoneal macrophages and were completely eliminated by 48 hpi.Citation96 Ers is a member of the Crp/Fnr family and showed 69% amino acid similarity to Srv, a PrfA-like regulator of S. pyogenes implicated in virulence. The Ers protein is important for survival within macrophages, in relation to oxidative challenge in E. faecalis.Citation97 Mice infected via peritoneal injection with Δers survived for up to 100 hpi whereas mice infected with WT survived for only 70 hpi.Citation98 E. faecalis expresses oxidative stress mechanisms such as hypR,Citation99 tpx,Citation100 spx,Citation101 msrA,Citation102 and msrBCitation102 that allow them to survive the intracellular environment of macrophages. The oxidative stress response transcriptional regulator HypR (hydrogen peroxide regulator) protects E. faecalis from oxidative challenge caused by hydrogen peroxide.Citation99 HypR directly controls the transcription of hypR itself, the ahpCF (alkyl hydroperoxide reductase) operon as well as tpx (thiol peroxidase). AhpC is a peroxide-reducing protein and is representative of a very large ubiquitous family of cysteine-based peroxidases, now designated as peroxiredoxins. AhpF is a flavoprotein and acts as the AhpC reductase.Citation103 Tpx, also a member of the peroxiredoxin family, plays an important role in protection against the oxidative burst produced by mouse peritoneal macrophages.Citation100 Besides intracellular macrophage survival, HprR and Tpx both contribute to virulence by causing lethality in peritoneally infected mice.Citation99,100 The major global stress regulator Spx of E. faecalis is highly conserved among low-GC Gram-positive bacteria and was first identified as a suppressor of ClpP and ClpX phenotypes in Bacillus subtilis.Citation104 It promotes colonization of the peritoneum and spleen, contributing to mortality in a mouse model of foreign body-associated peritonitis.Citation101 During the host immune response to an infection, E. faecalis that are engulfed by phagocytes must deal with reactive oxygen species within phagosomes and neutralize the environment for survival. E. faecalis survives by biosynthesizing superoxide dismutasesCitation105 and peroxidases,Citation100 upregulating oxidative stress response pathways,Citation99 or by using oxidative repair strategies.Citation102 Methionine sulfoxide reductases A and B (encoded by msrA and msrB) are antioxidant repair enzymes synthesized to reduce oxidized methionine residues (MetSO) to methionine.Citation102 Improved killing of E. faecalis ΔmsrAΔmsrB, compared with ΔmsrA or ΔmsrB, by activated mouse peritoneal macrophages demonstrates the essentiality of both msrAB for surviving phagocytosis.Citation102 The PerR (peroxide regulator) of B. subtilis functions as a transcriptional regulator involved in oxidative stress tolerance and homologs have been found in other Gram-positive bacteria including E. faecalis.Citation106 PerR exerts weak regulation on oxidative response mechanisms and does not play an important role in E. faecalis survival within macrophages.Citation106 In the mouse model of peritonitis, 60% of mice survived infection with E. faecalis ΔperR at 70 hpi compared with 0% survival after WT infection.Citation106 The Enterococcal leucine-rich operon (elrA-E) encodes proteins that interact with host cells and is positively regulated by an upstream elrR gene product.Citation107,108 During mouse peritonitis infection, E. faecalis ΔelrRΔelrA was attenuated the most compared with ΔelrA followed by ΔelrR (ΔelrRΔelrA>ΔelrA>ΔelrR), displaying reduced dissemination into liver and spleen and decreased IL-6 pro-inflammatory cytokine production in peritoneal fluid.Citation82,107,108 Deletion of elrA did not have any impact on macrophage phagocytosis but intracellular survival decreased at 24 hpi.Citation107 Overexpression of Elr inhibits adhesion and internalization into macrophages and increases killing of mice infected intraperitoneally, representing a mechanism for evading phagocytosis while promoting disease pathogenesis.Citation82 Altogether, elrR not only regulates elrA expression, as seen with the additive attenuation of virulence by ΔelrRΔelrA, but ElrB-E may contribute to virulence even in the absence of elrA.Citation108 Further work is required to define the role of combined or individual ElrB-E proteins in host pathogen interactions.
Bacterial polysaccharides confer adhesion, invasion, resistance to phagocytosis, and invoke host inflammatory responses.Citation84,109 E. faecalis Enterococcal polysaccharide antigen (epa) encodes a locus of 18 genes (epaA-R) responsible for polysaccharide biosynthesis.Citation109 Each mutant of E. faecalis epaB, epaE, epaM and epaN was attenuated in a mouse peritonitis model by assessing survival rates at 100 hpi.Citation84,109 Future studies to understand the implication of Epa in host pathogen interaction should include peritoneal macrophage survival assays as well as assessing infected tissue burden to determine dissemination. Cytokine profiling may also provide information about immunogenic properties of these proteins during infection.
The host immune system functions to recognize and activate responses to foreign molecules associated with invading bacteria, commonly termed as pathogen-associated molecular patterns (PAMPs). PAMPs from Gram-positive bacteria including peptidoglycan, lipoproteins, bacterial DNA, and LTA activate host immune defenses via the Toll-like receptor (TLR) pathway.Citation83 E. faecalis defective in bgsA does not synthesize the major cell membrane glycolipid diglycosyldiacylglycerol (DGlcDAG), resulting in a higher lipoprotein content in the cell membrane.Citation83 Mice challenged with ΔbgsA displayed increased lethality without a difference in bacterial burden (in blood, kidney and peritoneal lavage fluid) compared with WT.Citation83 Induction of proinflammatory cytokines (TNF-α and IL-6) and chemokine MIP-2, accompanied with increased TNF-α concentrations and increased leukocyte influx into the peritoneal lavage fluid, are indicative of an acute innate response detected in the plasma of infected mice 1 h following infection by ΔbgsA.Citation83 Absence of bgsA enhances kidney colonization in the mouse model of UTI and reduces colonization in the rat model of IE.Citation58,110 Overall, E. faecalis BgsA maintains lipoprotein levels on the cell membrane and subverts host responses by inhibiting immune activation in vivo.Citation83
Glucose starvation of E. faecalis induces general stress protein Gls24 expression, causing high mortality in the mouse peritonitis model.Citation68,69 Loss of gls24 resulted in a reduced LD50.Citation68 Antibody protection using anti-Gls24 rabbit serum was achieved by immunizing mice before and during infection. Therefore, Gls24 is expressed during the mouse model of peritonitis and is a potential target for immunotherapy. Homologs of E. faecalis gls24 and glsB present in E. faecium (gls24-like and glsB-like) displayed redundancy and had no role in virulence when inactivated individually, although disruption of both loci reduced mortality of mice during peritonitis.Citation111
E. faecium clinical strains belonging to clonal cluster 17 isolated in the US and Colombia harbor a large plasmid, pHylEfm (> 145 kb) that transfers readily from clinical strains to E. faecium hosts via conjugation and which promotes intestinal colonization in a mouse GI model.Citation7 Infection with a strain that acquired pHylEfm, encoding hylEfm, results in increased mortality in a mouse peritonitis model.Citation7,8 The hylEfm gene encodes a putative glycosyl hydrolase predicted to be a virulence determinant.Citation112 Strains bearing a deletion of the hylEfm-region on the pHylEfm plasmid (pHylEfmTX16Δ7,534) are attenuated in peritonitis.Citation112 Complementation of hylEfm or hylEfm plus its downstream gene did not restore virulence, indicating that hyl alone nor in combination with its downstream gene are responsible for peritonitis virulence.Citation112 Therefore, the precise genes encoded on pHylEfmTX16 that mediate virulence in murine peritonitis remain unknown.
Bloodstream and systemic infection
Bloodstream infections are the 10th leading cause of death in the US. Incidence of bloodstream infection ranges from 1% in ICU patients to 36% in bone-marrow transplant patients.Citation113 Bloodstream infections caused by Gram-positive bacteria are highly prevalent and 45% are Enterococci-associated.Citation114,115 Bloodstream and systemic infections are primarily modeled using mice due to the ease of handling small animals. Mice are infected by administering the bacterial inoculum via IV injection into the tail vein; and at the time of sacrifice, bacteremia is determined by bacterial burden in the blood, whereas systemic infection or bacterial translocation is assessed by CFU in the liver, kidneys and spleen.
Enterococcal survival and in vivo colonization rely on stress response mechanisms such as E. faecalis sigma factor SigV that regulates gene expression in response to stress conditions such as heat, acid, ethanol and lysozyme resistance.Citation116,117 In vitro, E. faecalis O-acetyl transferase (oatA) and D-alanylation of LTA (dltA) both had additive effects to lysozyme resistance, while the absence of sigV, oatA and dltA results in greater lysozyme susceptibility. Absence of sigV during systemic infection in mice resulted in attenuation of bacterial translocation, reducing colonization of kidneys and livers, while ΔdltA and Δoat did not.Citation116 A slight additive effect in attenuation was observed in Δoat-dltA-sigV. Oxidative stress mechanisms mediated by msrA and msrB mentioned above for peritonitis have also been assessed in the context of systemic infections.Citation102 In the systemic infection model, E. faecalis attenuation in kidneys and liver were observed for single and double mutants with slightly more pronounced attenuation in livers for the double mutant. The greater attenuation in livers may be associated with higher oxidative stress in that infected organ environment, resulting in lower bacterial burden in the absence of oxidative stress response pathways. In addition, extracytoplasmic foldases EF0685 and EF1534 of E. faecalis are peptidylprolyl cis/trans isomerases (PPIases) predicted to participate in folding extracellular proteins and the absence of either gene results in sensitivity to high salts, resistance to quinolones, ampicillin susceptibility, resistance to oxidative stress, and attenuated persistence in kidneys of mice displaying bacteremia.Citation118 Moreover, adhesion to Caco-2/TC7 is not altered, although cytotoxicity is reduced only in the Δef0685 mutant.Citation118 Absence of ef0685 or ef1534 reduced kidney, but not liver, colonization in the mouse model of bacteremia.Citation118 Lastly, nutrient adaption is crucial for E. faecalis survival in the host environment. Muller and colleagues showed induction of metabolism-related genes during peritonitis. Genes associated with glycerol metabolism were examined in the systemic mouse infection model to assess organ colonization.Citation87 The E. faecalis ΔgldAΔglpK double mutant, but not single gene mutants, were attenuated for kidney and liver colonization in mice at 7 dpi, suggesting that E. faecalis requires the activation of glycerol metabolism during bacteremia.Citation87
QS plays an important role in detecting signaling molecules and contributes to density-dependent virulence traits of pathogenic bacteria during adaptation to the environment. Signaling molecule autoinducer-2 (AI-2) disperses E. faecalis V583ΔABC (a derivative of V583 cured of plasmids A, B and C) biofilm and upregulates prophage 5-related genes in E. faecalis.Citation119 A probiotic E. faecalis strain exposed to culture supernatants from E. faecalis V583ΔABC generated virulent transduced strains, inducing TNF-α release in macrophages, increased Caco-2 cell adherence, and virulence in a mouse bacteremia model and rat endocarditis model.Citation119 This finding suggests that virulence genes can be shared through phage-mediated transfer, and that AI-2-mediated dispersal may generate a population with increased virulence. Ciprofloxacin-mediated phage release is also relevant in the GI environment which may be relevant after antibiotic treatment. Antibiotic-mediated phage transduction generates virulent Enterococci from commensal strains and could explain commensal Enterococci transition into opportunistic pathogens within the GI tract. Other virulence factors that demonstrate a role in more than one in vivo infection model include BgsA and BgsB which mediate kidney colonization of the mouse model of UTI, colonization of endocarditic lesions during IE in rats, and virulence in the mouse model of bacteremia.Citation58,110,120,121 Mutants deficient in either gene are impaired in colonization and more readily cleared from the bloodstream.Citation120,121 Both gene products are involved in LTA biosynthesis by synthesising major cell membrane glycolipids; ΔbgsA is deficient in DGlcDAG while ΔbgsB is deficient of both monoglucosyldiacylglycerol (MGlcDAG) and DGlcDAG.Citation120,121 BgsA and BgsB do not compensate each other, and MGlcDAG alone is insufficient for complementing ΔbgsA virulence defects.Citation120,121 BgsA and BgsB are potential drug targets to induce cell membrane instability for treatment of Enterococcal infections. The E. faecalis transcriptional regulator SlyA suppresses virulence in a mouse model of bacteremia and survival within peritoneal macrophages.Citation122 SlyA regulates more than 100 genes, and absence of this protein could influence virulence expression. Increased virulence of ΔslyA indicates a role in mediating transition between commensal to pathogenic state. Furthermore, post-transcriptional processing by the E. faecalis RNA-binding protein CspR (cold shock protein RNA binding protein) mediates persistence within murine peritoneal macrophages and kidneys of systemically infected mice.Citation123
It was previously shown that S. aureus can express lactate dehydrogenase (ldh-1) to withstand nitrosative stress, establishing a relationship between lactate metabolism and oxidative stress, which confers resistance of S. aureus to host innate immune defenses. Rana and colleagues found that E. faecalis lactate dehydrogenase gene paralogs (ldh-1 and ldh-2) protect E. faecalis against environmental stresses and also contributes to virulence in the mouse model of bacteremia, exhibiting persistence in liver and kidney colonization.Citation124 Gene redundancy is observed in vivo where inactivation of ldh-1 or ldh-2 does not alter virulence, whereas the double mutant is attenuated.Citation124 The ldh-1 gene, but not ldh-2, was essential in survival against stress conditions tested in vitro.Citation124 However, LDH-1-mediated survival of E. faecalis under hydrogen peroxide stress should be recapitulated in a mouse peritoneal macrophage assay or a mouse model of chronic wounds. As impaired wound healing correlates with an oxidatively stressed microenvironment with enhanced concentrations of reactive oxygen and nitrogen species, this could be appropriate for assessing LDH-1 function.Citation125
Similar to studies with E. faecalis, the pathogenesis of E. faecium has also been examined during mouse systemic infection. The global regulator AsrR (antibiotic and stress response regulator) represses virulence during systemic infection in mice by reducing colonization of kidneys and livers. AsrR is a stress-sensor that is inactive in the presence of hydrogen peroxide. The AsrR regulon is composed of 181 genes involved in pathogenesis, antibiotic and antimicrobial peptide resistance, oxidative stress, biofilm formation, adhesion to epithelial cells, and adaptive responses via AsrR-mediated deregulation of uvr and mutS2 which promotes DNA mutation and increased transfer frequency of conjugative transposon Tn916, respectively.Citation126 AsrR represses virulence traits such as biofilm formation, adhesion to intestinal epithelial cells, and deletion of asrR leads to persistence in tissues at 7 dpi during mouse systemic infection.Citation126 The putative capsular polysaccharide biosynthesis protein (capD) is made up of 336 amino acids and it putatively catalyzes N-linked glycosylation.Citation127 Deletion of capD resulted in decreased bacterial burden in blood and livers. Esp of E. faecium is a key virulence factor for persistence in murine UTI and bacteremia, perhaps due to its ability to mediate immune evasion.Citation128,129 E. faecium Esp also mediates kidney colonization.Citation126,127,129 However, passive immunization using antibodies against E. faecium Esp do not protect mice from bacteremia, but may prove to be effective during other infections where Esp is highly expressed.Citation129
Gastrointestinal infection and colitis
GI microbiota consisting of a consortium of bacterial species including Enterococci, E. coli, Streptococci, Lactobacilli, Bifidobacteria, Bacteroides, Eubacterium, and Clostridium plays an important role in disease prevention by mediating colonization resistance (CR), and dysbiosis of this consortium is associated with GI conditions ranging from cancer, obesity, malnutrition, diabetes and inflammatory bowel disease (IBD).Citation130 Modifications in dietary habit, health status or intake of medication influences the intestinal microbial composition.Citation130 IBD can result from (i) an aberrant mucosal immune response to gut commensals, (ii) dysbiosis alone, (iii) disruption of intestinal barrier function, or a combination of (ii) and (iii).Citation131 In these situations, commensal Enterococci may transition into opportunistic pathogens to cause chronic IBD after acute E. faecalis infection, as reported in germ-free and IL-10 knockout mice but not in humans.Citation132 Causes of other GI-related Enterococcal infections include bacterial translocation into organs and systemic infection from complications following colorectal surgery,Citation133,134 and development of colorectal cancer from high concentrations of extracellular superoxide emitted by E. faecalis in the colon.Citation135,136
Mice are the most frequently used animal model to assess Enterococcal virulence in the GI tract, and colonization is initiated by administering Enterococci via oral gavage or in the drinking water.Citation8,22,137-144 In contrast, rats have been used for studying post-surgical complication-induced Enterococcus infections, as their larger size is more amenable for surgical manipulation in these studies.Citation133,134 Infection model systems include immunocompromised mice (e.g., germ-free, gnotobiotic or IL-10 knockout), as well as the administration of antibiotics (e.g., streptomycin, ceftriaxone, clindamycin) to conventional mice before infection.Citation140,142 The utility of germ-free mice in mimicking any natural condition in humans is debatable. In mice, streptomycin perturbs many indigenous microbiota including E. coli, Enterococci, Streptococci, Lactobacilli and Bifidobacteria while Bacteroides spp, Eubacterium, and Clostridium are not affected.Citation141 Ceftriaxone antagonizes E. coli, Lactobacilli, Bifidobacteria, Clostridia and Bacteroides spp, while clindamycin decreases Streptococci and anaerobic bacteria in humans; neither ceftriaxone nor clindamycin inhibit Enterococci.Citation145,146
Bioluminescence imaging (BLI) is a powerful tool for monitoring disease spread in intact animals during infection. Colonization by E. faecalis variants expressing the luxABCDE cassette under the control of either the cytolysin or gelatinase promoter (detected using BLI) in a streptomycin pre-treated GI mouse model suggested greater expression in the lower GI tract (i.e., cecum and colon) than the upper GI tract measured by BL intensity.Citation141 Cytolysin promoter activation, but not gelatinase promoter activation, was detected at high levels during GI infection, suggesting that expression of the former may be important for successful intestinal colonization.Citation141 Drawbacks of using BLI in this setting include detection limit (105 CFU/fecal pellet), lower BL signal in intact animals compared with dissected single organs, low signal emission from tissues/cavities with limited oxygen concentrations, and plasmid stability. A more complete understanding of these virulence factors in GI colonization will require more sensitive methods of expression analyses.
Vancomycin-resistant Enterococci (VRE) contributes to a third of multidrug-resistant Enterococcal systemic infections in clinical settings.Citation22,147 In healthcare settings, patients pre-colonized with VRE can experience VRE overgrowth following antibiotic-induced disruption of the gut microbiota, giving rise to translocation of VRE from areas of high bacterial load in the gut to the bloodstream.Citation22,137 To prevent translocation of VRE to the blood, much research have focused on understanding the effects of antibiotic therapy on gut microbiota. In healthy individuals, resident bacteria of the GI tract stimulates the secretion of RegIIIγ, a C-type lectin secreted by intestinal epithelial and Paneth cells that is important for eliminating Gram-positive bacteria, via the TLR–MyD88 signaling pathway.Citation148,149 Antibiotic treatment results in RegIIIγ downregulation.Citation143 Antibiotic disruption of gut microbiota and mucosal innate immune response via RegIIIγ alteration allows VRE (E. faecium) proliferation by compromising CR.Citation143 Administrating resiquimod (R848), a synthetic ligand for TLR7-mediated induction of IL-22 and IL-23, to reactivate RegIIIγ secretion or introducing obligate anaerobic commensal bacteria containing Barnesiella species can re-establish CR against E. faecium (the presence of Barnesiella correlates with prevention of E. faecium gut colonization and bacteremia).Citation22,144 To gain a competitive edge for Enterococci in the intestinal niche, both E. faecalis and E. faecium are particularly receptive to taking up transferable elements, resulting in evolved strains gaining bacteriocin, pheromone, antibiotic resistance, and other virulence traits.Citation8,137,147 Commensal E. faecalis that harbor the pPD1 plasmid can express bacteriocin 21 can displace other E. faecalis colonizing the gut.Citation137 Gilmore and colleagues found that commensal E. faecalis of the intestinal environment secrete a heptapeptide pheromone that can cause lethal crosstalk between inherited mobile elements within E. faecalis V583, mediating CR by preventing incompatible Enterococci from inhabiting the GI tract.Citation147 These studies suggest multiple therapeutic strategies to combat intestinal colonization of resistant Enterococci, from introduction of probiotic E. faecalis harboring non-conjugative plasmids encoding bacteriocins, transferable colonizing ability among E. faecium recipient strains, pheromone-induced killing of multidrug-resistant E. faecalis strains, administrating resiquimod to reactivate RegIIIγ secretion, and introducing commensals belonging to the Barnesiella genus to make use of the host microbiota or host immune defenses to limit Enterococcal proliferation.Citation8,137,143,144,147
Antibiotic-mediated depletion of gut microbiota is often used in intestinal colonization studies to eliminate susceptible bacteria, in turn promoting colonization and outgrowth of resistant organisms. A mannose family PTS highly prevalent in clinical isolates of E. faecium aids in intestinal colonization in ceftriaxone-treated specific-pathogen-free (SPF) mice.Citation142 ptsD is predicted to encode the enzyme IID subunit of the PTS, and the absence of ptsD significantly impairs E. faecium colonization of the murine intestinal tract. In vitro growth of E. faecium ΔptsD was not affected when tested under a range of different conditions (65 different carbohydrate substrates, sensitivity to cefoxitin and ceftriaxone, or in BHI or BHI-supplemented with cefoxitin).Citation142 However, competitive infection in the mouse model of intestinal colonization revealed fewer mutant bacteria load in feces, small intestines, cecum, and colon of mice at 10 dpi compared with WT.Citation142 The carbohydrate substrate for PtsD in the GI tract was not identified; however, identification of this carbohydrate may inform future therapeutic strategies to manage infections associated with drug-resistant E. faecium. The E. faecalis genome also encodes a one-component signaling system encoded by the prkC gene that maintains cell integrity/morphology; resistance to cell wall-targeting antibiotics, sodium dodecyl sulfate, and bile-related components in vitro; as well as contributing to cecum persistence after intestinal colonization of conventional mice.Citation138 The selective advantage for E. faecalis survival and persistence in the intestine is likely PrkC-mediated bile resistance.Citation138 Using an intestinal model of clindamycin-treated mice that promotes Enterococci overgrowth, E. faecalis glycosyltransferase (epaX) encoded within the epa locus resulted in attenuated intestinal colonization.Citation140 Loss of EpaX function impairs sugar incorporation into cell wall polysaccharides, resulting in alteration of cell wall architecture and sensitivity to sodium deoxycholate (bile salts).Citation140 In vivo functionality of EpaX is crucial for intestinal colonization,Citation140 and the epa locus (epaA-epaR) also mediates virulence in a murine peritonitis model.Citation109 Colitis is an outcome of IBD that causes serious medical implications in immunocompromised patients. Recapitulating this human infection, E. faecalis-associated colitis in IL-10−/− mice showed that GelE partially compromises the intestinal barrier through E-cadherin degradation.Citation132 The absence of the virulence factors glucosyltransferase (epaB) and prolipoprotein diacylglyceryl transferase (lgt) did not impact on GelE activity, implying that EpaB and Lgt do not cause colitis via E-cadherin cleavage in the IL-10−/− mouse model.Citation132 Although they did not differ in colonization in the IL-10−/− mouse model of colitis, epaB and lgt mutants are defective for causing colitis, with a stronger attenuation seen in the absence of lgt. The absence of lgt is independent of colonic mucus penetration; however, innate immune cell activation is dependent on the presence of cell surface-associated lipoproteins. In contrast, absence of epaB leads to impaired penetration of the intestinal mucus layer and EpaB is less immunogenic than Lgt, mediating partial intestinal inflammation in the host.Citation132 While E. faecalis and E. faecium Esp do not appear to be essential for intestinal colonization in mice,Citation139,150 EbrB is important since E. faecium ΔebrB displays attenuated persistence in feces, small intestines, cecum of infected mice, as well as reduced biofilms in vitro.Citation79 RT-qPCR of in vitro grown E. faecium and its isogenic ebrB mutant demonstrates EbrB-mediated regulation of 3 genes downstream of esp, suggesting EbrB-mediated esp expression may indirectly involve their regulation.Citation79
Bacterial translocation due to post-surgical complication from surgical intervention often results in bacteremia, sepsis and in severe cases, death.Citation134 Colorectal anastomotic leak is the most significant post-surgical complication of intestinal anastomosis (reconnection of intestines following removal of an intestinal segment) where intestinal content leakage leads to mortality and morbidity. High collagenase-producing E. faecalis strains were associated with anastomotic leak via GelE- and SprE-mediated intestinal collagen-depletion followed by activation of tissue matrix metalloproteinase 9 (MMP9) that cleaves host ECM.Citation133 These models mimic complications arising from GI disorders (such as cirrhosis, hepatic ischemia and intestinal stasis or IBD) causing gut-associated bacteria to translocate into systemic organs and tissues.
Iron-rich diets have been correlated with increased incidence of colorectal cancer due to exposure to high iron leading to accelerated catalysis of superoxide to hydroxyl radical, inducing oxidative stress on the colon epithelium.Citation151 Accelerated oxidative stress from high concentrations of hydrogen peroxide and reactive oxygen species can arise via the generation of extracellular superoxide by intestinal bacteria.Citation135,136 Most intestinal bacteria produce small amounts of intracellular superoxide but Enterococci produce superoxide extracellularly.Citation152 Menaquinones are involved in superoxide production and E. faecalis appear unique among Enterococci and Streptococci in their ability to produce both menaquinone and extracellular superoxide.Citation153 Demethylmenaquinone (DMK) synthesis and superoxide production are dependent on the function of the menBEDF operon whereby inactivation of the menB gene in E. faecalis reduced hydroxyl radical levels in the rat intestinal colonization model.Citation136 High oxidative stress on the colonic epithelium is related to genomic instability of intestinal tumor cells, as more than 80% of sporadic colon cancers were due to genetic mutations.Citation135 Further studies are required to establish the association between oxidative stress within the intestinal tract milieu with colorectal cancer.
Urinary tract infection and catheter-associated urinary tract infection
Women are at a higher risk of UTI for several reasons, including an anatomically shorter urethra and proximity of the urethral opening to the microbial communities of the GI and vaginal tracts. In addition, age and sexual activities can increase a woman's susceptibility to UTI.Citation154,155 UTI is one of the most common bacterial infection and affects individuals from all age groups, accounting for approximately 8.1 million doctor visits per year.Citation156 According to the Centers for Disease Control and Prevention, an estimated 93,300 UTIs were reported in 2011 and the majority of these cases were associated with instrumentation of the UT.Citation157 A variety of uropathogens can cause both UTI and CAUTI. Uropathogens that cause UTI include UPEC (75%) Klebsiella oxytoca (6%), Staphylococcus saprophyticus (6%), E. faecalis (5%), group B Streptococcus (GBS) (3%), Proteus mirabilis (2%), Pseudomonas aeruginosa (1%), S. aureus (1%) and Candida spp (1%). Organisms associated with CAUTI include UPEC (65%), Enterococcus spp (11%), K. oxytoca (8%), Candida spp (7%), S. aureus (3%), P. mirabilis (2%), P. aeruginosa (2%) and GBS (2%).Citation154
Urinary tract infection
To investigate Enterococcal uropathogenicity, variations of a mouse model of ascending UTI for studying Gram-negative UTI has been used. Typically, female mice are inoculated transurethrally with 105−108 bacteria in a 50–200 μl volume, the higher volumes giving rise to vesicoureteral reflux (VUR) of bacteria into the kidney.Citation70,71,73,74,102,128,158-164 Among vertebrate models, mice have been favored for studying UTI as they are the smallest animal to possess similar anatomy and immunological responses to humans for an in vivo recapitulation of the infection.Citation4
Since 2000, several virulence factors that mediate UTI by E. faecalis have been reported. Individual deletion mutants of the following genes result in attenuation compared with isogenic parental strains: esp,Citation161 srtC,Citation164 ebpA and ebpC,Citation74,163 ace,Citation71,160 epaB,Citation158 msrA and msrB,Citation102 sigV,Citation116 efbA,Citation73 and grvR/etaR.Citation70 Unlike the srtC mutant, deletion of srtA did not indicate a strong role in colonization of the UT. Moreover, a double mutant of both srtA and srtC showed no further reduction of colonization when compared with a single srtC deletion mutant suggesting that polymerized pili are essential for UTI, whether they remain retained on the membrane in a srtA mutant or become wall-associated in WT strains.Citation164,165
In contrast, the expression of several factors appears to limit UTI colonization, including the dltA and oatA, such that their deletion promotes colonization of the UT.Citation116,166 DltA was found to suppress UT colonization in one study, while no contribution was found in another. Reasons for these disparate observations may include differing expression of dltA in different E. faecalis strains used as well as protocol variations.Citation116,166 The actual role of D-alanylated LTA within the bladder or with urothelial cell remains to be determined. In addition, E. faecalis deficient in either bgsA or bgsB also display enhanced colonization of mouse kidneys.Citation110
A noteworthy aspect of this model is that E. faecalis has greater tropism for the kidneys, making it useful to study factors implicated in Enterococcal-mediated pyelonephritis.Citation4,73,110,162 Inoculum volumes of 100–200 μl are used to induce VUR; both Kau et al. and Singh et al. reported inconsistent recovery of E. faecalis from infected organs of mice administered with a 50 μl inoculum.Citation162,163 Therefore, studies of virulence factors where no contribution to upper or lower UT colonization was found, such as for E. faecalis AS and E. faecalis Enterococcal binding substance (Ebs) should be revisited by increasing inoculum volumes.Citation167
A variation of this model, designed to recapitulate UTI pathogenesis in Type I diabetics, was described by Rosen and colleagues in which pancreatic islet cells are depleted by the β-cell toxin streptozocin in C57BL/6 mice. Streptozocin-treated diabetic mice are more susceptible to infection by E. faecalis which is consistent with higher prevalence of Enterococcal-associated UTI in diabetic women compared with nondiabetics.Citation168
Although E. faecium is commonly isolated from nosocomial UTIs, very few UTI studies have been performed in comparison to E. faecalis.Citation76,159 Like E. faecalis UTI, E. faecium surface proteins Esp and EmpABC pili (previously EbpABCfm) mediate colonization of the mouse UT,Citation76,128,159 and E. faecium also display kidney tropism.Citation159
Catheter-associated urinary tract infection
Enterococcal species are among the top 3 most common causes of HAI,Citation169 and CAUTI accounts for more than 40% of all nosocomial infections arising from acute-care hospitals and extended-care facilities.Citation169,170 Among CAUTI, Enterococcal species are the second most common bacterial species detected.Citation6 CAUTI in mice is achieved by first transurethrally placing a 0.5 cm piece of silicone tubing into the bladder, before subsequent transurethral inoculation of bacteria in a 50 μl volume harboring 104−107 CFU.Citation171 In this model, the catheter serves as a substrate for biofilm formation by uropathogens and acts as a reservoir for continual re-seeding of the infection.
Sterile catheterization of the bladder elicits a marked increase in mammalian gene expression associated with defense responses and cellular migration, production of cytokines and chemoattractants that induce immune cell maturation, and recruitment of neutrophils and monocytes into the bladder.Citation172 Infection at the same time as catheterization, before catheter-dependent sterile inflammation and immune infiltration, occurs at an E. faecalis ID50 of 104 CFU.Citation172 In contrast, infection after catheterization and in the presence of catheter-mediated inflammation requires a 3 log-fold higher ID to establish UTI.Citation172,173 Furthermore, major histological changes including edema in the mouse bladder and secretion of host fibrinogen occur in response to a urinary catheter.Citation173,174 Fibrinogen released during the inflammatory response accumulates on the catheter, mediating adhesion of E. faecalis via EbpA.Citation174 Microscopic examination of urinary catheters from hospitalized human subjects with indwelling catheters ranging from 1 h to 59 d demonstrated that fibrinogen accumulation on urinary catheters is associated with dwell times, independent of catheter material.Citation175 Moreover, E. faecalis utilizes fibrinogen as a carbon source to replicate and form catheter-associated biofilms in vitro in female human urine.Citation174 E. faecalis biofilms formed on the catheter surface contribute to persistence in the bladder because they are recalcitrant to immune clearance, shear force from urination, and antibiotic treatment.Citation154,171,173,176 E. faecalis CAUTI is preceded by adhesion to the catheter via ebpA,Citation174,176 ebpB,Citation176 ahrC,Citation38 eep,Citation38 srtC,Citation176 and srtA.Citation171 Importantly, an EbpA-based vaccine can limit or prevent E. faecalis-associated CAUTI by inhibiting the interaction between EbpA and fibrinogen.Citation174 Therefore, this model has proved to be a valuable tool for the identification of virulence determinants and subsequent development of targeted therapeutics against Enterococcal infections.
Despite the many advantages of using mice to study the role of E. faecalis virulence factors in CAUTI, one limitation of this experimental model does not recapitulate typical human urinary catheters, which extend from the bladder to the urethral opening. However, this can be achieved in rats by inserting a 24 gauge polyurethane IV catheter (0.8–1 cm) into the urethra whereby one end of the catheter tip is located inside of the bladder and the distal end located outside of the urethral meatus, with the catheter secured in place by suturing it to the vaginal opening and hiding the knot inside the vagina.Citation177 Although this model has not been used to study the role of E. faecalis virulence, it would be clinically relevant for studies of bacterial ascension during CAUTI.
Endophthalmitis
Endophthalmitis is a vision-threatening nosocomial infection emerging from postoperative procedures in hospitals. Enterococci are only rarely associated with endophthalmitis, accounting for only 1.23% of acute endophthalmitis cases.Citation178 Using Enterococci as the infectious agent in endophthalmitis models is deemed useful in elucidating Enterococcal virulence factors contributing to endophthalmitis, with added advantages of assessing retinal function without sacrificing animals and correlating outcomes with disease progression.Citation4 Rabbits and rats have been used for modeling Gram-positive bacterial endophthalmitis but rabbits are preferred due to technical reasons.Citation4,129 In the rabbit model, New Zealand White rabbits receive general anesthesia and their eyes are dilated to expose the injection site. Subsequently, the ocular surface and its surrounding tissues are disinfected before aspirating fluid from the anterior chamber below the cornea to relieve intraocular pressure. E. faecalis are introduced via injection into the mid vitreous cavity of the eye 3 mm behind the limbus, avoiding damage to the lens.Citation54,55,178
In 2 separate studies, individual mutants of fsrB, gelE, or sprE, and a double mutant of gelE and sprE were compared with WT during endophthalmitis in rabbits.Citation54,55 Virulence was evaluated by loss of retinal function and the study showed that ΔfsrB was the most attenuated followed by ΔgelEΔsprE. Single mutant strains ΔgelE or ΔsprE were as virulent as their WT counterparts, suggesting redundancy of these genes in the pathogenesis of endophthalmitis.Citation54,55 Severity of infection caused by the fsrB mutant was the lowest with retinal layers still intact and an absence of immune infiltration, whereas WT infected eyes suffered the most severe outcomes with compromised structural integrity and cellular infiltrate.Citation54,55 Surprisingly, intraocular bacterial burdens for all deletion mutants were similar to WT levels suggesting that these genes do not impact infectivity or fitness in this model but that disease severity is dependent on specific virulence traits.Citation54,55 The contribution of FsrB to rabbit endophthalmitis is consistent with observations on the role of Fsr in other mammalian models, suggesting it is important for E. faecalis virulence in both local and systemic infections.Citation51,53,56
Foreign body subdermal abscess
To mimic abscess formation in the presence of a foreign material (e.g., a splinter), the first in vivo application of a RIVET system was performed by Frank and colleagues using a subdermal abscess model in rabbits to investigate E. faecalis adaptation to the host environment during infection.Citation47,179 Subdermal chambers were created by aseptic implantation of a sterilized hollow, perforated polyethylene golf ball into a surgically created subcutaneous pocket on the flanks of anesthetized rabbits. During 6 weeks of post-surgical recovery, the perforated golf ball surface becomes encapsulated with fibrous tissue and filled with serous fluid containing immune cells.Citation47 To perform the infection, a small volume of serous fluid from the subdermal chamber is replaced with the same volume of bacteria. During the course of infection, E. faecalis co-exists with a stable immune cell population encompassing approximately 10% granulocytes (or PMNs) and 90% mononuclear cells (lymphocytes and monocytes), while being subjected to a changing host response from additional immune cells that enter the chamber during the course of infection.Citation47 This in vivo model is highly amenable to time course experiments since bacterial gene expression studies, metagenomic analyses during polymicrobial infections, and changes in host immune responses can be followed in the same animal.
Using this model, 249 putative promoters were identified that were induced during subdermal abscess infection by E. faecalis including promoters for genes encoding transport and binding proteins, energy metabolism, cell envelope, DNA metabolism, regulatory functions, protein fate, hypothetical proteins, and others. More than one-third of these promoters are predicted to generate antisense transcripts and this was the first report of in vivo-expressed antisense RNA in E. faecalis.Citation47 Overlapping putative virulence genes from the in vivo RIVET screen with genes associated with in vitro biofilm formation were subsequently validated in a rabbit IE model and showed that both the eep and proB mutants were both attenuated.Citation47 This work led Frank and colleagues to assess differential gene expression using microarray, whereby cellular adaptation was induced at 2 hpi while cellular pathways required for growth and replication were downregulated at 8 hpi.Citation179 Downregulated genes were consistent with bacteria entering a stringent response state, similar to stress conditions such as nutrient deprivation and antibiotic treatment.Citation179 Upon activation of the stringent response, accumulation of the metabolite (p)ppGpp modulates transcription of stress-related genes, some of those identified at 8 hpi were consistent with genes reported from the transcriptome of E. coli response to amino acid starvation and E. faecalis exposure to mupirocin.Citation179 Subdermal chambers infected with deletion mutants in the bifunctional synthetase/hydrolase RSH (or RelA) and small synthetase RelQ, involved in stress responses and maintenance of (p)ppGpp levels, were evaluated for survival in subdermal chambers up to 96 hpi and both of ΔrelQ and ΔrshΔrelQ were attenuated compared with WT. These findings suggest that persistence in vivo is dependent on intracellular (p)ppGpp concentrations rather than activation of stringent response.Citation179
Surgical site infection
SSI occurs in 2–5% of patients undergoing inpatient surgery, arising either at the incision site or in deeper tissue or surrounding organs at the incision site.Citation22 SSIs are common in immunocompromised patients and are associated with up to 7–11 additional postoperative hospital-days, which carries a large economic burden.Citation180 Patients with SSIs are at 2–11 times higher risk of death compared with surgical patients without SSIs.Citation22 Enterococci were the 2nd most commonly isolated organism reported for SSIs in the US between 2006–2007.Citation6,181 A mouse cutaneous infection that models SSI and evaluates phagocyte-mediated bacterial clearance from the infection site was used to examine the role of E. faecalis capsular polysaccharides in SSIs.Citation182 cpsI, encodes capsular polysaccharide I, is one of the genes in the cps locus necessary to generate the carbohydrate for capsule polysaccharide.Citation182 At 4 dpi, persistence within draining abdominal lymph nodes of subcutaneously-injected mice are dependent on the cpsI capsule of E. faecalis.Citation182
Endodontic infection
Although an uncommon dental pathogen, E. faecalis has been isolated from persistent root canal and periapical infections.Citation183 The link between E. faecalis and periodontal disease has been investigated using change-mediated antigen technology (CMAT) and a canine experimental animal model of endodontic infection to identify proteins upregulated in E. faecalis during endodontic infection. CMAT involves generation of hyperimmune antisera against E. faecalis harvested from infected clinical samples of pulp and endodontic infection from humans to identify in vivo-induced proteins or antigens during the course of infection. One of the limitations of this approach for human studies is the low number of E. faecalis cells that can be recovered. Hence, a beagle dog was used in a pilot study to screen for proteins expressed by E. faecalis specifically under in vivo conditions. In an anesthetized canine, perforations in the dental pulp tissue are made and E. faecalis is injected into the pulp cavity to induce endodontic infection with radiography monitoring. Infected premolars are extracted and hemisectioned, infected pulp and periapical tissues removed, and bacterial cell pellets are saved for hyperimmune antisera production in rabbits.Citation184 This study identified 16 in vivo-induced protein antigens involved in housekeeping functions, metabolism, and cellular processes. Potential virulence factors include a copper resistance protein that could confer resistance to metallic copper-coated antimicrobial surfaces and several putative membrane proteins of unknown function.Citation184 Given the lack of sensitivity in this pilot screen due to an underrepresentation of E. faecalis proteins thought to be induced in vivo, a more sensitive approach such as RNA-sequencing may better identify Enterococcal-related endodontic virulence factors in the future.
Zebrafish
While vertebrate models of Enterococcal infection have been informative with regards to pathogenic strategies, they are limited by the fact that they are not practical to large-scale studies and are costly. Over the past decade, Zebrafish (Danio rerio) have gained popularity as an alternative vertebrate model to study human bacterial pathogens due to their affordability and amenability to scaling up. In addition, due to their physical transparency, in vivo imaging of complex host-pathogen interactions, including developmental and dissemination studies can be easily performed in vivo.Citation185 Zebrafish are infected by inoculating bacteria into the bloodstream of embryos at varying hours post fertilization (hpf). The study of E. faecalis in zebrafish was first performed by Prajsnar and colleagues, where they found that 2 E. faecalis strains, OG1RF and V583, were virulent in zebrafish in a dose-dependent manner.Citation186 Mutants lacking virulence factors, epaB, or gelE, sprE and fsrB (Fsr-regulon) displayed attenuated virulence in the zebrafish larvae.Citation186 In another study, zebrafish were used in parallel with mice in a colitis model. They found that colonic heme oxygenase-1 (HO-1) protects mice from colitis by degrading heme and produces carbon monoxide as a by-product. The production of HO-1 enzyme can be induced by enteric microbiota to enhance bactericidal activity of macrophages against E. coli, E. faecalis, and S. typhimurium.Citation187 Despite the numerous advantages that the zebrafish model can provide, due to high degree of evolutionary divergence, translating the host response to mammalian infection can be difficult. In addition, successive inbreeding often leads to infertility in the lower vertebrates, making it difficult to create truly inbred zebrafish lines.Citation188
Polymicrobial infections
The prevalence of polymicrobial E. faecalis infections highlights the need for more in vivo studies to recapitulate clinical scenarios. Risk factors for polymicrobial infection include extremes of age, being immunocompromised, having pre-existing health conditions, experiencing prolonged hospital stays (especially in ICUs), or undergoing surgical intervention.Citation9,14,15,189,190 More often than not, the etiology of these infections and the virulence mechanisms involved are largely unknown and although clinical reports may associate polymicrobial infections with poor outcomes, studies by Lagnaf et al. and García-Granja et al. dispute this claim.Citation9,15,19,191 Early Enterococcal polymicrobial infection studies found that co-infecting mice with P. aeruginosa and E. faecalis in the model of ascending UTI aggravates pyelonephritis and persistence of P. aeruginosa in the kidneys despite β-lactam antibiotic treatment during co-infection.Citation192 An experimental polymicrobial wound infection in mice showed impaired wound healing at 8 dpi using a mixed inoculum of E. faecalis, P. aeruginosa, S. aureus, and Finegoldia magna. Polymicrobial biofilms from 4 day old wounds were less susceptible to bleach and gentamicin treatment compared with their monomicrobial counterparts.Citation193 It was also demonstrated recently that E. faecalis metabolic cues augment E. coli virulence and growth in a mouse model of polymicrobial wound infection.Citation27 Proximity to E. faecalis-within biofilms results in the induction of the E. coli siderophore enterobactin, enabling E. coli to overcome iron limitation in vitro. E. faecalis-mediated E. coli biofilm growth augmentation in a mixed species wound infection suggests a role for E. faecalis ornithine and E. coli enterobactin siderophore biosynthesis during polymicrobial wound infections.Citation27
Peritonitis-associated abscess formation is often a result of strong host immune response to implanted foreign bodies, postoperative procedures, or bacterial infection.Citation47,194 Bacterial-induced intra-abdominal abscesses are usually polymicrobial, and Bacteroides fragilis is the most common anaerobe isolated from abscesses associated with intra-abdominal sepsis together with Enterococcus spp, E. coli, S. aureus, Peptostreptococcus spp, Clostridia, and Prevotella sppCitation194 While most peritonitis infections are modeled as monomicrobial infections, here we describe 2 reports of polymicrobial peritonitis infection studies, which are more representative of human infections. B. fragilis capsular polysaccharides strongly invoke immunologic responses to cause abscess formation in rats.Citation195 Abscess formation is mediated by T-cell activation and may be blocked by a CTLA4Ig fusion protein.Citation81 Rats infected with B. fragilis alone, Bacteroides distasonis plus E. faecium, or S. aureus alone showed lower incidence rates of abscess formation when administered with CTLA4Ig, indicating that abscess formation is mediated by the activation of T-cells via the CD28-B7 pathway.Citation81 Together with in vitro data, the pathway by which T-cells are activated during abscess formation was confirmed to be via CD28-B7–2.Citation81 E. faecium expresses surface polysaccharides (zwitterionic bacterial polysaccharides; Zps) while B. distasonis does not; hence, it remains to be tested whether synergistic interactions in polymicrobial infections may be an outcome of abscess caused by E. faecium Zps.Citation81 Recent emergence in glycopeptide antibiotic resistance (i.e., vancomycin) renders vancomycin-resistant E. faecalis infections difficult to treat. Vancomycin resistance is encoded by vanA or vanB genes and their influence on E. faecalis virulence is not known. A polymicrobial inoculum of E. coli, B. fragilis, plus each strain of E. faecalis (susceptible, VanA+ or VanB+) introduced in a rat peritonitis model demonstrated stronger colonization by E. faecalis strains at 24 and 72 hpi.Citation85 Immune cell infiltration and proinflammatory cytokine (IL-6 and TNF-α) levels in peritoneal fluid at 6 hpi were similar in all infections; this immune response has been suggested to be contributed by both E. coli and B. fragilis.Citation85 Nevertheless, increased serum α1-acid glycoprotein concentrations at 72 hpi detected in vancomycin-resistant E. faecalis infections reflects inflammation and immunomodulation by resistant E. faecalis.Citation85
In the hospital setting, both pre-colonization by VRE and surgery are risk factors for VRE colonization and infection in critical care patients.Citation14,189,190 Polymicrobial peritonitis is one of the common infections originating from a VRE colonized GI tract; that may or may not lead to poorer prognosis.Citation189,196-203 Interestingly, mice colonized with VR E. faecium for 14 d in the GI tract followed by intestinal perforation (CLP; cecal ligation and puncture), suffered leakage of intestinal contents into the peritoneal cavity, but were able to accelerate bacterial clearance at 48 hours post-CLP compared with those not pre-colonized by VR E. faecium.Citation204 This was accompanied by diminished peritoneal and plasma inflammatory response (TNF-α, IL-6 and MCP-1), and lower neutrophil-attracting and -activating chemokines (KC, MIP-2 and LIX) at 48 hours post-CLP in VR E. faecium colonized mice.Citation204
Invertebrates
In the last decade, invertebrate models have been increasingly used to study Enterococcal infections and have been developed to complement mammalian host models. C. elegans, the Greater wax moth caterpillar Galleria mellonella, and Drosophila melanogaster provide advantages for pathogenesis studies because they are smaller in size, easy to maintain, economical, ethically expedient, and are generally inexpensive. Furthermore, as they are amenable to large-scale screening, both C. elegans and D. melanogaster have been used for high-throughput screening (HTS) of antimicrobial compounds; or mutant libraries for pathogenic determinants of a variety of microbes, including E. faecalis and E. faecium.
Caenorhabditis elegans
C. elegans is a well-characterized and common invertebrate model organism that is used to study a variety of host-pathogen interactions. C. elegans are the smallest in size (adults are ∼1mm) among the 3 invertebrate models and has a rapid life cycle (∼3.5 d at 20°C). Its transparent body and genetic tractability make the organism well-suited for experimental observation under the microscope and observation of host-pathogen interactions in vivo. The ease of scaling up for biochemical analyses, and for HTS, makes C. elegans a favorable option over G. mellonella and D. melanogaster because they are easiest to maintain in large-scale quantities. As infection is performed ad libitum, C. elegans are an excellent choice to investigate gut microbial-host interaction. However, a limitation of using C. elegans as an invertebrate model is their lack of a cellular and humoral response system. Although C. elegans express Toll pathways homologous to the mammalian system, there is no clear evidence that they play a role in defense response in this organism.Citation205 Hence, most C. elegans infection studies are often paired with a mammalian model organism.Citation53,206,207
Studies describing Enterococcal virulence using the C. elegans infection model are assessed by monitoring the amount of time taken to kill the host. Garsin and colleagues noted that even though both E. faecalis and E. faecium can accumulate to high titers in the intestinal lumen causing gut distention, only E. faecalis is able to kill the nematodes efficiently when they are infected at high titers, while causing persistence when infected with low titers. Despite the inability of E. faecium to kill adult hermaphrodites, both E. faecalis and E. faecium can kill eggs and hatchlings of C. elegans.Citation206 However, Moy and colleagues recently showed that certain strains of E. faecium, when grown anaerobically, are able to kill adult hermaphrodites due to the high level of hydrogen peroxide production.Citation208
Two well-established Enterococcal virulence factors, cytolysin and the QS Fsr system, are important in killing adult C. elegans.Citation52,53,206,207,213 However, together these factors only account for approximately 50% of host killing, indicating that additional bacterial factors must contribute to E. faecalis virulence in C. elegans. A recent study revealed that phage03-like gene clusters are associated with significant nematocidal activity in non-cytolytic and gelatinase-negative E. faecalis. This phage03-like element acts as an efficient vehicle for HGT and is enriched in nosocomial and clinical isolates. Absence of the phage03-like element results in attenuation of E. faecalis virulence in both C. elegans and G. mellonella larvae.Citation209 Other E. faecalis virulence-associated genes validated using C. elegans include paiA,Citation207 epaB,Citation132 lgt,Citation132 scrB (sucrose-6-phosphate hydrolase),Citation206,207 and genes encoding enzymes likely involved in DNA damage control and repair such as recQ and phrB.Citation207
Taking advantage of the genetic tractability of C. elegans, gene orthologs whose functions were known to be involved in metabolism and IBD in humans were selected for knock-down by RNAi and screened for their roles in gut colonization, fat metabolism and maintaining epithelial junction integrity. C. elegans genes identified as having such protective roles include nhr-49, dlg-1, and ajm-1. This study also shed light on the association between fatty acid metabolism, innate immunity, and maintenance of epithelial junction integrity that can affect nematode survival rate.Citation210 In addition to acting as a pathogen to C. elegans, a recent study showed that E. faecalis Symbioflor® can be used as a probiotic to increase worms' lifespan when co-infected with enterohemorrhagic E. coli (EHEC). E. faecalis Symbioflor® was able to downregulate several virulence-associated genes of EHEC when they are co-fed in C. elegans.Citation211 Another study by King and colleagues showed that a mildly pathogenic, resident strain of E. faecalis OG1RF evolved rapidly within the C. elegans host to protect C. elegans against infection by pathogenic S. aureus.Citation212
Galleria mellonella
The G. mellonella larvae infection model has recently been used to investigate Enterococcal infections. Unlike C. elegans, G. mellonella has a more complex immune system, based on both cellular and humoral systems that correlate to mammalian systems.Citation4 Similar to other invertebrate models, G. mellonella can be reared at low cost and are readily available from commercial sources. The main advantage of G. mellonella over C. elegans or D. melanogaster is that infections can be performed at 37°C, thus more effectively simulating human body temperatures, whereas 25°C is the maximum growth temperature for C. elegans and D. melanogaster.Citation213 Among the 3 invertebrate models, G. mellonella is the largest in size and can be directly injected with bacteria or drugs, allowing exact quantification of the experimental inoculum or drug dosage administered.Citation213 Thus, this model organism has been used to assess the safety and efficacy of antimicrobial compounds against several microbial infections including Enterococcus spp, and may have potential in future HTS studies.Citation214
Infection studies are performed with G. mellonella caterpillars at the final-instar stage of development and infected with bacterial culture or saline as a negative control.Citation215 The larvae are kept on petri dishes without food at 37°C, and survival can be monitored over time. Like in C. elegans, presence of E. faecalis GelE and the Fsr system correlates with killing of G. mellonella larvae, whereas E. faecium displayed limited virulence.Citation215 Absence of Ace in E. faecalis also limited the ability of E. faecalis to kill G. mellonella larvae.Citation71 After 24 hpi, fewer than 10% of G. mellonella infected with WT E. faecalis survived, whereas 80% of larvae infected with the Δace mutant strain survived.Citation71 PPIases encoded by ef_0685 and ef_1534 play a role in bacterial fitness and, by extension, in the virulence of E. faecalis. Δef0685 mutant displayed strong attenuated virulence traits while Δef1534 mutant displayed weak attenuated virulence traits in G. mellonella.Citation118 asrR, encoding an antibiotic and stress response regulator, promotes greater persistence of E. faecium in G. mellonella colonization. Other E. faecalis virulence factors that contribute to G. mellonella killing include MsrA and MsrB,Citation102 SlyA,Citation122 EpaB,Citation132 the cytolysin promoter (Pcyl),Citation141 polyamine N-acetyltransferase-like enzyme (PmvE),Citation216 ef_3196/7 2-component system,Citation86 lysozyme-induced peptidoglycan N-acetylglucosamine deacetylase (PgdA),Citation217 phage03-like element,Citation209 ef0377,Citation87 and cspR.Citation123,126 Significant limitations of using G. mellonella include the lack of a fully sequenced genome or molecular tools to genetically manipulate this organism.Citation213,218,219
Drosophila melanogaster
The GI tract of D. melanogaster is naturally colonized by several Enterococcal species, including both E. faecalis and E. faecium, which makes Drosophila an excellent model to study Enterococcal gut infections. In addition to a fully sequenced genome, the availability of Drosophila genetic mutant libraries makes manipulation of the host achievable. Unlike C. elegans, inoculation of bacteria into Drosophila is achieved via pricking the animal with a needle previously dipped into a concentrated culture of bacteria.Citation220
Infection studies using D. melanogaster are routinely performed using anaesthetized male flies followed by injection of the bacteria inoculum with a pulled glass capillary needle using a micro-injector. The first study reporting E. faecalis virulence in D. melanogaster was in 2001.Citation220 Since then, Teixeira and colleagues used D. melanogaster to examine virulence associated with the fsr quorum sensing system. D. melanogaster infected with E. faecalis mutants in the Fsr system and/or in Fsr-regulated genes gelE and sprE all survived longer than flies infected with the wild type parental strain.Citation221 In response to GelE and SprE induction, lrgAB, the 2-component system genes lytRS, and genes encoding proteins involved in cell wall metabolism, transport, and regulatory functions are all induced. Differential expression of any of these Fsr regulon genes may explain attenuated phenotypes of fsr and protease mutants in D. melanogaster.Citation221 Enterococcal cytolysin is also an important virulence factor in D. melanogaster. Additionally, Cox and Gilmore demonstrated the importance of cytolysin by showing lethality after high-level colonization as compared with a noncytolytic control.Citation222
Plants
In addition to vertebrate and invertebrate models, plant models have been used to analyze the virulence of Enterococcal infections. Despite the evolutionary distance between plants and animals, there is evidence that some virulence mechanisms and factors involved in bacterial pathogenesis may be similar between these 2 kingdoms.Citation223 Enterococci have been reported to cause diseases in plants.Citation224 A range of plants have been used as models to study bacterial pathogenesis. Arabidopsis thaliana is commonly used and was first introduced to examine P. aeruginosa virulence in phyto-pathogenesis. A. thaliana infections can be initiated via the root, leaf, or soil. In root pathogenesis assays, bacteria are added into solid medium containing 25 day old A. thaliana plants and incubated at 30°C with a photoperiod of 16 h light and 8 h dark. To assay pathogenicity in the leaf, bacteria are injected directly into the leaf using the blunt end of the hypodermic needle. For soil infiltration, soil is flooded with bacteria culture and disease symptoms, such as leaf lesions, can be observed both macro- and microscopically.Citation224
The first study of E. faecalis infection using A. thaliana was performed on a leaf infection model by Jha and colleagues in which they discovered that absence of the E. faecalis FsrB QS system results in attenuated virulence in A. thaliana.Citation224 A. thaliana infection begins with bacterial attachment to the leaf, followed by entry through the stomata or wounded surface. Attachment, followed by replication takes place shortly after entry into the intercellular space, causing damage to the plant cell walls and membrane, ultimately leading to rotting and eventual cell death. Three different E. faecalis strains (OG1RF, V583 and FA2–2) were capable of infecting the leaves and roots of A. thaliana, causing plant mortality by 7 dpi.Citation224 Strains lacking either fsrB or sprE resulted in an inability to colonize the root.Citation224 Unlike zebrafish and C. elegans, E. faecalis strains lacking gelE are not attenuated in the Arabidopsis model of infection. Although this study is the only report examining E. faecalis virulence using A. thaliana, it shows that E. faecalis can use a similar subset of virulence determinants that are used to elicit disease in animals, invertebrates in plants as well. Therefore, the results of plant infection studies can be relevant and useful to animal pathogenesis. Screening for putative virulence factors of animal pathogenic bacteria can be easily achieved in plant as it is less time-consuming and less tedious. Other plant models, such as rice, maize, eucalyptus, wheat, tomato have not been used to study Enterococcal infections but may be exploited in future for differential infection by E. faecalis that may provide leads to novel antimicrobial drugs. These plant models may also potentially be useful as a surrogate to overcome inherent limitations of other eukaryotic systems.Citation225
Conclusion
In this review we (i) summarized the current vertebrate and invertebrate host systems that are available to model E. faecalis and E. faecium virulence (), (ii) examined conserved and niche-specific virulence mechanisms for E. faecalis and E. faecium in vivo, and (iii) presented evidence that polymicrobial infections involving E. faecalis and E. faecium are prevalent in some niches (e.g., intestinal tracts and peritoneal abscesses) and are understudied due to the historical assumption that most infections are monomicrobial.
Emphasis on vertebrate host models to investigate Enterococcal virulence during mono- and polymicrobial infections has greatly advanced the elucidation of virulence factors relevant to infection and the consequence of virulence factor expression on host immune responses. Going forward, combining these model systems with high throughput ‘omics approaches will increase the speed of discovery of new Enterococcal pathways important for colonization and infection. To date, screening for in vivo gene expression by Enterococci has been most widely applied.Citation47,86,179 RNA sequencing (RNA-seq) and transposon sequencing (Tn-seq) provide unbiased approaches to study large-scale gene expression and fitness profiling, and can be combined to probe bacterial virulence in different infection models.Citation226
Understanding Enterococcal virulence factors and their mechanism of action in the host may help identify future antibacterial or anti-virulence therapeutic targets. The best-described virulence factors and mechanisms, identified using model systems for IE, GI colonization and CAUTI, are depicted in . During IE, vegetation formation is promoted by aggregation and colonization by AS-expressing E. faecalis. Host fibrin enveloping E. faecalis restricts host defenses, while GelE plays a role in digesting fibrin to accelerate dissemination (). In the GI tract, CR and host immune responses are critical in combating Enterococcal infections. Synergistic interaction between commensals is key in preventing pathogenic bacterial overgrowth the intestinal tract (). The intestinal environment is by far the best-described model for studying mechanistic interactions between infecting bacteria (VRE), the gut microbiome, and the host. The intestinal model can be used to examine how antibiotics increase host susceptibility to VRE infection, and how host CR can be re-established to combat VRE infection. In addition, applications on using bacteriocin or heptapeptide pheromone secretion to limit VRE can be studied. This model is also useful for determining the host and bacterial factors E. faecalis rely on to drive intestinal leakage, and whether high-iron levels in the gut mediate oxidative stress in the intestinal milieu as a consequence of E. faecalis superoxide production ().Citation22,137,143,144,147 During CAUTI, catheterization of the bladder results in cellular damage, a robust host response driving immune cell infiltration and host fibrinogen secretion that coats the catheter, ultimately forming the substrate for E. faecalis Ebp pilus adhesion ().Citation171-175
Figure 4. Role of E. faecalis and E. faecium virulence genes in pathogenesis. Cartoon depicting virulence factors involved during (A) systemic dissemination and infective endocarditis, (B) intestinal colonization, and (C) catheter-associated urinary tract infection and community-acquired urinary tract infection
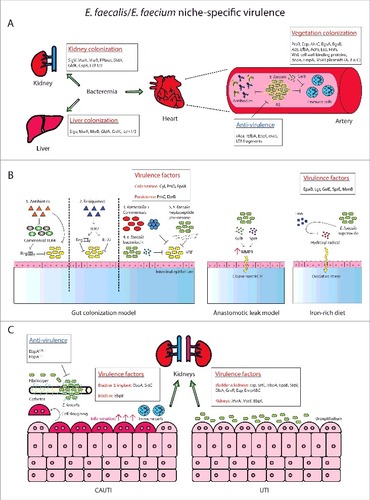
Given the multi-factorial mechanisms of Enterococcal pathogenesis, new treatment strategies should similarly be multi-pronged. Currently, most UTI studies focus on bladder infection, but our understanding of Enterococcal-mediated kidney infection is poorly understood. Future studies should also include identifying commensals of the UT, whether the UT microbiome mediates CR to limit Enterococcal colonization, or whether changes of UT commensal populations after catheterization promote CAUTI. Since oral antibiotics complicate gut infections, alternatives to treat bacterial infections should be steered toward non-antibiotic approaches: triggering host responses or by introducing helpful members of the microbiota. To curb problems with colonic oxidative stress arising from high-iron diets, orally introduced Lactobacillus rhamnosus and L. paracasei may help in antagonizing E. faecium and E. coli already present in the gut, in turn relieving the oxidative stress and mucosal barrier injury observed in mice.Citation227 In the UT, Wullt and Svanborg showed that E. coli 83972 (a prototype asymptomatic bacteriuria/ABU strain) protects patients with complicated recurrent UTI and reduced the frequency of symptomatic UTI by establishing protective ABU.Citation228 Clearly, emerging antibiotic resistance calls for alternative therapies to treat bacterial infections. This is where prior knowledge of virulence factors informs the design of anti-virulence strategies to limit, but not kill pathogenic bacteria, in turn relying on host immune defenses for bacterial clearance. For those that are immunocompromised, anti-virulence approaches combined with antibiotics could be an effective solution. These strategies are designed to reduce selective pressure on virulent bacteria to minimize evolution of multidrug tolerant bacteria, increasing success rates for disease treatment. Several examples of anti-virulence therapy targets include include: recombinant Ace,Citation63 recombinant EfbA,Citation64 EbpA monoclonal antibodies,Citation62 LTA fragments reducing host susceptibility to Enterococcal IE,Citation59 partial protection when immunized with IgG Fab fragments (from rabbit antibodies raised against purified AS) against IE,Citation42 and an EbpA-based vaccine that can limit or prevent E. faecalis-associated CAUTI.Citation174 Enterococcal SrtA that plays a crucial role in anchoring cell wall-associated virulence proteins (such as Ebp) is also an attractive candidate for anti-virulence therapy.Citation229
Enterococci rarely cause disease in immunocompetent individuals. Instead, susceptible hosts include those who are immunocompromised or suffering comorbidities. In animals, predisposed states for infection are created to recapitulate clinical scenarios. For example, vegetation induction via heart valve catheterization (rabbits and rats), intestinal perforation (rats and mice), antibiotic-induced dysbiosis (mice), and CAUTI (mice). On the other hand, germfree, gnotobiotic, immunodeficient mice are readily available to facilitate studies that delve deeper into bacterial infections of immunocompromised hosts, making them valuable models for virulence studies.Citation230-232 Alternatively, when knockout mice are not easily accessible, immunodeficiency can be elicited in mice by administrating cyclophosphamide (neutrophil depletion),Citation233 anti-G-CSF antibodies (G-CSF cytokine neutralization),Citation234 or antibodies specific for C-C chemokine receptor 3 (eosinophil depletion).Citation235 Only by depleting immunological factors individually or in combination will we be able to identify key contributors of Enterococcal immune modulation. To better understand Enterococcal infections in hosts that are having or are susceptible to genetic diseases, we propose investigating mice with genetic disorders such as diabetes, IBD and even those with cancer susceptibilities to address virulence and therapeutic strategies for SSI (and wounds), GI complications (e.g., Crohn's disease), and iron-rich diet-associated colon cancer studies, respectively.Citation236,237
The future for in vivo polymicrobial Enterococcal studies should be expanded for models where mixed species infections are most common such as in chronic wounds in diabetic (db/db) hosts, similar to what has been used to study P. aeruginosa as well as CAUTI.Citation238 E. faecalis, P. aeruginosa and S. aureus are the top 3 organisms found in polymicrobial surgical wounds, chronic foot ulcers, diabetic foot wounds, skin injury or burn wounds,Citation239-242 and these interactions should be modeled in vivo. Uropathogens commonly isolated with E. faecalis during UTI and CAUTI include UPEC, P. aeruginosa and P. mirabilis.Citation15,21 These should be of main concern and the primary species of interest for studies on polymicrobial CAUTI. Other relevant models that should be examined include the use of males and aged hosts, as both groups display increased susceptibility to CAUTI.
E. faecalis is associated with a variety of medical device-related and biofilm-associated hospital infections such as CAUTI, central venous catheter bloodstream infections, VAP and orthopedic implant infections.Citation11 Osteomyelitis is a bacteria-mediated inflammation of the bone that can be acute or chronic. Infection of the bone is usually a post-surgical complication due to implantation of a prosthetic device. E. faecalis is associated with 2.5% of prosthetic joint infections (PJIs)Citation3 but very limited work has been performed to understand these infections. The rat model of Enterococcal PJI is initiated by pre-incubating wire implants with E. faecalis to allow biofilm formation on the surface. Infected wires are then surgically implanted into the proximal tibia of rats and bacterial enumeration performed on the wires and bone at subsequent timepoints. One study has been reported since 2000 but found no role of ahrC or eep in biofilm-mediated acute foreign body osteomyelitis, suggesting that biofilm formation may not be sufficient for the pathogenesis of Enterococcal PJI.Citation3,38,47 The well-established non-implant mouse model of osteomyelitis for Staphylococci may be more physiologically relevant for PJI and could be modified for Enterococcal studies such that a wire implant modeling the prostheses is implanted before bacterial inoculation.Citation243 Larger animals such as rats and rabbits are more amenable to surgical procedures compared with smaller animals and are ideal for modeling device- and biofilm-associated infections, mimicking SSIs. However, the main drawback is the lack of genetically modified rats or rabbits available. Pigs and nonhuman primates better represent human anatomy and physiology to better mimic human disease,Citation244,245 and can be suitable models to study IE and CAUTI, but cost and housing availability are often limiting factors. Larger animals allow large volume of blood sampling, echocardiography, and may be amenable to BLI for long-term studies in the same animal. Examination of experimental catheter coatings to prevent CAUTI are better performed in pigs or primates than in the mouse model, because Foley catheters with closed-system urine drainage bags are available for these species. Silicone catheters used in mice possess a limited surface area for antimicrobial coating and in low concentrations the coating may be ineffective. The advantages and disadvantages of each animal model discussed in this review are compared in .
The utility of non-mammalian models should not be underestimated as these can be important for large-scale anti-infection and therapeutic screens. These models are ideal for simple survival studies for mono- or polymicrobial infection which can be difficult to carry out in mammalian hosts due to ethical considerations. When selecting the appropriate non-mammalian model, incubation temperature should be kept optimal for Enterococcal growth (37°C) as some virulence factors may require a physiologic temperature for gene expression, hence C. elegans and D. melanogaster may not be appropriate for these reasons. Although mammalian models (especially mouse models) are ideally positioned to address knowledge gaps during infections in a variety of infection niches, it is technically demanding to handle large numbers of animals for large-scale studies. Therefore, throughput combination of non-mammalian models for initial and large-scale screening, followed by confirmatory experiments in an animal model, may be ideal.
There remain many unanswered questions regarding mechanisms of Enterococcal colonization and pathogenesis. While there is a growing appreciation of Enterococcal virulence factors, the same factor may not necessary play the same role in different animal models or in different niches within the same animal. Hence, there is no one-model-fits-all that can address all mechanisms of Enterococcal colonization and opportunistic infection. It is also clear that Enterococcal pathogenesis is multifactorial and there are virulence determinants that play redundant roles in certain infection sites. Moreover, some in vitro phenotypes meant as surrogate models for infection do not correlate with virulence in vivo, re-enforcing the need to utilize multiple host-based model systems to elucidate the contribution of specific virulence factors to infection.
Abbreviations
| ABU | = | Asymptomatic bacteriuria |
| AI-2 | = | Autoinducer-2 |
| AS | = | Aggregation substance |
| BHI | = | Brain heart infusion |
| BLI | = | Bioluminescence imaging |
| CAUTI | = | Catheter-associated urinary tract infection |
| CFU | = | Colony-forming unit |
| CLABSI | = | Central-line associated bloodstream infection |
| CLP | = | Cecal ligation and puncture |
| CMAT | = | Change-mediated antigen technology |
| CR | = | Colonization resistance |
| DGlcDAG | = | Diglycosyldiacylglycerol |
| DMK | = | Demethylmenaquinones |
| Dpi | = | Days post infection |
| ECM | = | Extracellular matrix |
| EHEC | = | Enterohemorrhagic E. coli |
| GBS | = | Group B Streptococcus |
| GI | = | Gastrointestinal |
| HAI | = | Hospital-acquired infection |
| HGT | = | Horizontal gene transfer |
| Hpc | = | Hours post catheterization |
| Hpf | = | Hours post fertilization |
| Hpi | = | Hours post infection |
| HTS | = | High-throughput screening |
| IBD | = | Inflammatory bowel disease |
| ICU | = | Intensive-care unit |
| ID | = | Infective dose |
| IE | = | Infective endocarditis |
| IP | = | Intraperitoneal |
| IV | = | Intravenous |
| LD | = | Lethal dose |
| LTA | = | Lipoteichoic acid |
| Mab | = | Monoclonal antibody |
| MGlcDAG | = | Monoglucosyldiacylglycerol |
| MSCRAMM | = | Microbial surface components recognizing adhesive matrix molecules |
| PAMP | = | Pathogen-associated molecular pattern |
| PAI | = | Pathogenicity island |
| PJI | = | Prosthetic joint infection |
| PPIase | = | Peptidylprolyl cis/trans isomerase |
| PTS | = | Phosphotransferase system |
| QS | = | Quorum sensing |
| RIVET | = | Recombinase-based in vivo expression technology |
| RNA-seq | = | RNA sequencing |
| RT-qPCR | = | Reverse-transcription quantitative polymerase chain reaction |
| SPF | = | Specific-pathogen-free |
| SSI | = | Surgical site infection |
| TLR | = | Toll-like receptor |
| Tn-seq | = | Transposon sequencing |
| UPEC | = | uropathogenic E. coli |
| UTI | = | Urinary tract infection |
| VAP | = | Ventilator-associated pneumonia |
| VRE | = | Vancomycin-resistant Enterococci |
| VUR | = | Vesicoureteral reflux |
| WT | = | Wildtype |
| Zps | = | Zwitterionic bacterial polysaccharides |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We gratefully acknowledge Dr. Eliza Soh for critical reading of this manuscript.
Funding
Work in the Kline laboratory related to this article is supported by the National Research Foundation and Ministry of Education Singapore under its Research Center of Excellence Program, the NRF under its Singapore NRF Fellowship program (NRF-NRFF2011-11), and by the Ministry of Education Singapore under its Tier 2 program (MOE2014-T2–1–129 and MOE2014-T2–2–124). H.M.S. Goh is supported by MOE2014-T2–1–129.
References
- Lebreton F, Willems RJL, Gilmore MS. Enterococcus diversity, origins in nature, and gut colonization. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, eds. Enterococci: From commensals to leading causes of drug resistant infection. Boston, 2014
- Weiner LM, Webb AK, Limbago B, Dudeck MA, Patel J, Kallen AJ, Edwards JR, Sievert DM. Antimicrobial-Resistant Pathogens Associated With Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011-2014. Infect Control Hosp Epidemiol 2016; 37(11):1288-301
- Frank KL, Vergidis P, Brinkman CL, Greenwood Quaintance KE, Barnes AM, Mandrekar JN, Schlievert PM, Dunny GM, Patel R. Evaluation of the Enterococcus faecalis biofilm-associated virulence factors AhrC and Eep in rat foreign body osteomyelitis and in vitro biofilm-associated antimicrobial resistance. PLoS One 2015; 10:e0130187; PMID:26076451; https://doi.org/https://doi.org/10.1371/journal.pone.0130187
- Garsin DA, Frank KL, Silanpaa J, Ausubel FM, Hartke A, Shankar N, Murray BE. Pathogenesis and models of Enterococcal infection. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, eds. Enterococci: From commensals to leading causes of drug resistant infection. Boston, 2014
- Jett BD, Huycke MM, Gilmore MS. Virulence of Enterococci. Clinical microbiology reviews 1994; 7:462-78; PMID:7834601; https://doi.org/https://doi.org/10.1128/CMR.7.4.462
- Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect Control Hosp Epidemiol 2008; 29:996-1011; PMID:18947320; https://doi.org/https://doi.org/10.1086/591861
- Arias CA, Panesso D, Singh KV, Rice LB, Murray BE. Cotransfer of antibiotic resistance genes and a hylEfm-containing virulence plasmid in Enterococcus faecium. Antimicrobial Agents Chemotherapy 2009; 53:4240-6; PMID:19667280; https://doi.org/https://doi.org/10.1128/AAC.00242-09
- Rice LB, Lakticova V, Carias LL, Rudin S, Hutton R, Marshall SH. Transferable capacity for gastrointestinal colonization in Enterococcus faecium in a mouse model. J Infect Dis 2009; 199:342-9; PMID:19049434; https://doi.org/https://doi.org/10.1086/595986
- Pammi M, Zhong D, Johnson Y, Revell P, Versalovic J. Polymicrobial bloodstream infections in the neonatal intensive care unit are associated with increased mortality: a case-control study. BioMed Central Infect Dis 2014; 14:390; PMID:25022748; https://doi.org/https://doi.org/10.1186/1471-2334-14-390
- Gold HS. Vancomycin-resistant Enterococci: mechanisms and clinical observations. Clin Infect Dis 2001; 33:210-9; PMID:11418881; https://doi.org/https://doi.org/10.1086/321815
- Higuita NIA, Huycke MM. Enterococcal disease, epidemiology, and implications for treatment. In: Gilmore MS, Clewell DB, Ike Y, eds. Enterococci: From commensals to leading causes of drug resistant infection [Internet]. Boston: Massachusetts Eye and Ear Infirmary, 2014
- Pinholt M, Ostergaard C, Arpi M, Bruun NE, Schonheyder HC, Gradel KO, Sogaard M, Knudsen JD. Incidence, clinical characteristics and 30-day mortality of Enterococcal bacteraemia in Denmark 2006-2009: a population-based cohort study. Clin Microbiol Infect 2014; 20:145-51; PMID:23647880; https://doi.org/https://doi.org/10.1111/1469-0691.12236
- Patterson JE, Sweeney AH, Simms M, Carley N, Mangi R, Sabetta J, Lyons RW. An analysis of 110 serious enterococcal infections. Epidemiology, antibiotic susceptibility, and outcome. Medicine (Baltimore) 1995; 74:191-200
- Maki DG, Agger WA. Enterococcal bacteremia: clinical features, the risk of endocarditis, and management. Medicine (Baltimore) 1988; 67:248-69; PMID:3134590; https://doi.org/https://doi.org/10.1097/00005792-198807000-00005
- Croxall G, Weston V, Joseph S, Manning G, Cheetham P, McNally A. Increased human pathogenic potential of Escherichia coli from polymicrobial urinary tract infections in comparison to isolates from monomicrobial culture samples. J Med Microbiol 2011; 60:102-9; PMID:20947667; https://doi.org/https://doi.org/10.1099/jmm.0.020602-0
- Fourcade C, Canini L, Lavigne JP, Sotto A. A comparison of monomicrobial versus polymicrobial Enterococcus faecalis bacteriuria in a French University Hospital. Eur J Clin Microbiol Infect Dis 2015; 34:1667-73; https://doi.org/https://doi.org/10.1007/s10096-015-2403-0
- Gjodsbol K, Christensen JJ, Karlsmark T, Jorgensen B, Klein BM, Krogfelt KA. Multiple bacterial species reside in chronic wounds: a longitudinal study. Int Wound J 2006; 3:225-31; PMID:16984578; https://doi.org/https://doi.org/10.1111/j.1742-481X.2006.00159.x
- Citron DM, Goldstein EJ, Merriam CV, Lipsky BA, Abramson MA. Bacteriology of moderate-to-severe diabetic foot infections and in vitro activity of antimicrobial agents. J Clin Microbiol 2007; 45:2819-28; PMID:17609322; https://doi.org/https://doi.org/10.1128/JCM.00551-07
- Garcia-Granja PE, Lopez J, Vilacosta I, Ortiz-Bautista C, Sevilla T, Olmos C, Sarria C, Ferrera C, Gomez I, San Roman JA. Polymicrobial Infective Endocarditis: Clinical Features and Prognosis. Medicine (Baltimore) 2015; 94:e2000; PMID:26656328; https://doi.org/https://doi.org/10.1097/MD.0000000000002000
- Alexopoulou A, Vasilieva L, Agiasotelli D, Siranidi K, Pouriki S, Tsiriga A, Toutouza M, Dourakis SP. Extensively drug-resistant bacteria are an independent predictive factor of mortality in 130 patients with spontaneous bacterial peritonitis or spontaneous bacteremia. World J Gastroenterol 2016; 22:4049-56; PMID:27099449; https://doi.org/https://doi.org/10.3748/wjg.v22.i15.4049
- Jacobsen SM, Stickler DJ, Mobley HL, Shirtliff ME. Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis. Clin Microbiol Rev 2008; 21:26-59; PMID:18202436; https://doi.org/https://doi.org/10.1128/CMR.00019-07
- Abt MC, Buffie CG, Susac B, Becattini S, Carter RA, Leiner I, Keith JW, Artis D, Osborne LC, Pamer EG. TLR-7 activation enhances IL-22-mediated colonization resistance against vancomycin-resistant enterococcus. Sci Transl Med 2016; 8:327ra25; PMID:26912904; https://doi.org/https://doi.org/10.1126/scitranslmed.aad6663
- Galvan EM, Mateyca C, Ielpi L. Role of interspecies interactions in dual-species biofilms developed in vitro by uropathogens isolated from polymicrobial urinary catheter-associated bacteriuria. Biofouling 2016; 32:1067-77; PMID:27642801; https://doi.org/https://doi.org/10.1080/08927014.2016.1231300
- Ditu LM, Chifiriuc MC, Bezirtzoglou E, Voltsi C, Bleotu C, Pelinescu D, Mihaescu G, Lazar V. Modulation of virulence and antibiotic susceptibility of enteropathogenic Escherichia coli strains by Enterococcus faecium probiotic strain culture fractions. Anaerobe 2011; 17:448-51; PMID:21723403; https://doi.org/https://doi.org/10.1016/j.anaerobe.2011.05.019
- Hammer ND, Cassat JE, Noto MJ, Lojek LJ, Chadha AD, Schmitz JE, Creech CB, Skaar EP. Inter- and intraspecies metabolite exchange promotes virulence of antibiotic-resistant Staphylococcus aureus. Cell Host Microbe 2014; 16:531-7; https://doi.org/https://doi.org/10.1016/j.chom.2014.09.002
- Kaplan A, Kaplan CW, He X, McHardy I, Shi W, Lux R. Characterization of aid1, a novel gene involved in Fusobacterium nucleatum interspecies interactions. Microbial Ecol 2014; 68:379-87; PMID:24643713; https://doi.org/https://doi.org/10.1007/s00248-014-0400-y
- Keogh D, Tay WH, Ho YY, Dale JL, Chen S, Umashankar S, Williams RBH, Chen SL, Dunny G, Kline KA. Enterococcal metabolite cues facilitate interspecies niche modulation and polymicrobial infection. Cell Host Microbe 2016; 20(4):493-503
- Hughes ER, Winter SE. Enterococcus faecalis: E. coli's siderophore-inducing sidekick. Cell Host Microbe 2016; 20:411-2; https://doi.org/https://doi.org/10.1016/j.chom.2016.09.018
- Zackular JP, Moore JL, Jordan AT, Juttukonda LJ, Noto MJ, Nicholson MR, Crews JD, Semler MW, Zhang Y, Ware LB, et al. Dietary zinc alters the microbiota and decreases resistance to Clostridium difficile infection. Nat Med 2016; 22:1330-4; PMID:27668938; https://doi.org/https://doi.org/10.1038/nm.4174
- Lavigne JP, Nicolas-Chanoine MH, Bourg G, Moreau J, Sotto A. Virulent synergistic effect between Enterococcus faecalis and Escherichia coli assayed by using the Caenorhabditis elegans model. PLoS One 2008; 3:e3370; PMID:18843374; https://doi.org/https://doi.org/10.1371/journal.pone.0003370
- Cruz MR, Graham CE, Gagliano BC, Lorenz MC, Garsin DA. Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect Immunity 2013; 81:189-200; PMID:23115035; https://doi.org/https://doi.org/10.1128/IAI.00914-12
- Rasmussen RV, Karchmer AW, Bruun NE, Fowler VG. Intravascular infection. In: Engleberg NC, DiRita VJ, Dermody TS, eds. Schaechter's Mechanisms of Microbial Disease. Baltimore: Lippincott Williams & Wilkins, Wolters Kluwer, 2013:670-88
- Cabell CH, Abrutyn E, Karchmer AW. Cardiology patient page. Bacterial endocarditis: the disease, treatment, and prevention. Circulation 2003; 107:e185-7; PMID:12777321
- Giannitsioti E, Skiadas I, Antoniadou A, Tsiodras S, Kanavos K, Triantafyllidi H, Giamarellou H. Nosocomial vs. community-acquired infective endocarditis in Greece: changing epidemiological profile and mortality risk. Clin Microbiol Infect 2007; 13:763-9; PMID:17488327; https://doi.org/https://doi.org/10.1111/j.1469-0691.2007.01746.x
- Murdoch DR, Corey GR, Hoen B, Miro JM, Fowler VG, Jr., Bayer AS, Karchmer AW, Olaison L, Pappas PA, Moreillon P, et al. Clinical presentation, etiology, and outcome of infective endocarditis in the 21st century: the International Collaboration on Endocarditis-Prospective Cohort Study. Arch Intern Med 2009; 169:463-73; PMID:19273776; https://doi.org/https://doi.org/10.1001/archinternmed.2008.603
- Murray BE. The life and times of the Enterococcus. Clin Microbiol Rev 1990; 3:46-65; PMID:2404568; https://doi.org/https://doi.org/10.1128/CMR.3.1.46
- Santoro J, Levison ME. Rat model of experimental endocarditis. Infect Immunity 1978; 19:915-8; PMID:417032
- Frank KL, Guiton PS, Barnes AM, Manias DA, Chuang-Smith ON, Kohler PL, Spaulding AR, Hultgren SJ, Schlievert PM, Dunny GM. AhrC and Eep are biofilm infection-associated virulence factors in Enterococcus faecalis. Infect Immunity 2013; 81:1696-708; PMID:23460519; https://doi.org/https://doi.org/10.1128/IAI.01210-12
- McCormick JK, Hirt H, Waters CM, Tripp TJ, Dunny GM, Schlievert PM. Antibodies to a surface-exposed, N-terminal domain of aggregation substance are not protective in the rabbit model of Enterococcus faecalis infective endocarditis. Infect Immunity 2001; 69:3305-14; PMID:11292753; https://doi.org/https://doi.org/10.1128/IAI.69.5.3305-3314.2001
- Thurlow LR, Thomas VC, Narayanan S, Olson S, Fleming SD, Hancock LE. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis. Infect Immunity 2010; 78:4936-43; PMID:20713628; https://doi.org/https://doi.org/10.1128/IAI.01118-09
- Hirt H, Schlievert PM, Dunny GM. In vivo induction of virulence and antibiotic resistance transfer in Enterococcus faecalis mediated by the sex pheromone-sensing system of pCF10. Infect Immunity 2002; 70:716-23; PMID:11796604; https://doi.org/https://doi.org/10.1128/IAI.70.2.716-723.2002
- Schlievert PM, Chuang-Smith ON, Peterson ML, Cook LC, Dunny GM. Enterococcus faecalis endocarditis severity in rabbits is reduced by IgG Fabs interfering with aggregation substance. PLoS One 2010; 5:e13194; PMID:20957231; https://doi.org/https://doi.org/10.1371/journal.pone.0013194
- Chuang ON, Schlievert PM, Wells CL, Manias DA, Tripp TJ, Dunny GM. Multiple functional domains of Enterococcus faecalis aggregation substance Asc10 contribute to endocarditis virulence. Infect Immunity 2009; 77:539-48; PMID:18955479; https://doi.org/https://doi.org/10.1128/IAI.01034-08
- McCormick JK, Tripp TJ, Dunny GM, Schlievert PM. Formation of vegetations during infective endocarditis excludes binding of bacterial-specific host antibodies to Enterococcus faecalis. J Infect Dis 2002; 185:994-7; PMID:11920326; https://doi.org/https://doi.org/10.1086/339604
- Eckert C, Lecerf M, Dubost L, Arthur M, Mesnage S. Functional analysis of AtlA, the major N-acetylglucosaminidase of Enterococcus faecalis. J Bacteriol 2006; 188:8513-9; PMID:17041059; https://doi.org/https://doi.org/10.1128/JB.01145-06
- Bravetti AL, Mesnage S, Lefort A, Chau F, Eckert C, Garry L, Arthur M, Fantin B. Contribution of the autolysin AtlA to the bactericidal activity of amoxicillin against Enterococcus faecalis JH2-2. Antimicrobial Agents Chemotherapy 2009; 53:1667-9; PMID:19188384; https://doi.org/https://doi.org/10.1128/AAC.00692-08
- Frank KL, Barnes AM, Grindle SM, Manias DA, Schlievert PM, Dunny GM. Use of recombinase-based in vivo expression technology to characterize Enterococcus faecalis gene expression during infection identifies in vivo-expressed antisense RNAs and implicates the protease Eep in pathogenesis. Infect Immunity 2012; 80:539-49; PMID:22144481; https://doi.org/https://doi.org/10.1128/IAI.05964-11
- Sleator RD, Gahan CG, Hill C. Identification and disruption of the proBA locus in Listeria monocytogenes: role of proline biosynthesis in salt tolerance and murine infection. Applied Environmental Microbiol 2001; 67:2571-7; PMID:11375165; https://doi.org/https://doi.org/10.1128/AEM.67.6.2571-2577.2001
- Urban S. Making the cut: central roles of intramembrane proteolysis in pathogenic microorganisms. Nat Rev Microbiol 2009; 7:411-23; PMID:19421188
- Leuck AM, Johnson JR, Dunny GM. A widely used in vitro biofilm assay has questionable clinical significance for Enterococcal endocarditis. PLoS One 2014; 9:e107282; PMID:25255085; https://doi.org/https://doi.org/10.1371/journal.pone.0107282
- Qin X, Singh KV, Weinstock GM, Murray BE. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect Immunity 2000; 68:2579-86; PMID:10768947; https://doi.org/https://doi.org/10.1128/IAI.68.5.2579-2586.2000
- Wang B, Muir TW. Regulation of virulence in Staphylococcus aureus: Molecular mechanisms and remaining puzzles. Cell Chem Biol 2016; 23:214-24; PMID:26971873; https://doi.org/https://doi.org/10.1016/j.chembiol.2016.01.004
- Sifri CD, Mylonakis E, Singh KV, Qin X, Garsin DA, Murray BE, Ausubel FM, Calderwood SB. Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect Immunity 2002; 70:5647-50; PMID:12228293; https://doi.org/https://doi.org/10.1128/IAI.70.10.5647-5650.2002
- Engelbert M, Mylonakis E, Ausubel FM, Calderwood SB, Gilmore MS. Contribution of gelatinase, serine protease, and fsr to the pathogenesis of Enterococcus faecalis endophthalmitis. Infect Immunity 2004; 72:3628-33; PMID:15155673; https://doi.org/https://doi.org/10.1128/IAI.72.6.3628-3633.2004
- Mylonakis E, Engelbert M, Qin X, Sifri CD, Murray BE, Ausubel FM, Gilmore MS, Calderwood SB. The Enterococcus faecalis fsrB gene, a key component of the fsr quorum-sensing system, is associated with virulence in the rabbit endophthalmitis model. Infect Immunity 2002; 70:4678-81; PMID:12117982; https://doi.org/https://doi.org/10.1128/IAI.70.8.4678-4681.2002
- Singh KV, Nallapareddy SR, Nannini EC, Murray BE. Fsr-independent production of protease(s) may explain the lack of attenuation of an Enterococcus faecalis fsr mutant versus a gelE-sprE mutant in induction of endocarditis. Infect Immunity 2005; 73:4888-94; PMID:16041002; https://doi.org/https://doi.org/10.1128/IAI.73.8.4888-4894.2005
- Nannini EC, Teng F, Singh KV, Murray BE. Decreased virulence of a gls24 mutant of Enterococcus faecalis OG1RF in an experimental endocarditis model. Infect Immunity 2005; 73:7772-4; PMID:16239583; https://doi.org/https://doi.org/10.1128/IAI.73.11.7772-7774.2005
- Haller C, Berthold M, Wobser D, Kropec A, Lauriola M, Schlensak C, Huebner J. Cell-wall glycolipid mutations and their effects on virulence of E. faecalis in a rat model of infective endocarditis. PloS One 2014; 9:e91863; PMID:24637922; https://doi.org/https://doi.org/10.1371/journal.pone.0091863
- Laverde D, Wobser D, Romero-Saavedra F, Hogendorf W, van der Marel G, Berthold M, Kropec A, Codee J, Huebner J. Synthetic teichoic acid conjugate vaccine against nosocomial Gram-positive bacteria. PLoS One 2014; 9:e110953; PMID:25333799; https://doi.org/https://doi.org/10.1371/journal.pone.0110953
- Nallapareddy SR, Singh KV, Murray BE. Contribution of the collagen adhesin Acm to pathogenesis of Enterococcus faecium in experimental endocarditis. Infect Immunity 2008; 76:4120-8; PMID:18591236; https://doi.org/https://doi.org/10.1128/IAI.00376-08
- Paganelli FL, Huebner J, Singh KV, Zhang X, van Schaik W, Wobser D, Braat JC, Murray BE, Bonten MJ, Willems RJ, et al. Genome-wide screening identifies PTS permease BepA to be involved in Enterococcus faecium endocarditis and biofilm formation. J Infect Dis 2016; 214(2):189-95; PMID:26984142
- Pinkston KL, Singh KV, Gao P, Wilganowski N, Robinson H, Ghosh S, Azhdarinia A, Sevick-Muraca EM, Murray BE, Harvey BR. Targeting pili in Enterococcal pathogenesis. Infect Immunity 2014; 82:1540-7; PMID:24452680; https://doi.org/https://doi.org/10.1128/IAI.01403-13
- Singh KV, Nallapareddy SR, Sillanpaa J, Murray BE. Importance of the collagen adhesin Ace in pathogenesis and protection against Enterococcus faecalis experimental endocarditis. PLoS Pathogens 2010; 6:e1000716; PMID:20072611; https://doi.org/https://doi.org/10.1371/journal.ppat.1000716
- Singh KV, La Rosa SL, Somarajan SR, Roh JH, Murray BE. The fibronectin-binding protein EfbA contributes to pathogenesis and protects against infective endocarditis caused by Enterococcus faecalis. Infect Immunity 2015; 83:4487-94; PMID:26351286; https://doi.org/https://doi.org/10.1128/IAI.00884-15
- Somarajan SR, Roh JH, Singh KV, Weinstock GM, Murray BE. CcpA is important for growth and virulence of Enterococcus faecium. Infect Immunity 2014; 82:3580-7; PMID:24914215; https://doi.org/https://doi.org/10.1128/IAI.01911-14
- Somarajan SR, La Rosa SL, Singh KV, Roh JH, Hook M, Murray BE. The fibronectin-binding protein Fnm contributes to adherence to extracellular matrix components and virulence of Enterococcus faecium. Infect Immunity 2015; 83:4653-61; PMID:26371130; https://doi.org/https://doi.org/10.1128/IAI.00885-15
- Heikens E, Singh KV, Jacques-Palaz KD, van Luit-Asbroek M, Oostdijk EA, Bonten MJ, Murray BE, Willems RJ. Contribution of the enterococcal surface protein Esp to pathogenesis of Enterococcus faecium endocarditis. Microb Infect 2011; 13:1185-90; PMID:21911077; https://doi.org/https://doi.org/10.1016/j.micinf.2011.08.006
- Teng F, Nannini EC, Murray BE. Importance of gls24 in virulence and stress response of Enterococcus faecalis and use of the Gls24 protein as a possible immunotherapy target. J Infect Dis 2005; 191:472-80; PMID:15633107; https://doi.org/https://doi.org/10.1086/427191
- Giard JC, Rince A, Capiaux H, Auffray Y, Hartke A. Inactivation of the stress- and starvation-inducible gls24 operon has a pleiotrophic effect on cell morphology, stress sensitivity, and gene expression in Enterococcus faecalis. J Bacteriol 2000; 182:4512-20; PMID:10913085; https://doi.org/https://doi.org/10.1128/JB.182.16.4512-4520.2000
- Roh JH, Singh KV, La Rosa SL, Cohen AL, Murray BE. The two-component system GrvRS (EtaRS) regulates ace expression in Enterococcus faecalis OG1RF. Infect Immunity 2015; 83:389-95; PMID:25385790; https://doi.org/https://doi.org/10.1128/IAI.02587-14
- Lebreton F, Riboulet-Bisson E, Serror P, Sanguinetti M, Posteraro B, Torelli R, Hartke A, Auffray Y, Giard JC. Ace, which encodes an adhesin in Enterococcus faecalis, is regulated by Ers and is involved in virulence. Infect Immunity 2009; 77:2832-9; PMID:19433548; https://doi.org/https://doi.org/10.1128/IAI.01218-08
- Rich RL, Kreikemeyer B, Owens RT, LaBrenz S, Narayana SV, Weinstock GM, Murray BE, Hook M. Ace is a collagen-binding MSCRAMM from Enterococcus faecalis. J Biol Chem 1999; 274:26939-45; PMID:10480905; https://doi.org/https://doi.org/10.1074/jbc.274.38.26939
- Torelli R, Serror P, Bugli F, Paroni Sterbini F, Florio AR, Stringaro A, Colone M, De Carolis E, Martini C, Giard JC, et al. The PavA-like fibronectin-binding protein of Enterococcus faecalis, EfbA, is important for virulence in a mouse model of ascending urinary tract infection. J Infect Dis 2012; 206:952-60; PMID:22782954; https://doi.org/https://doi.org/10.1093/infdis/jis440
- Sillanpaa J, Chang C, Singh KV, Montealegre MC, Nallapareddy SR, Harvey BR, Ton-That H, Murray BE. Contribution of individual Ebp pilus subunits of Enterococcus faecalis OG1RF to pilus biogenesis, biofilm formation and urinary tract infection. PLoS One 2013; 8:e68813; PMID:23874774; https://doi.org/https://doi.org/10.1371/journal.pone.0068813
- Galloway-Pena JR, Liang X, Singh KV, Yadav P, Chang C, La Rosa SL, Shelburne S, Ton-That H, Hook M, Murray BE. The identification and functional characterization of WxL proteins from Enterococcus faecium reveal surface proteins involved in extracellular matrix interactions. J Bacteriol 2015; 197:882-92; PMID:25512313; https://doi.org/https://doi.org/10.1128/JB.02288-14
- Montealegre MC, Singh KV, Somarajan SR, Yadav P, Chang C, Spencer R, Sillanpaa J, Ton-That H, Murray BE. Role of the Emp pilus subunits of Enterococcus faecium in biofilm formation, adherence to host extracellular matrix components, and experimental infection. Infect Immunity 2016; 84:1491-500; PMID:26930703; https://doi.org/https://doi.org/10.1128/IAI.01396-15
- Hienz SA, Schennings T, Heimdahl A, Flock JI. Collagen binding of Staphylococcus aureus is a virulence factor in experimental endocarditis. J Infect Dis 1996; 174:83-8; PMID:8656018; https://doi.org/https://doi.org/10.1093/infdis/174.1.83
- Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev 1999; 63:174-229; PMID:10066836
- Top J, Paganelli FL, Zhang X, van Schaik W, Leavis HL, van Luit-Asbroek M, van der Poll T, Leendertse M, Bonten MJ, Willems RJ. The Enterococcus faecium Enterococcal biofilm regulator, EbrB, regulates the esp operon and is implicated in biofilm formation and intestinal colonization. PLoS One 2013; 8:e65224; PMID:23741484; https://doi.org/https://doi.org/10.1371/journal.pone.0065224
- Nemzek JA, Hugunin KM, Opp MR. Modeling sepsis in the laboratory: merging sound science with animal well-being. Comparative Med 2008; 58:120-8; PMID:18524169
- Tzianabos AO, Chandraker A, Kalka-Moll W, Stingele F, Dong VM, Finberg RW, Peach R, Sayegh MH. Bacterial pathogens induce abscess formation by CD4+ T-cell activation via the CD28-B7-2 costimulatory pathway. Infect Immunity 2000; 68:6650-5; PMID:11083777; https://doi.org/https://doi.org/10.1128/IAI.68.12.6650-6655.2000
- Cortes-Perez NG, Dumoulin R, Gaubert S, Lacoux C, Bugli F, Martin R, Chat S, Piquand K, Meylheuc T, Langella P, et al. Overexpression of Enterococcus faecalis elr operon protects from phagocytosis. BioMed Central Microbiol 2015; 15:112; PMID:26003173
- Theilacker C, Diederich AK, Otto A, Sava IG, Wobser D, Bao Y, Hese K, Broszat M, Henneke P, Becher D, et al. Enterococcus faecalis glycolipids modulate lipoprotein-content of the bacterial cell membrane and host immune response. PLoS One 2015; 10:e0132949; PMID:26172831; https://doi.org/https://doi.org/10.1371/journal.pone.0132949
- Xu Y, Singh KV, Qin X, Murray BE, Weinstock GM. Analysis of a gene cluster of Enterococcus faecalis involved in polysaccharide biosynthesis. Infect Immunity 2000; 68:815-23; PMID:10639451; https://doi.org/https://doi.org/10.1128/IAI.68.2.815-823.2000
- Plantefeve G, Dupont H, Hubert V, Garry L, Pous C, Carbon C, Montravers P. Impact of elements containing glycopeptide resistance genes on expression of virulence in Enterococcus faecalis peritonitis: a pilot study with rats. Antimicrobial Agents Chemotherapy 2003; 47:1560-4; PMID:12709322; https://doi.org/https://doi.org/10.1128/AAC.47.5.1560-1564.2003
- Hanin A, Sava I, Bao Y, Huebner J, Hartke A, Auffray Y, Sauvageot N. Screening of in vivo activated genes in Enterococcus faecalis during insect and mouse infections and growth in urine. PLoS One 2010; 5:e11879; PMID:20686694; https://doi.org/https://doi.org/10.1371/journal.pone.0011879
- Muller C, Cacaci M, Sauvageot N, Sanguinetti M, Rattei T, Eder T, Giard JC, Kalinowski J, Hain T, Hartke A. The intraperitoneal transcriptome of the opportunistic pathogen Enterococcus faecalis in mice. PLoS One 2015; 10:e0126143; PMID:25978463; https://doi.org/https://doi.org/10.1371/journal.pone.0126143
- Haas W, Shepard BD, Gilmore MS. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature 2002; 415:84-7; PMID:11780122; https://doi.org/https://doi.org/10.1038/415084a
- Low YL, Jakubovics NS, Flatman JC, Jenkinson HF, Smith AW. Manganese-dependent regulation of the endocarditis-associated virulence factor EfaA of Enterococcus faecalis. J Medical Microbiol 2003; 52:113-9; PMID:12543916; https://doi.org/https://doi.org/10.1099/jmm.0.05039-0
- Ike Y, Hashimoto H, Clewell DB. Hemolysin of Streptococcus faecalis subspecies zymogenes contributes to virulence in mice. Infect Immunity 1984; 45:528-30; PMID:6086531
- Singh KV, Coque TM, Weinstock GM, Murray BE. In vivo testing of an Enterococcus faecalis efaA mutant and use of efaA homologs for species identification. FEMS Immunol Medical Microbiol 1998; 21:323-31; PMID:9753005; https://doi.org/https://doi.org/10.1111/j.1574-695X.1998.tb01180.x
- Dupont H, Montravers P, Mohler J, Carbon C. Disparate findings on the role of virulence factors of Enterococcus faecalis in mouse and rat models of peritonitis. Infect Immunity 1998; 66:2570-5; PMID:9596718
- Teng F, Wang L, Singh KV, Murray BE, Weinstock GM. Involvement of PhoP-PhoS homologs in Enterococcus faecalis virulence. Infect Immunity 2002; 70:1991-6; PMID:11895963; https://doi.org/https://doi.org/10.1128/IAI.70.4.1991-1996.2002
- Coburn PS, Baghdayan AS, Dolan GT, Shankar N. An AraC-type transcriptional regulator encoded on the Enterococcus faecalis pathogenicity island contributes to pathogenesis and intracellular macrophage survival. Infect Immunity 2008; 76:5668-76; PMID:18824537; https://doi.org/https://doi.org/10.1128/IAI.00930-08
- Ruiz-Cruz S, Espinosa M, Goldmann O, Bravo A. Global regulation of gene expression by the MafR protein of Enterococcus faecalis. Frontiers Microbiol 2015; 6:1521; PMID:26793169
- Giard JC, Riboulet E, Verneuil N, Sanguinetti M, Auffray Y, Hartke A. Characterization of Ers, a PrfA-like regulator of Enterococcus faecalis. FEMS Immunol Medical Microbiol 2006; 46:410-8; PMID:16553815; https://doi.org/https://doi.org/10.1111/j.1574-695X.2005.00049.x
- Reid SD, Montgomery AG, Musser JM. Identification of srv, a PrfA-like regulator of group A Streptococcus that influences virulence. Infect Immunity 2004; 72:1799-803; PMID:14977990; https://doi.org/https://doi.org/10.1128/IAI.72.3.1799-1803.2004
- Riboulet-Bisson E, Sanguinetti M, Budin-Verneuil A, Auffray Y, Hartke A, Giard JC. Characterization of the Ers regulon of Enterococcus faecalis. Infect Immunity 2008; 76:3064-74; PMID:18426870; https://doi.org/https://doi.org/10.1128/IAI.00161-08
- Verneuil N, Rince A, Sanguinetti M, Auffray Y, Hartke A, Giard JC. Implication of hypR in the virulence and oxidative stress response of Enterococcus faecalis. FEMS Microbiol Lett 2005; 252:137-41; PMID:16216443; https://doi.org/https://doi.org/10.1016/j.femsle.2005.08.043
- La Carbona S, Sauvageot N, Giard JC, Benachour A, Posteraro B, Auffray Y, Sanguinetti M, Hartke A. Comparative study of the physiological roles of three peroxidases (NADH peroxidase, alkyl hydroperoxide reductase and thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis. Mol Microbiol 2007; 66:1148-63; https://doi.org/https://doi.org/10.1111/j.1365-2958.2007.05987.x
- Kajfasz JK, Mendoza JE, Gaca AO, Miller JH, Koselny KA, Giambiagi-Demarval M, Wellington M, Abranches J, Lemos JA. The Spx regulator modulates stress responses and virulence in Enterococcus faecalis. Infect Immunity 2012; 80:2265-75; https://doi.org/https://doi.org/10.1128/IAI.00026-12
- Zhao C, Hartke A, La Sorda M, Posteraro B, Laplace JM, Auffray Y, Sanguinetti M. Role of methionine sulfoxide reductases A and B of Enterococcus faecalis in oxidative stress and virulence. Infect Immunity 2010; 78:3889-97; PMID:20566694; https://doi.org/https://doi.org/10.1128/IAI.00165-10
- Poole LB. Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch Biochem Biophys 2005; 433:240-54; PMID:15581580; https://doi.org/https://doi.org/10.1016/j.abb.2004.09.006
- Nakano MM, Hajarizadeh F, Zhu Y, Zuber P. Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol Microbiol 2001; 42:383-94; PMID:11703662; https://doi.org/https://doi.org/10.1046/j.1365-2958.2001.02639.x
- Verneuil N, Maze A, Sanguinetti M, Laplace JM, Benachour A, Auffray Y, Giard JC, Hartke A. Implication of (Mn)superoxide dismutase of Enterococcus faecalis in oxidative stress responses and survival inside macrophages. Microbiology 2006; 152:2579-89; PMID:16946253; https://doi.org/https://doi.org/10.1099/mic.0.28922-0
- Verneuil N, Rince A, Sanguinetti M, Posteraro B, Fadda G, Auffray Y, Hartke A, Giard JC. Contribution of a PerR-like regulator to the oxidative-stress response and virulence of Enterococcus faecalis. Microbiology 2005; 151:3997-4004; PMID:16339944; https://doi.org/https://doi.org/10.1099/mic.0.28325-0
- Brinster S, Posteraro B, Bierne H, Alberti A, Makhzami S, Sanguinetti M, Serror P. Enterococcal leucine-rich repeat-containing protein involved in virulence and host inflammatory response. Infect Immunity 2007; 75:4463-71; PMID:17620355; https://doi.org/https://doi.org/10.1128/IAI.00279-07
- Dumoulin R, Cortes-Perez N, Gaubert S, Duhutrel P, Brinster S, Torelli R, Sanguinetti M, Posteraro B, Repoila F, Serror P. Enterococcal Rgg-like regulator ElrR activates expression of the elrA operon. J Bacteriol 2013; 195:3073-83; PMID:23645602; https://doi.org/https://doi.org/10.1128/JB.00121-13
- Teng F, Singh KV, Bourgogne A, Zeng J, Murray BE. Further characterization of the epa gene cluster and Epa polysaccharides of Enterococcus faecalis. Infect Immunity 2009; 77:3759-67; PMID:19581393; https://doi.org/https://doi.org/10.1128/IAI.00149-09
- Diederich AK, Wobser D, Spiess M, Sava IG, Huebner J, Sakiotanc T. Role of glycolipids in the pathogenesis of Enterococcus faecalis urinary tract infection. PLoS One 2014; 9:e96295; PMID:24806450; https://doi.org/https://doi.org/10.1371/journal.pone.0096295
- Choudhury T, Singh KV, Sillanpaa J, Nallapareddy SR, Murray BE. Importance of two Enterococcus faecium loci encoding Gls-like proteins for in vitro bile salts stress response and virulence. J Infect Dis 2011; 203:1147-54; PMID:21451003; https://doi.org/https://doi.org/10.1093/infdis/jiq160
- Panesso D, Montealegre MC, Rincon S, Mojica MF, Rice LB, Singh KV, Murray BE, Arias CA. The hylEfm gene in pHylEfm of Enterococcus faecium is not required in pathogenesis of murine peritonitis. BioMed Central Microbiol 2011; 11:20; PMID:21266081
- Collin BA, Leather HL, Wingard JR, Ramphal R. Evolution, incidence, and susceptibility of bacterial bloodstream isolates from 519 bone marrow transplant patients. Clin Infect Dis 2001; 33:947-53; PMID:11528564; https://doi.org/https://doi.org/10.1086/322604
- de Fatima Silva Lopes M, Ribeiro T, Abrantes M, Figueiredo Marques JJ, Tenreiro R, Crespo MT. Antimicrobial resistance profiles of dairy and clinical isolates and type strains of Enterococci. Int J Food Microbiol 2005; 103:191-8; PMID:16083821; https://doi.org/https://doi.org/10.1016/j.ijfoodmicro.2004.12.025
- Willems RJ, Bonten MJ. Glycopeptide-resistant Enterococci: deciphering virulence, resistance and epidemicity. Curr Opin Infect Dis 2007; 20:384-90; PMID:17609597; https://doi.org/https://doi.org/10.1097/QCO.0b013e32818be63d
- Le Jeune A, Torelli R, Sanguinetti M, Giard JC, Hartke A, Auffray Y, Benachour A. The extracytoplasmic function sigma factor SigV plays a key role in the original model of lysozyme resistance and virulence of Enterococcus faecalis. PLoS One 2010; 5:e9658; PMID:20300180; https://doi.org/https://doi.org/10.1371/journal.pone.0009658
- Benachour A, Muller C, Dabrowski-Coton M, Le Breton Y, Giard JC, Rince A, Auffray Y, Hartke A. The Enterococcus faecalis sigV protein is an extracytoplasmic function sigma factor contributing to survival following heat, acid, and ethanol treatments. J Bacteriol 2005; 187:1022-35; PMID:15659680; https://doi.org/https://doi.org/10.1128/JB.187.3.1022-1035.2005
- Reffuveille F, Connil N, Sanguinetti M, Posteraro B, Chevalier S, Auffray Y, Rince A. Involvement of peptidylprolyl cis/trans isomerases in Enterococcus faecalis virulence. Infect Immunity 2012; 80:1728-35; PMID:22331431; https://doi.org/https://doi.org/10.1128/IAI.06251-11
- Rossmann FS, Racek T, Wobser D, Puchalka J, Rabener EM, Reiger M, Hendrickx AP, Diederich AK, Jung K, Klein C, et al. Phage-mediated dispersal of biofilm and distribution of bacterial virulence genes is induced by quorum sensing. PLoS Pathogens 2015; 11:e1004653; PMID:25706310; https://doi.org/https://doi.org/10.1371/journal.ppat.1004653
- Theilacker C, Sava I, Sanchez-Carballo P, Bao Y, Kropec A, Grohmann E, Holst O, Huebner J. Deletion of the glycosyltransferase bgsB of Enterococcus faecalis leads to a complete loss of glycolipids from the cell membrane and to impaired biofilm formation. BioMed Central Microbiol 2011; 11:67; PMID:21470413
- Theilacker C, Sanchez-Carballo P, Toma I, Fabretti F, Sava I, Kropec A, Holst O, Huebner J. Glycolipids are involved in biofilm accumulation and prolonged bacteraemia in Enterococcus faecalis. Mol Microbiol 2009; 71:1055-69; PMID:19170884; https://doi.org/https://doi.org/10.1111/j.1365-2958.2008.06587.x
- Michaux C, Sanguinetti M, Reffuveille F, Auffray Y, Posteraro B, Gilmore MS, Hartke A, Giard JC. SlyA is a transcriptional regulator involved in the virulence of Enterococcus faecalis. Infect Immunity 2011; 79:2638-45; PMID:21536798; https://doi.org/https://doi.org/10.1128/IAI.01132-10
- Michaux C, Martini C, Shioya K, Ahmed Lecheheb S, Budin-Verneuil A, Cosette P, Sanguinetti M, Hartke A, Verneuil N, Giard JC. CspR, a cold shock RNA-binding protein involved in the long-term survival and the virulence of Enterococcus faecalis. J Bacteriol 2012; 194:6900-8; PMID:23086208; https://doi.org/https://doi.org/10.1128/JB.01673-12
- Rana NF, Sauvageot N, Laplace JM, Bao Y, Nes I, Rince A, Posteraro B, Sanguinetti M, Hartke A. Redox balance via lactate dehydrogenase is important for multiple stress resistance and virulence in Enterococcus faecalis. Infect Immunity 2013; 81:2662-8; PMID:23649090; https://doi.org/https://doi.org/10.1128/IAI.01299-12
- Dhall S, Do D, Garcia M, Wijesinghe DS, Brandon A, Kim J, Sanchez A, Lyubovitsky J, Gallagher S, Nothnagel EA, et al. A novel model of chronic wounds: importance of redox imbalance and biofilm-forming bacteria for establishment of chronicity. PLoS One 2014; 9:e109848; PMID:25313558; https://doi.org/https://doi.org/10.1371/journal.pone.0109848
- Lebreton F, van Schaik W, Sanguinetti M, Posteraro B, Torelli R, Le Bras F, Verneuil N, Zhang X, Giard JC, Dhalluin A, et al. AsrR is an oxidative stress sensing regulator modulating Enterococcus faecium opportunistic traits, antimicrobial resistance, and pathogenicity. PLoS Pathogens 2012; 8:e1002834; PMID:22876178; https://doi.org/https://doi.org/10.1371/journal.ppat.1002834
- Ali L, Spiess M, Wobser D, Rodriguez M, Blum HE, Sakiotanc T. Identification and functional characterization of the putative polysaccharide biosynthesis protein (CapD) of Enterococcus faecium U0317. Infect Genetics Evolution 2016; 37:215-24; https://doi.org/https://doi.org/10.1016/j.meegid.2015.11.020
- Leendertse M, Heikens E, Wijnands LM, van Luit-Asbroek M, Teske GJ, Roelofs JJ, Bonten MJ, van der Poll T, Willems RJ. Enterococcal surface protein transiently aggravates Enterococcus faecium-induced urinary tract infection in mice. J Infect Dis 2009; 200:1162-5; PMID:19702507; https://doi.org/https://doi.org/10.1086/605609
- Sava IG, Heikens E, Kropec A, Theilacker C, Willems R, Huebner J. Enterococcal surface protein contributes to persistence in the host but is not a target of opsonic and protective antibodies in Enterococcus faecium infection. J Medical Microbiol 2010; 59:1001-4; PMID:20522627; https://doi.org/https://doi.org/10.1099/jmm.0.020578-0
- Guinane CM, Cotter PD. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Therapeutic Adv Gastroenterol 2013; 6:295-308; PMID:23814609; https://doi.org/https://doi.org/10.1177/1756283X13482996
- Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest 2007; 117:514-21; PMID:17332878; https://doi.org/https://doi.org/10.1172/JCI30587
- Ocvirk S, Sava IG, Lengfelder I, Lagkouvardos I, Steck N, Roh JH, Tchaptchet S, Bao Y, Hansen JJ, Huebner J, et al. Surface-associated lipoproteins link Enterococcus faecalis virulence to colitogenic activity in IL-10-deficient mice independent of their expression levels. PLoS Pathogens 2015; 11:e1004911; PMID:26067254; https://doi.org/https://doi.org/10.1371/journal.ppat.1004911
- Shogan BD, Belogortseva N, Luong PM, Zaborin A, Lax S, Bethel C, Ward M, Muldoon JP, Singer M, An G, et al. Collagen degradation and MMP9 activation by Enterococcus faecalis contribute to intestinal anastomotic leak. Sci Transl Med 2015; 7:286ra68; PMID:25947163; https://doi.org/https://doi.org/10.1126/scitranslmed.3010658
- van der Heijden KM, van der Heijden IM, Galvao FH, Lopes CG, Costa SF, Abdala E, D'Albuquerque LA, Levin AS. Intestinal translocation of clinical isolates of vancomycin-resistant Enterococcus faecalis and ESBL-producing Escherichia coli in a rat model of bacterial colonization and liver ischemia/reperfusion injury. PLoS One 2014; 9:e108453; PMID:25255079; https://doi.org/https://doi.org/10.1371/journal.pone.0108453
- Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 2002; 23:529-36; PMID:11895869; https://doi.org/https://doi.org/10.1093/carcin/23.3.529
- Huycke MM, Moore D, Joyce W, Wise P, Shepard L, Kotake Y, Gilmore MS. Extracellular superoxide production by Enterococcus faecalis requires demethylmenaquinone and is attenuated by functional terminal quinol oxidases. Mol Microbiol 2001; 42:729-40; PMID:11722738; https://doi.org/https://doi.org/10.1046/j.1365-2958.2001.02638.x
- Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P, Cao Y, Bousounis P, Kristich CJ, Salzman NH. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 2015; 526:719-22; PMID:26479034; https://doi.org/https://doi.org/10.1038/nature15524
- Kristich CJ, Wells CL, Dunny GM. A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc Natl Acad Sci U S A 2007; 104:3508-13; PMID:17360674; https://doi.org/https://doi.org/10.1073/pnas.0608742104
- Pultz NJ, Shankar N, Baghdayan AS, Donskey CJ. Enterococcal surface protein Esp does not facilitate intestinal colonization or translocation of Enterococcus faecalis in clindamycin-treated mice. FEMS Microbiol Lett 2005; 242:217-9; PMID:15621440; https://doi.org/https://doi.org/10.1016/j.femsle.2004.11.006
- Rigottier-Gois L, Madec C, Navickas A, Matos RC, Akary-Lepage E, Mistou MY, Serror P. The surface rhamnopolysaccharide epa of Enterococcus faecalis is a key determinant of intestinal colonization. J Infect Dis 2015; 211:62-71; PMID:25035517; https://doi.org/https://doi.org/10.1093/infdis/jiu402
- La Rosa SL, Casey PG, Hill C, Diep DB, Nes IF, Brede DA. In vivo assessment of growth and virulence gene expression during commensal and pathogenic lifestyles of luxABCDE-tagged Enterococcus faecalis strains in murine gastrointestinal and intravenous infection models. Applied Environ Microbiol 2013; 79:3986-97; PMID:23603680; https://doi.org/https://doi.org/10.1128/AEM.00831-13
- Zhang X, Top J, de Been M, Bierschenk D, Rogers M, Leendertse M, Bonten MJ, van der Poll T, Willems RJ, van Schaik W. Identification of a genetic determinant in clinical Enterococcus faecium strains that contributes to intestinal colonization during antibiotic treatment. J Infect Dis 2013; 207:1780-6; PMID:23447698; https://doi.org/https://doi.org/10.1093/infdis/jit076
- Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 2008; 455:804-7; PMID:18724361; https://doi.org/https://doi.org/10.1038/nature07250
- Ubeda C, Bucci V, Caballero S, Djukovic A, Toussaint NC, Equinda M, Lipuma L, Ling L, Gobourne A, No D, et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immunity 2013; 81:965-73; PMID:23319552; https://doi.org/https://doi.org/10.1128/IAI.01197-12
- Rafii F, Sutherland JB, Cerniglia CE. Effects of treatment with antimicrobial agents on the human colonic microflora. Therapeutics Clin Risk Management 2008; 4:1343-58; PMID:19337440; https://doi.org/https://doi.org/10.2147/TCRM.S4328
- Kristich CJ, Rice LB, Arias CA. Enterococcal infection-treatment and antibiotic resistance. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, eds. Enterococci: From commensals to leading causes of drug resistant infection. Boston, 2014
- Gilmore MS, Rauch M, Ramsey MM, Himes PR, Varahan S, Manson JM, Lebreton F, Hancock LE. Pheromone killing of multidrug-resistant Enterococcus faecalis V583 by native commensal strains. Proc Natl Acad Sci U S A 2015; 112:7273-8; PMID:26039987; https://doi.org/https://doi.org/10.1073/pnas.1500553112
- Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J Exp Med 2007; 204:1891-900; PMID:17635956; https://doi.org/https://doi.org/10.1084/jem.20070563
- Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 2006; 313:1126-30; PMID:16931762; https://doi.org/https://doi.org/10.1126/science.1127119
- Heikens E, Leendertse M, Wijnands LM, van Luit-Asbroek M, Bonten MJ, van der Poll T, Willems RJ. Enterococcal surface protein Esp is not essential for cell adhesion and intestinal colonization of Enterococcus faecium in mice. BioMed Central Microbiol 2009; 9:19; PMID:19178704
- Babbs CF. Free radicals and the etiology of colon cancer. Free Radical Biol Med 1990; 8:191-200; PMID:2185144; https://doi.org/https://doi.org/10.1016/0891-5849(90)90091-V
- Huycke MM, Joyce W, Wack MF. Augmented production of extracellular superoxide by blood isolates of Enterococcus faecalis. J Infect Dis 1996; 173:743-6; PMID:8627044; https://doi.org/https://doi.org/10.1093/infdis/173.3.743
- Baum RH, Dolin MI. Isolation of 2-solanesyl-1,4-naphthoquinone from Streptococcus faecalis, 10Cl. J Biol Chem 1965; 240:3425-33; PMID:14321383
- Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 2015; 13:269-84; PMID:25853778; https://doi.org/https://doi.org/10.1038/nrmicro3432
- Mody L, Juthani-Mehta M. Urinary tract infections in older women: a clinical review. JAMA 2014; 311:844-54; PMID:24570248; https://doi.org/https://doi.org/10.1001/jama.2014.303
- Schappert SM, Rechtsteiner EA. Ambulatory medical care utilization estimates for 2006. National health statistics reports; No 8. Hyattsville, MD: National Center for Health Statistics; 2008.
- Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, et al. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 2014; 370:1198-208; PMID:24670166; https://doi.org/https://doi.org/10.1056/NEJMoa1306801
- Singh KV, Lewis RJ, Murray BE. Importance of the epa locus of Enterococcus faecalis OG1RF in a mouse model of ascending urinary tract infection. J Infect Dis 2009; 200:417-20; PMID:19545208; https://doi.org/https://doi.org/10.1086/600124
- Sillanpaa J, Nallapareddy SR, Singh KV, Prakash VP, Fothergill T, Ton-That H, Murray BE. Characterization of the ebp(fm) pilus-encoding operon of Enterococcus faecium and its role in biofilm formation and virulence in a murine model of urinary tract infection. Virulence 2010; 1:236-46; PMID:20676385; https://doi.org/https://doi.org/10.4161/viru.1.4.11966
- Nallapareddy SR, Singh KV, Sillanpaa J, Zhao M, Murray BE. Relative contributions of Ebp pili and the collagen adhesin ace to host extracellular matrix protein adherence and experimental urinary tract infection by Enterococcus faecalis OG1RF. Infect Immunity 2011; 79:2901-10; PMID:21505082; https://doi.org/https://doi.org/10.1128/IAI.00038-11
- Shankar N, Lockatell CV, Baghdayan AS, Drachenberg C, Gilmore MS, Johnson DE. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect Immunity 2001; 69:4366-72; PMID:11401975; https://doi.org/https://doi.org/10.1128/IAI.69.7.4366-4372.2001
- Kau AL, Martin SM, Lyon W, Hayes E, Caparon MG, Hultgren SJ. Enterococcus faecalis tropism for the kidneys in the urinary tract of C57BL/6J mice. Infect Immunity 2005; 73:2461-8; PMID:15784592; https://doi.org/https://doi.org/10.1128/IAI.73.4.2461-2468.2005
- Singh KV, Nallapareddy SR, Murray BE. Importance of the endocarditis- and biofilm-associated pilus (ebp) locus in the pathogenesis of Enterococcus faecalis ascending urinary tract infection. J Infect Dis 2007; 195:1671-7; PMID:17471437; https://doi.org/https://doi.org/10.1086/517524
- Kemp KD, Singh KV, Nallapareddy SR, Murray BE. Relative contributions of Enterococcus faecalis OG1RF sortase-encoding genes, srtA and bps (srtC), to biofilm formation and a murine model of urinary tract infection. Infect Immunity 2007; 75:5399-404; PMID:17785477; https://doi.org/https://doi.org/10.1128/IAI.00663-07
- Nielsen HV, Flores-Mireles AL, Kau AL, Kline KA, Pinkner JS, Neiers F, Normark S, Henriques-Normark B, Caparon MG, Hultgren SJ. Pilin and sortase residues critical for endocarditis- and biofilm-associated pilus biogenesis in Enterococcus faecalis. J Bacteriol 2013; 195:4484-95; PMID:23913319; https://doi.org/https://doi.org/10.1128/JB.00451-13
- Wobser D, Ali L, Grohmann E, Huebner J, Sakinc T. A novel role for D-alanylation of lipoteichoic acid of Enterococcus faecalis in urinary tract infection. PLoS One 2014; 9:e107827; PMID:25296179; https://doi.org/https://doi.org/10.1371/journal.pone.0107827
- Johnson JR, Clabots C, Hirt H, Waters C, Dunny G. Enterococcal aggregation substance and binding substance are not major contributors to urinary tract colonization by Enterococcus faecalis in a mouse model of ascending unobstructed urinary tract infection. Infect Immunity 2004; 72:2445-8; PMID:15039379; https://doi.org/https://doi.org/10.1128/IAI.72.4.2445-2448.2004
- Rosen DA, Hung CS, Kline KA, Hultgren SJ. Streptozocin-induced diabetic mouse model of urinary tract infection. Infect Immunity 2008; 76:4290-8; PMID:18644886; https://doi.org/https://doi.org/10.1128/IAI.00255-08
- National Nosocomial Infections Surveillance (NNIS). System Report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control 2004; 32:470-85; PMID:15573054; https://doi.org/https://doi.org/10.1016/j.ajic.2004.10.001
- Maki DG, Tambyah PA. Engineering out the risk for infection with urinary catheters. Emerg Infect Dis 2001; 7:342-7; PMID:11294737; https://doi.org/https://doi.org/10.3201/eid0702.010240
- Guiton PS, Hung CS, Hancock LE, Caparon MG, Hultgren SJ. Enterococcal biofilm formation and virulence in an optimized murine model of foreign body-associated urinary tract infections. Infect Immunity 2010; 78:4166-75; PMID:20696830; https://doi.org/https://doi.org/10.1128/IAI.00711-10
- Rousseau M, Goh HMS, Holec S, Albert ML, Williams RBH, Ingersoll MA, Kline KA. Bladder catheterization increases susceptibility to infection that can be prevented by prophylactic antibiotic treatment. JCI Insight 2016; 1:e88178; PMID:27699248; https://doi.org/https://doi.org/10.1172/jci.insight.88178
- Guiton PS, Hannan TJ, Ford B, Caparon MG, Hultgren SJ. Enterococcus faecalis overcomes foreign body-mediated inflammation to establish urinary tract infections. Infect Immunity 2012; 81:329-39; PMID:23132492; https://doi.org/https://doi.org/10.1128/IAI.00856-12
- Flores-Mireles AL, Pinkner JS, Caparon MG, Hultgren SJ. EbpA vaccine antibodies block binding of Enterococcus faecalis to fibrinogen to prevent catheter-associated bladder infection in mice. Sci Transl Med 2014; 6:254ra127; PMID:25232179; https://doi.org/https://doi.org/10.1126/scitranslmed.3009384
- Flores-Mireles AL, Walker JN, Bauman TM, Potretzke AM, Schreiber HLt, Park AM, Pinkner JS, Caparon MG, Hultgren SJ, Desai A. Fibrinogen release and deposition on urinary catheters placed during urologic procedures. J Urol 2016; 196(2):416-21; PMID:26707506
- Nielsen HV, Guiton PS, Kline KA, Port GC, Pinkner JS, Neiers F, Normark S, Henriques-Normark B, Caparon MG, Hultgren SJ. The metal ion-dependent adhesion site motif of the Enterococcus faecalis EbpA pilin mediates pilus function in catheter-associated urinary tract infection. MBio 2012; 3:e00177-12; PMID:22829678; https://doi.org/https://doi.org/10.1128/mBio.00177-12
- Kim HY, Choe HS, Lee DS, Yoo JM, Lee SJ. A novel rat model of catheter-associated urinary tract infection. Int Urol Nephrol 2015; 47:1259-63; PMID:26122120; https://doi.org/https://doi.org/10.1007/s11255-015-1038-5
- Ozcimen M, Kurtoglu MG, Goktas S, Omeroglu E, Sakarya Y, Alpfidan I, Ozcimen S, Sakarya R, Yener HI, Demir LS, et al. Daptomycin versus vancomycin in an Enterococcus faecalis endophthalmitis rabbit model. Curr Eye Res 2016; 41:232-9; PMID:25658242; https://doi.org/https://doi.org/10.3109/02713683.2015.1004722
- Frank KL, Colomer-Winter C, Grindle SM, Lemos JA, Schlievert PM, Dunny GM. Transcriptome analysis of Enterococcus faecalis during mammalian infection shows cells undergo adaptation and exist in a stringent response state. PLoS One 2014; 9:e115839; PMID:25545155; https://doi.org/https://doi.org/10.1371/journal.pone.0115839
- Anderson DJ, Podgorny K, Berrios-Torres SI, Bratzler DW, Dellinger EP, Greene L, Nyquist AC, Saiman L, Yokoe DS, Maragakis LL, et al. Strategies to prevent surgical site infections in acute care hospitals: 2014 update. Infect Control Hospital Epidemiol 2014; 35 Suppl 2:S66-88; PMID:25376070; https://doi.org/https://doi.org/10.1017/S0899823X00193869
- Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infect Control Hospital Epidemiol 2013; 34:1-14; PMID:23221186; https://doi.org/https://doi.org/10.1086/668770
- Hancock LE, Gilmore MS. The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. Proc Natl Acad Sci US A 2002; 99:1574-9; PMID:11830672; https://doi.org/https://doi.org/10.1073/pnas.032448299
- Sundqvist G, Figdor D, Persson S, Sjogren U. Microbiologic analysis of teeth with failed endodontic treatment and the outcome of conservative re-treatment. Oral Surg Oral Med Oral Pathol Oral Radiol Endodontol 1998; 85:86-93; https://doi.org/https://doi.org/10.1016/S1079-2104(98)90404-8
- Lee SW, Shet UK, Park SW, Lim HP, Yun KD, Kang SS, Kim SE. Identification of Enterococcus faecalis antigens specifically expressed in vivo. Restorative Dentistry Endodontics 2015; 40:306-11; PMID:26587417; https://doi.org/https://doi.org/10.5395/rde.2015.40.4.306
- Meijer AH, Spaink HP. Host-pathogen interactions made transparent with the zebrafish model. Curr Drug Targets 2011; 12:1000-17; PMID:21366518; https://doi.org/https://doi.org/10.2174/138945011795677809
- Prajsnar TK, Renshaw SA, Ogryzko NV, Foster SJ, Serror P, Mesnage S. Zebrafish as a novel vertebrate model to dissect enterococcal pathogenesis. Infect Immunity 2013; 81:4271-9; PMID:24002065; https://doi.org/https://doi.org/10.1128/IAI.00976-13
- Onyiah JC, Sheikh SZ, Maharshak N, Steinbach EC, Russo SM, Kobayashi T, Mackey LC, Hansen JJ, Moeser AJ, Rawls JF, et al. Carbon monoxide and heme oxygenase-1 prevent intestinal inflammation in mice by promoting bacterial clearance. Gastroenterology 2013; 144:789-98; PMID:23266559; https://doi.org/https://doi.org/10.1053/j.gastro.2012.12.025
- Trede NS, Langenau DM, Traver D, Look AT, Zon LI. The use of zebrafish to understand immunity. Immunity 2004; 20:367-79; PMID:15084267; https://doi.org/https://doi.org/10.1016/S1074-7613(04)00084-6
- Sitges-Serra A, Lopez MJ, Girvent M, Almirall S, Sancho JJ. Postoperative Enterococcal infection after treatment of complicated intra-abdominal sepsis. Br J Surg 2002; 89:361-7; PMID:11872065; https://doi.org/https://doi.org/10.1046/j.0007-1323.2001.02023.x
- Caballero-Granado FJ, Becerril B, Cisneros JM, Cuberos L, Moreno I, Pachon J. Case-control study of risk factors for the development of Enterococcal bacteremia. Eur J Clin Microbiol Infect Dis 2001; 20:83-90; PMID:11305477
- Lagnf AM, Zasowski EJ, Claeys KC, Casapao AM, Rybak MJ. Comparison of clinical outcomes and risk factors in polymicrobial versus monomicrobial Enterococcal bloodstream infections. Am J Infect Control 2016; 44:917-21; PMID:27079241; https://doi.org/https://doi.org/10.1016/j.ajic.2016.02.017
- Tsuchimori N, Hayashi R, Shino A, Yamazaki T, Okonogi K. Enterococcus faecalis aggravates pyelonephritis caused by Pseudomonas aeruginosa in experimental ascending mixed urinary tract infection in mice. Infect Immunity 1994; 62:4534-41; PMID:7927719
- Dalton T, Dowd SE, Wolcott RD, Sun Y, Watters C, Griswold JA, Rumbaugh KP. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One 2011; 6:e27317; PMID:22076151; https://doi.org/https://doi.org/10.1371/journal.pone.0027317
- Brook I, Frazier EH. Microbiology of subphrenic abscesses: a 14-year experience. Am Surgeon 1999; 65:1049-53; PMID:10551755
- Tzianabos AO, Onderdonk AB, Rosner B, Cisneros RL, Kasper DL. Structural features of polysaccharides that induce intra-abdominal abscesses. Science 1993; 262:416-9; PMID:8211161; https://doi.org/https://doi.org/10.1126/science.8211161
- Montravers P, Gauzit R, Muller C, Marmuse JP, Fichelle A, Desmonts JM. Emergence of antibiotic-resistant bacteria in cases of peritonitis after intraabdominal surgery affects the efficacy of empirical antimicrobial therapy. Clin Infect Dis 1996; 23:486-94; PMID:8879770; https://doi.org/https://doi.org/10.1093/clinids/23.3.486
- Nichols RL, Muzik AC. Enterococcal infections in surgical patients: the mystery continues. Clin Infect Dis 1992; 15:72-6; PMID:1617075; https://doi.org/https://doi.org/10.1093/clinids/15.1.72
- Patel R. Clinical impact of vancomycin-resistant Enterococci. J Antimicrobial Chemotherapy 2003; 51 Suppl 3:iii13-21; PMID:12801938
- Willems RJ, Top J, van Santen M, Robinson DA, Coque TM, Baquero F, Grundmann H, Bonten MJ. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis 2005; 11:821-8; PMID:15963275; https://doi.org/https://doi.org/10.3201/1106.041204
- Burnett RJ, Haverstock DC, Dellinger EP, Reinhart HH, Bohnen JM, Rotstein OD, Vogel SB, Solomkin JS. Definition of the role of Enterococcus in intraabdominal infection: analysis of a prospective randomized trial. Surgery 1995; 118:716-21; discussion 21-3; PMID:7570327; https://doi.org/https://doi.org/10.1016/S0039-6060(05)80040-6
- Hopkins JA, Lee JC, Wilson SE. Susceptibility of intra-abdominal isolates at operation: a predictor of postoperative infection. Am Surgeon 1993; 59:791-6
- Safdar N, Maki DG. The commonality of risk factors for nosocomial colonization and infection with antimicrobial-resistant Staphylococcus aureus, Enterococcus, gram-negative bacilli, Clostridium difficile, and Candida. Annals Internal Med 2002; 136:834-44; https://doi.org/https://doi.org/10.7326/0003-4819-136-11-200206040-00013
- Sotto A, Lefrant JY, Fabbro-Peray P, Muller L, Tafuri J, Navarro F, Prudhomme M, De La Coussaye JE. Evaluation of antimicrobial therapy management of 120 consecutive patients with secondary peritonitis. J Antimicrobial Chemotherapy 2002; 50:569-76; PMID:12356803; https://doi.org/https://doi.org/10.1093/jac/dkf167
- Leendertse M, Willems RJ, Oei GA, Florquin S, Bonten MJ, van der Poll T. Intestinal Enterococcus faecium colonization improves host defense during polymicrobial peritonitis. J Infect Dis 2009; 200:735-44; PMID:19604046; https://doi.org/https://doi.org/10.1086/603542
- Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan MW, Ray KP, Solari R, Johnson CD, Ewbank JJ. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol 2001; 11:809-21; PMID:11516642; https://doi.org/https://doi.org/10.1016/S0960-9822(01)00241-X
- Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV, Murray BE, Calderwood SB, Ausubel FM. A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci U S A 2001; 98:10892-7; PMID:11535834; https://doi.org/https://doi.org/10.1073/pnas.191378698
- Maadani A, Fox KA, Mylonakis E, Garsin DA. Enterococcus faecalis mutations affecting virulence in the Caenorhabditis elegans model host. Infect Immunity 2007; 75:2634-7; PMID:17307944; https://doi.org/https://doi.org/10.1128/IAI.01372-06
- Moy TI, Mylonakis E, Calderwood SB, Ausubel FM. Cytotoxicity of hydrogen peroxide produced by Enterococcus faecium. Infect Immunity 2004; 72:4512-20; PMID:15271910; https://doi.org/https://doi.org/10.1128/IAI.72.8.4512-4520.2004
- La Rosa SL, Snipen LG, Murray BE, Willems RJ, Gilmore MS, Diep DB, Nes IF, Brede DA. A genomic virulence reference map of Enterococcus faecalis reveals an important contribution of phage03-like elements in nosocomial genetic lineages to pathogenicity in a Caenorhabditis elegans infection model. Infect Immunity 2015; 83:2156-67; PMID:25776747; https://doi.org/https://doi.org/10.1128/IAI.02801-14
- Sim S, Hibberd ML. Caenorhabditis elegans susceptibility to gut Enterococcus faecalis infection is associated with fat metabolism and epithelial junction integrity. BioMed Central Microbiol 2016; 16:6; PMID:26769134
- Neuhaus K, Lamparter MC, Zolch B, Landstorfer R, Simon S, Spanier B, Ehrmann MA, Vogel RF. Probiotic Enterococcus faecalis Symbioflor(R) down regulates virulence genes of EHEC in vitro and decrease pathogenicity in a Caenorhabditis elegans model. Arch Microbiol 2016; doi:https://doi.org/10.1007/s00203-016-1291-8; PMID:27655246
- King KC, Brockhurst MA, Vasieva O, Paterson S, Betts A, Ford SA, Frost CL, Horsburgh MJ, Haldenby S, Hurst GD. Rapid evolution of microbe-mediated protection against pathogens in a worm host. ISME J 2016; 10:1915-24; PMID:26978164; https://doi.org/https://doi.org/10.1038/ismej.2015.259
- Glavis-Bloom J, Muhammed M, Mylonakis E. Of model hosts and man: using Caenorhabditis elegans, Drosophila melanogaster and Galleria mellonella as model hosts for infectious disease research. Adv Exp Med Biol 2012; 710:11-7; PMID:22127881
- Luther MK, Arvanitis M, Mylonakis E, LaPlante KL. Activity of daptomycin or linezolid in combination with rifampin or gentamicin against biofilm-forming Enterococcus faecalis or E. faecium in an in vitro pharmacodynamic model using simulated endocardial vegetations and an in vivo survival assay using Galleria mellonella larvae. Antimicrobial Agents Chemotherapy 2014; 58:4612-20; PMID:24867993; https://doi.org/https://doi.org/10.1128/AAC.02790-13
- Gaspar F, Teixeira N, Rigottier-Gois L, Marujo P, Nielsen-LeRoux C, Crespo MT, Lopes Mde F, Serror P. Virulence of Enterococcus faecalis dairy strains in an insect model: the role of fsrB and gelE. Microbiology 2009; 155:3564-71; PMID:19696101; https://doi.org/https://doi.org/10.1099/mic.0.030775-0
- Martini C, Michaux C, Bugli F, Arcovito A, Iavarone F, Cacaci M, Paroni Sterbini F, Hartke A, Sauvageot N, Sanguinetti M, et al. The polyamine N-acetyltransferase-like enzyme PmvE plays a role in the virulence of Enterococcus faecalis. Infect Immunity 2015; 83:364-71; PMID:25385793; https://doi.org/https://doi.org/10.1128/IAI.02585-14
- Benachour A, Ladjouzi R, Le Jeune A, Hebert L, Thorpe S, Courtin P, Chapot-Chartier MP, Prajsnar TK, Foster SJ, Mesnage S. The lysozyme-induced peptidoglycan N-acetylglucosamine deacetylase PgdA (EF1843) is required for Enterococcus faecalis virulence. JBacteriol 2012; 194:6066-73; PMID:22961856; https://doi.org/https://doi.org/10.1128/JB.00981-12
- Mylonakis E. Galleria mellonella and the study of fungal pathogenesis: making the case for another genetically tractable model host. Mycopathologia 2008; 165:1-3; PMID:18060516; https://doi.org/https://doi.org/10.1007/s11046-007-9082-z
- Olsen RJ, Watkins ME, Cantu CC, Beres SB, Musser JM. Virulence of serotype M3 Group A Streptococcus strains in wax worms (Galleria mellonella larvae). Virulence 2011; 2:111-9; PMID:21258213; https://doi.org/https://doi.org/10.4161/viru.2.2.14338
- Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 2001; 414:756-9; PMID:11742401; https://doi.org/https://doi.org/10.1038/414756a
- Teixeira N, Varahan S, Gorman MJ, Palmer KL, Zaidman-Remy A, Yokohata R, Nakayama J, Hancock LE, Jacinto A, Gilmore MS, et al. Drosophila host model reveals new Enterococcus faecalis quorum-sensing associated virulence factors. PLoS One 2013; 8:e64740; PMID:23734216; https://doi.org/https://doi.org/10.1371/journal.pone.0064740
- Cox CR, Gilmore MS. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect Immunity 2007; 75:1565-76; PMID:17220307; https://doi.org/https://doi.org/10.1128/IAI.01496-06
- Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. Common virulence factors for bacterial pathogenicity in plants and animals. Science 1995; 268:1899-902; PMID:7604262; https://doi.org/https://doi.org/10.1126/science.7604262
- Jha AK, Bais HP, Vivanco JM. Enterococcus faecalis mammalian virulence-related factors exhibit potent pathogenicity in the Arabidopsis thaliana plant model. Infect Immunity 2005; 73:464-75; PMID:15618186; https://doi.org/https://doi.org/10.1128/IAI.73.1.464-475.2005
- Kirzinger MW, Nadarasah G, Stavrinides J. Insights into cross-kingdom plant pathogenic bacteria. Genes (Basel) 2011; 2:980-97; PMID:24710301
- Turner KH, Everett J, Trivedi U, Rumbaugh KP, Whiteley M. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genetics 2014; 10:e1004518; PMID:25057820; https://doi.org/https://doi.org/10.1371/journal.pgen.1004518
- Sun J, Hu XL, Le GW, Shi YH. Inhibition of Fe-induced colon oxidative stress by Lactobacilli in mice. World J Microbiol Biotechnol 2013; 29:209-16; https://doi.org/https://doi.org/10.1007/s11274-012-1172-5
- Wullt B, Svanborg C. Deliberate establishment of asymptomatic bacteriuria-a novel strategy to prevent recurrent UTI. Pathogens 2016; 5:52; https://doi.org/https://doi.org/10.3390/pathogens5030052
- Cascioferro S, Totsika M, Schillaci D. Sortase A: an ideal target for anti-virulence drug development. Microbial Pathogenesis 2014; 77:105-12; PMID:25457798; https://doi.org/https://doi.org/10.1016/j.micpath.2014.10.007
- Gentry-Weeks C, Estay M, Loui C, Baker D. Intravenous mouse infection model for studying the pathology of Enterococcus faecalis infections. Infect Immunity 2003; 71:1434-41; PMID:12595461; https://doi.org/https://doi.org/10.1128/IAI.71.3.1434-1441.2003
- Buer J, Balling R. Mice, microbes and models of infection. Nat Rev Genetics 2003; 4:195-205; PMID:12610524; https://doi.org/https://doi.org/10.1038/nrg1019
- Moore RJ, Stanley D. Experimental design considerations in microbiota/inflammation studies. Clin Transl Immunol 2016; 5:e92; PMID:27525065; https://doi.org/https://doi.org/10.1038/cti.2016.41
- Zuluaga AF, Salazar BE, Rodriguez CA, Zapata AX, Agudelo M, Vesga O. Neutropenia induced in outbred mice by a simplified low-dose cyclophosphamide regimen: characterization and applicability to diverse experimental models of infectious diseases. BioMed Central Infect Diseases 2006; 6:55; PMID:16545113; https://doi.org/https://doi.org/10.1186/1471-2334-6-55
- Ingersoll MA, Kline KA, Nielsen HV, Hultgren SJ. G-CSF induction early in uropathogenic Escherichia coli infection of the urinary tract modulates host immunity. Cell Microbiol 2008; 10:2568-78; PMID:18754853; https://doi.org/https://doi.org/10.1111/j.1462-5822.2008.01230.x
- Grimaldi JC, Yu NX, Grunig G, Seymour BW, Cottrez F, Robinson DS, Hosken N, Ferlin WG, Wu X, Soto H, et al. Depletion of eosinophils in mice through the use of antibodies specific for C-C chemokine receptor 3 (CCR3). J Leukocyte Biol 1999; 65:846-53; PMID:10380909
- Demant P. Cancer susceptibility in the mouse: genetics, biology and implications for human cancer. Nat Rev Genetics 2003; 4:721-34; PMID:12951573; https://doi.org/https://doi.org/10.1038/nrg1157
- Rosenthal N, Brown S. The mouse ascending: perspectives for human-disease models. Nat Cell Biol 2007; 9:993-9; PMID:17762889; https://doi.org/https://doi.org/10.1038/ncb437
- Zhao G, Hochwalt PC, Usui ML, Underwood RA, Singh PK, James GA, Stewart PS, Fleckman P, Olerud JE. Delayed wound healing in diabetic (db/db) mice with Pseudomonas aeruginosa biofilm challenge: a model for the study of chronic wounds. Wound Repair Regeneration 2010; 18:467-77; PMID:20731798; https://doi.org/https://doi.org/10.1111/j.1524-475X.2010.00608.x
- Giacometti A, Cirioni O, Schimizzi AM, Del Prete MS, Barchiesi F, D'Errico MM, Petrelli E, Scalise G. Epidemiology and microbiology of surgical wound infections. J Clin Microbiol 2000; 38:918-22; PMID:10655417
- Nair N, Biswas R, Gotz F, Biswas L. Impact of Staphylococcus aureus on pathogenesis in polymicrobial infections. Infect Immunity 2014; 82:2162-9; PMID:24643542; https://doi.org/https://doi.org/10.1128/IAI.00059-14
- Iduh UM, Chollom CS, Nuhu A, Spencer THI, Nura MB, Ashcroft OF, Faruku N. Nosocomial infections in post-operative wounds due to Staphylococcus aureus and Pseudomonas aeruginosa in Benue State Nigeria. African J Microbiol Res 2015; 9:1989-96; https://doi.org/https://doi.org/10.5897/AJMR2014.6809
- Mottola C, Mendes JJ, Cristino JM, Cavaco-Silva P, Tavares L, Oliveira M. Polymicrobial biofilms by diabetic foot clinical isolates. Folia Microbiologica (Praha) 2016; 61:35-43; https://doi.org/https://doi.org/10.1007/s12223-015-0401-3
- Cassat JE, Skaar EP. Recent advances in experimental models of osteomyelitis. Exp Rev Anti-Infective Therapy 2013; 11:1263-5; PMID:24215241; https://doi.org/https://doi.org/10.1586/14787210.2013.858600
- Meurens F, Summerfield A, Nauwynck H, Saif L, Gerdts V. The pig: a model for human infectious diseases. Trends Microbiol 2012; 20:50-7; https://doi.org/https://doi.org/10.1016/j.tim.2011.11.002
- Gardner MB, Luciw PA. Macaque models of human infectious disease. ILAR J 2008; 49:220-55; PMID:18323583; https://doi.org/https://doi.org/10.1093/ilar.49.2.220
- Flajnik MF. Comparative analyses of immunoglobulin genes: surprises and portents. Nat Rev Immunol 2002; 2:688-98; PMID:12209137; https://doi.org/https://doi.org/10.1038/nri889
- Heinz S, Haehnel V, Karaghiosoff M, Schwarzfischer L, Muller M, Krause SW, Rehli M. Species-specific regulation of Toll-like receptor 3 genes in men and mice. J Biol Chem 2003; 278:21502-9; PMID:12672806; https://doi.org/https://doi.org/10.1074/jbc.M301476200
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature 2002; 420:520-62; PMID:12466850; https://doi.org/https://doi.org/10.1038/nature01262
- Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 2007; 449:811-8; PMID:17943117; https://doi.org/https://doi.org/10.1038/nature06245
- Jiminez JA, Uwiera TC, Douglas Inglis G, Uwiera RR. Animal models to study acute and chronic intestinal inflammation in mammals. Gut Pathogens 2015; 7:29; PMID:26561503; https://doi.org/https://doi.org/10.1186/s13099-015-0076-y
- Hvidberg H, Struve C, Krogfelt KA, Christensen N, Rasmussen SN, Frimodt-Moller N. Development of a long-term ascending urinary tract infection mouse model for antibiotic treatment studies. Antimicrobial Agents Chemotherapy 2000; 44:156-63; PMID:10602738; https://doi.org/https://doi.org/10.1128/AAC.44.1.156-163.2000
- Mobley HLT, Warren JW, eds. Urinary tract infections: molecular pathogenesis and clinical management. Washington, D.C.: ASM Press, 1996
- Jabr RI, Fry CH. Urinary tract, kidney and bowel. In: Conn MP, ed. Animal models for the study of human disease. Beaverton, Oregon, USA: Academic Press, 2013:461-82
- Dai T, Kharkwal GB, Tanaka M, Huang YY, Bil de Arce VJ, Hamblin MR. Animal models of external traumatic wound infections. Virulence 2011; 2:296-315; PMID:21701256; https://doi.org/https://doi.org/10.4161/viru.2.4.16840
- Hershberger E, Coyle EA, Kaatz GW, Zervos MJ, Rybak MJ. Comparison of a rabbit model of bacterial endocarditis and an in vitro infection model with simulated endocardial vegetations. Antimicrobial Agents Chemotherapy 2000; 44:1921-4; PMID:10858355; https://doi.org/https://doi.org/10.1128/AAC.44.7.1921-1924.2000
- Hady WA, Bayer AS, Xiong YQ. Experimental endocarditis model of methicillin resistant Staphylococcus aureus (MRSA) in rat. J Vis Exp 2012; 64:e3863; PMID:22711072
- Popov D, Pavlov G. Sepsis models in experimental animals. Trakia J Sci 2013; 1:13-23
- Doi K, Leelahavanichkul A, Yuen PS, Star RA. Animal models of sepsis and sepsis-induced kidney injury. J Clin Invest 2009; 119:2868-78; PMID:19805915; https://doi.org/https://doi.org/10.1172/JCI39421
- Park SY, Kim KM, Lee JH, Seo SJ, Lee IH. Extracellular gelatinase of Enterococcus faecalis destroys a defense system in insect hemolymph and human serum. Infect Immunity 2007; 75:1861-9; PMID:17261598; https://doi.org/https://doi.org/10.1128/IAI.01473-06
- Bao Y, Sakinc T, Laverde D, Wobser D, Benachour A, Theilacker C, Hartke A, Huebner J. Role of mprF1 and mprF2 in the pathogenicity of Enterococcus faecalis. PLoS One 2012; 7:e38458; PMID:22723861; https://doi.org/https://doi.org/10.1371/journal.pone.0038458