ABSTRACT
Staphylococcus pseudintermedius is an opportunistic pathogen in dogs and the most frequent cause of canine pyoderma. Protein A, a potent virulence factor in S. aureus is encoded by the spa gene. S. pseudintermedius possesses genes seemingly analogous to spa, but the expression and the characteristics of their products have not been directly determined. The purpose of this study was to test isolates from major clonal groups for the presence of spa gene orthologs, quantitate their expression levels, and to characterize protein A in S. pseudintermedius.
From the data, it was observed that S. pseudintermedius isolates express genes analogous to spa in S. aureus. Isolates representing major clonal populations in the United States and Europe, ST68 and ST71 respectively, bound significantly higher amounts of canine IgG than isolates with other genetic backgrounds, suggesting that these isolates have a higher density of protein A on their surface. Also, canine IgG bound to protein A on S. pseudintermedius via its Fc region similar to protein A from S. aureus. The mRNA profile differed based on the bacterial sequence types and correlated to the density of protein A on the bacterial surface. Protein A was also found to be secreted during the exponential growth phase. Phagocytosis experiments with S. pseudintermedius show that blocking of protein A enhanced phagocytosis in whole blood, neutrophils and in DH82 canine macrophage-like cell line. Taken together, the results demonstrate that S. pseudintermedius produces protein A that shares S. aureus protein A's ability to bind the Fc region of immunoglobulins and may serve as a potential virulence factor by evading the host immune system.
Introduction
S. pseudintermedius is a Gram positive cocci that belongs to the Staphylococcus intermedius group (SIG).Citation1 It is a common commensal that colonizes the skin and mucous membranes of animals, particularly dogs.Citation2 However, under conditions where there is a breach in the external skin barrier or when the immune system of the host is compromised, S. pseudintermedius can cause severe infection.Citation2,Citation3 S. pseudintermedius is the leading cause of skin and ear infections, and post-operative wound infections in dogs. It is implicated in the pathogenesis of canine pyoderma.Citation2,Citation4 Recently, this organism has developed widespread resistance to methicillin.Citation5-11 Many methicillin-resistant S. pseudintermedius (MRSP) strains are also multidrug resistant.Citation12 For example, in one study by Sasaki et al., all 18 of the MRSP strains detected in their patients were resistant to erythromycin, clindamycin, trimethoprim-sulfamethoxazole, and levofloxacin.Citation13 The development of antimicrobial resistance has not only complicated treatment but is also a considerable threat since S. pseudintermedius has zoonotic potential, and this could risk the spread of antimicrobial resistance to human pathogens like S. aureus.Citation2-4,Citation14 The emergence of antimicrobial resistance has gradually shifted the focus of research in this field to searching for prophylactic methods of control for these pathogen.Citation3 Vaccine development, however, requires a deeper understanding of the surface properties and virulence mechanisms in various strains of S. pseudintermedius.Citation15
Several sequence types (STs) of S. pseudintermedius have been described and their population is genetically diverse.Citation16 ST68, which was considered the epidemic clone in the United States is now found in European countries as well. ST71, the epidemic clone in European countries is now widespread in the U.S as well.Citation14,Citation16-19 Along with ST68 and ST71, ST84 (which is more common in the U.S) and ST45, most commonly associated with chloramphenicol resistance, is prevalent in Asia.Citation14 These STs are considered the most dominant and successful clonal populations but the reason for their success over other strains has yet to be elucidated.
Staphylococci have numerous defenses that allow evasion of the host immune system including immunoglobulin binding proteins such as staphylococcal protein A (Spa).Citation20-22 Protein A, a highly potent virulence and immune evasion factor, is produced by many species of staphylococci.Citation23 Within S. aureus it is a 40–60 kDa cell-wall protein encoded by the spa geneCitation24,Citation25 and secreted during exponential growth phase.Citation26-28 It is synthesized by nearly all S. aureus isolates and binds to the Fc and F (ab’)2 regions of immunoglobulins,Citation29,Citation30 inhibits opsonophagocytic killing,Citation20,Citation23,Citation31 and serves as a B-cell superantigen.Citation32-34 Furthermore, marginal zone B-cells and B-1 cells undergo, preferentially, induced cell death with supraclonal depletion and immune tolerance upon exposure to protein A.Citation35 Thus, protein A may interfere with the efficacy of vaccines that depend on opsonophagocytic antibody production by binding to B-cells and inhibiting their function.Citation36 On the other hand, it has been demonstrated that antibodies directed against protein A have protected mice in experimental models of S. aureus infection.Citation37-39
Protein A-like immunoglobulin binding activity has been correlated with virulence in S. pseudintermedius; however, this conclusion was based on a semi-quantitative dot blot IgG- binding assay and did not take other immunoglobulin-binding proteins into consideration or demonstrate the presence of spa genes.Citation40 Genome sequencing studies have shown that S. pseudintermedius contains genes analogous to those encoding protein A in S. aureus, as well as genes encoding other IgG-binding proteins.Citation15 A putative spa gene was initially described by Moodley et al. in 2008 in the isolate ED99.Citation41 This gene was used to develop a species-specific spa typing protocol. The spa gene ortholog in ED99 is 1389 bp in length and has 68% nucleotide and 55% predicted amino acid identity to the homologous gene in the S. aureus reference strain 8325-4 in which protein A was originally described. The predicted protein contained a number of functional regions and conserved domains that were previously described in S. aureus, including four IgG-binding domains and a polymorphic X-region.Citation15,Citation41 Lack of IgG-binding domain region B (58 amino acids) partly accounted for the shorter protein in S. pseudintermedius ED99 compared to protein A in S. aureus.Citation41
Bannoehr et al. in 2011 suggested that there were two predicted orthologues of S. aureus spa gene in ED99, spsP (designated as spa2 and encodes the protein SpsP) and spsQ (designated as spa1 and encodes the protein SpsQ, which is analogous to the full length protein A in S. aureus).Citation15 The spsP gene encodes a protein that consists of 377 amino acids in ED99. This protein is 73% identical to SpsQ at the amino acid level. It has a predicted N-terminal sequence of 33 amino acids, followed by a repeat region consisting of three predicted IgG-binding domains. Each IgG-binding domain consists of 55 amino acids with 78%-85% identity between domains. The C-terminal region has a predicted X-region which shares 63% overall sequence similarity to the X-region of S. aureus. The spsQ gene encodes a protein that is 462 amino acids in ED99. It has a predicted N-terminal sequence of 33 amino acids, followed by a repeat region consisting of four IgG-binding domains. Domains one and two have 59 amino acids each, domain three has 55 amino acids and domain four has 52 amino acids. The domains share between 59%-86% identity at the protein level. The repeated IgG-binding domains are well conserved between SpsP and SpsQ, with 67% to 90% sequence identity in pairwise alignments. The C-terminal region consists of a predicted X-region which includes a 77 amino acid-long repeat sequence (Xr) and a constant region (Xc) with 70% similarity to the X-region of S.aureus.Citation41
Whole genome sequencing of three important S. pseudintermedius clinical isolatesCitation42 along with several unpublished S. pseudintermedius genomes from this lab have shown that while most S. pseudintermedius isolates have a full-length spsQ gene, they also less frequently, harbor the full length spsP gene (Riley, M.C., et al. unpublished data). It is not known, however, whether or not these genes are expressed and produce functional protein in S. pseudintermedius, or if there are differences in the level of protein A expression in different strains. The purpose of this study was to characterize protein A in S. pseudintermedius. In order to do this, 18 clinical S. pseudintermedius isolates with varying genetic backgrounds were selected and examined. The presence/absence of gene encoding protein A and its expression were determined by conventional and real-time PCR respectively. Gene expression profiles were correlated with surface protein expression and the ability of S. pseudintermedius protein A to bind canine IgG and its Fab and Fc fragments was determined. In addition, the ability of secreted protein A in S. pseudintermedius to function as a superantigen for canine B-cells was also investigated. Finally, the involvement of protein A in evasion of phagocytosis was examined. The results from this study suggest that S. pseudintermedius expresses genes analogous to protein A in S. aureus. Protein A is found to be both cell wall-associated and secreted. It binds to canine IgG via its Fc region and functions as a potent immune evasion factor similar to protein A in S. aureus.
Results
S. pseudintermedius genome contains genes analogous to spa in S. aureus
Whole genome sequences of isolates representing MLST sequence types ST68 (06-3228), ST71 (08-1661) and ST84 (NA45) were examined for the spa gene ortholog.Citation42 Gene-specific primers were designed from a consensus sequence after performing multiple sequence alignment, to amplify both the full-length spsQ gene in all the 18 isolates listed in . The PCR results showed that all but one isolate (57395, which was used as the negative control strain for spa in S. pseudintermedius) examined in this study contained the spsQ gene. The PCR product was ∼1200 base pairs and this was confirmed by agar gel electrophoresis. Further, sequence analysis of the spsQ gene from isolates representing ST68 and ST71 showed that they share 96% identity with that of the previously published spsQ gene from ED99, while the spsQ gene from NA45 shared about 53% identity with that of ED99. The spsQ gene sequences from 06–3228, 08–1661 and NA45 are available in GenBank.Citation42
Table 1. Strains of S. pseudintermedius with their sequence types and methicillin resistance.
Expression of spsQ gene in S. pseudintermedius
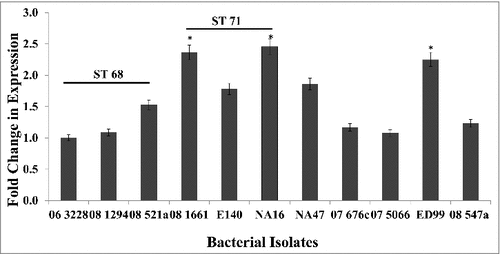
Reverse transcriptase qPCR was used to analyze the expression of spsQ in various S. pseudintermedius isolates listed in Table 1. Expression levels of spsQ varied among sequence types (). For ease and accuracy of data presentation, isolates for which the mRNA levels were extremely low or below the detection limits of the assay have not been included for graphical presentation. Isolates representing ST68 had lower expression (approximately 1.5 log reduction) compared to isolates from ST71, but in general, isolates from ST68 and ST71 had higher spsQ expression than isolates from other sequence types (). The spsQ expression levels in isolates NA12, NA45, KM241, 08–1791, 08–1721 and 07–1447 could not be quantified because they were below the detection limits of the assay. Isolate 57395, which was negative for spsQ gene by conventional PCR, did not produce detectable amounts of spsQ mRNA. spsQ expression levels were normalized against 16S rRNA as the endogenous control and the results were expressed as fold change in mRNA levels using the ΔΔCT method. To ensure equal amplification efficiencies of the target and control genes, four 10-fold serial dilutions of the target RNA were tested. The efficiencies of both the target gene and the endogenous control were comparable.
Figure 1. Expression levels of spsQ in S. pseudintermedius. qPCR was performed to measure the amount of spsQ mRNA in log phase bacterial cells. Relative gene expression was calculated using the 2−ΔΔCT method and expressed as fold change. 16S rRNA was used as the endogenous control. The values represent average from three independent experiments. (#P < 0.05 was considered significant). #spsQ mRNA levels in isolates NA12, NA45, KM241, 08–1791, 08–1721, 07–1447 and 57395 were below the detection limits of the assay.
Canine IgG reacts with protein a on the surface of s. pseudintermedius and binds to protein a via its Fc region
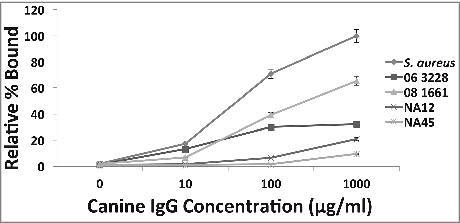
To determine if canine IgG binds to protein A on the surface of S. pseudintermedius, a binding experiment was performed. Canine IgG at concentrations 10 μg/ml, 100 μg/ml and 1000 μg/ml were incubated with log phase cultures of various strains of S. pseudintermedius. A dose response was observed for binding with increasing amounts of canine IgG (), suggesting that canine IgG recognizes and binds to protein A on S. pseudintermedius. The S. aureus isolate Cowan 1 was used as positive control to normalize the amount of binding. Data for isolates representing the major clonal populations ST68 (06-3228), ST71 (08-1661) and ST84 (NA45) are presented in . A similar binding trend was observed for all 18 strains examined in this study.
Figure 2. Binding of canine IgG to protein A on S. pseudintermedius (dose-response). A dose-response effect observed when varying concentrations of canine IgG ranging from 10 μg/ml-1000 μg/ml was reacted with S. pseudintermedius. The values represent average from three independent experiments. (P < 0.05 was considered significant). Data for isolates representing ST 68 (06-3228), ST 71 (08-1661) and ST 84 (NA45) have been presented.
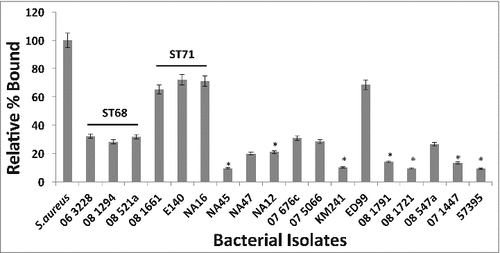
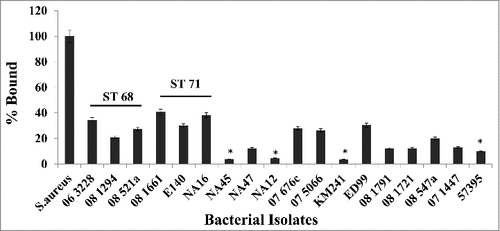
Isolates representing various sequence types (listed in Table 1) were reacted with 100 μg/ml of canine IgG and the amount of binding was measured by flow cytometry. It was observed that isolates representing ST68 and ST71 bound significantly more canine IgG than isolates from other sequence types suggesting that they have a higher density of cell-wall associated protein A on their surface (). Between ST68 and ST71, isolates from ST71 bound significantly higher amounts of canine IgG with 08–1661 (65.3%), E140 (72.1%) and NA16 (71.1%) compared to ST68 isolates 06–3228 (32.2%), 08–1284 (28.3%) and 08–521a (31.8%) (). The amount of binding was normalized against S. aureus Cowan 1 and expressed as the percent bound relative to the positive control. Isolates NA45, NA12, KM241, 08–1791, 08–1721 and 07–1447 for which the spsQ mRNA could not be quantified, bound little to no detectable canine IgG, suggesting that these isolates may have a lower density of protein A on their surface.
Figure 3. Canine IgG recognizes and binds to protein A on S. pseudintermedius. Isolates representing various sequence types of S. pseudintermedius were reacted with 100μg/ml of canine IgG. The amount of binding was determined by flow cytometry and expressed as percentage bound relative to the positive control S. aureus Cowan 1 strain. The values represent average from three independent experiments. (#P < 0.05 was considered significant).
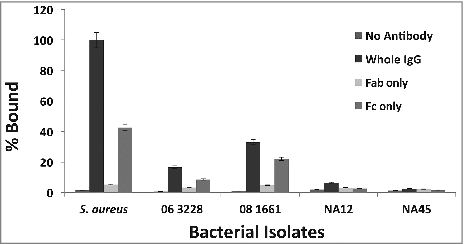
After determining that canine IgG recognizes and binds to protein A on S. pseudintermedius, the next step was to examine which fraction of canine IgG specifically binds to protein A. Protein A in S. aureus binds to IgG primarily via its Fc region.Citation37 It was hypothesized that protein A in S. pseudintermedius also binds IgG via its Fc region. In order to test this, canine IgG was digested with papain to generate Fab and Fc fragments and reacted with bacteria along with whole IgG as the control. The results showed that for isolate 06–3228, the percentage binding was 16.69% (whole IgG), 3.25% (Fab) and 8.57% (Fc). For isolate 08–1661, the percentage binding was 33.09% (whole IgG), 4.79% (Fab) and 22.12% (Fc) (). Isolate NA45 for which the spsQ mRNA could not be quantified also had low density of Protein A on its surface ( and ). Therefore, NA45 was not expected to show any difference in binding with Fab and Fc which was confirmed experimentally (). Taken together, these results suggest that protein A in S. pseudintermedius binds to canine IgG primarily via its Fc region. Data for isolates representing the predominant sequence types 06–3228 (ST68), 08–1661 (ST71) and NA45 (ST84) are presented. A similar binding pattern was observed for all the 18 strains examined in this study. The amount of binding was normalized against S. aureus Cowan 1 and expressed as percent bound relative to the positive control.
Figure 4. Protein A in S. pseudintermedius binds to canine IgG primarily via its Fc region. S. pseudintermedius isolates were treated with IgG and its papain digestion fragments, Fab and Fc. The amount of binding was determined by flow cytometry. The values represent average from three independent experiments. (#P < 0.05 was considered significant). Data for isolates representing ST 68 (06-3228), ST 71 (08-1661) and ST 84 (NA45) are presented.
Cell wall-associated protein A in S. pseudintermedius
The density of surface-anchored protein A on S. pseudintermedius was determined by flow cytometry. Since there was no commercially available antibody against protein A from S. pseudintermedius, FITC conjugated anti-protein A antibody raised against S. aureus protein A was used for this experiment. The amount of binding was normalized against S. aureus Cowan 1 and expressed as percentage bound relative to the positive control. Isolates representing ST68 and ST71 bound significantly more antibody compared to isolates representing other sequence types suggesting that both ST68s and ST71s express a higher density of protein A on their surface. Isolates 06–3228, 08–1294 and 08–521a (representing ST68) bound 34.47%, 20.94% and 27.54% of the labeled antibody respectively; while isolates 08–1661, E140 and NA16 (representing ST71) bound 40.77%, 29.92% and 38.26% of the labeled antibody respectively, relative to Cowan 1 (). Between isolates from ST68 and ST71, it was found that ST71s bound a higher percentage of the antibody, suggesting that ST71s have a higher density of protein A on their surface. Isolates NA12, NA45, KM241, 08–1791, 08–1721 and 07–1447 for which spsQ mRNA could not be quantified, had a very low density of protein A on their surface. Also, as expected, the spsQ-negative isolate 57395 had very low density of protein A on the surface (). Taken together, these results suggest that S. pseudintermedius expresses cell wall-associated protein A that is recognized by the anti-protein A antibody raised against S. aureus protein A.
Figure 5. Protein A on the surface of S. pseudintermedius. The density of cell wall-associated protein A in various strains of S. pseudintermedius was measured using FITC-conjugated anti-protein A antibody by flow cytometry. The values represent the average from three independent experiments. (#P < 0.05 was considered significant).
Anti-protein A antibody inhibits the binding of canine fc fragment to S. pseudintermedius
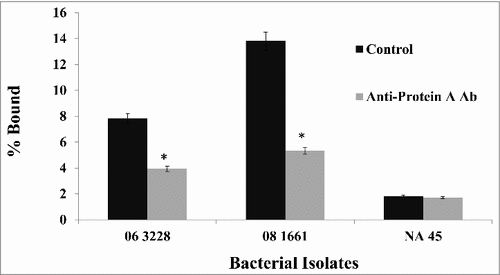
In order to confirm that both canine IgG and anti-Protein A antibody specifically bind to protein A, a competitive inhibition assay was performed. Bacterial cells were incubated with excess unlabeled chicken anti-protein A antibody prior to adding biotin-conjugated canine Fc. Anti-protein A antibody competitively inhibited the binding of canine Fc as determined by a decrease in fluorescence when detected by flow cytometry (). A titration experiment was performed to determine the optimal dilution of both the anti-protein A antibody and the canine Fc fraction that resulted in at least a 50% reduction in binding of canine Fc relative to its own control without anti-protein A antibody. Data for isolates ST68 (06-3228), ST71 (08-1661) and ST84 (NA45) are presented in . Comparable results were observed for all the 18 strains examined in this study. Since isolate NA45 showed very little binding to canine IgG and anti-protein A antibody ( and ), this isolate was not expected to show any inhibition and this is confirmed experimentally. This further strengthened the supposition that S. pseudintermedius expresses protein A on its surface that binds canine IgG and anti-Protein A antibody.
Figure 6. Binding of anti-protein A antibody prevents the binding of canine Fc to protein A on S. pseudintermedius. Bacterial isolates were incubated with an excess of chicken anti-protein A antibody prior to the addition of canine Fc. There was at least 50% reduction in the binding of canine Fc because of anti-protein A antibody binding. The values represent average from three independent experiments. (#P < 0.05 was considered significant). Data for isolates representing ST 68 (06-3228), ST 71 (08-1661) and ST 84 (NA45) are presented.
Protein A in bacterial culture supernatant
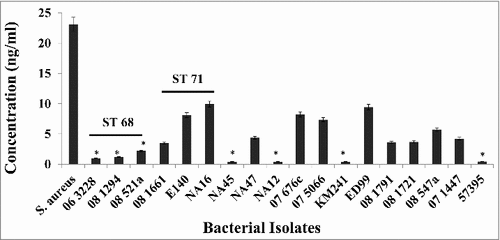
S. aureus isolates secrete protein A during log phase.Citation26,Citation28 A capture ELISA was performed to determine if S. pseudintermedius isolates also secrete protein A. Log phase culture supernatants from various bacterial isolates were tested. The results showed that isolates representing ST68 and ST71 secreted higher amounts of Protein A into the culture supernatant compared to isolates from other sequence types. S.aureus Cowan 1, used as the positive control, secreted 23.1 ng/ml of protein A. Isolates 06–3228, 08–1294 and 08–521 (representing ST68) secreted between 0.98 ng/ml-2.19 ng/ml of protein A. Isolates 06–1661, E140 and NA16 (representing ST71) secreted between 3.45 ng/ml-9.94 ng/ml of protein A (). Between isolates from ST68 and ST71, a significant difference was observed in the amount of secreted protein A, with ST71 isolates secreting higher amounts than ST68s. As expected, isolate 57395, which was negative for the spsQ gene by both conventional and quantitative real-time PCR, did not produce detectable amounts of protein A. Similar results were obtained for isolates NA12, NA45, KM241, 08–1791, 08–1721 and 07–1447. The lower limits of detection and quantification of the assay were 0.125 ng/ml and 0.3 ng/ml respectively. Taken together, these results suggest that S. pseudintermedius not only expresses protein A on its surface but also secretes it into the culture supernatant during log phase.
Figure 7. Production of extracellular protein A in S. pseudintermedius. The amount of extracellular protein A was measured using an antigen-capture ELISA. Commercially available protein A from S. aureus was used to generate the standard curve. The values represent the average from three independent experiments. (#P < 0.05 was considered significant).
Blocking protein A makes S. pseudintermedius more susceptible to phagocytosis in whole blood, canine neutrophils and DH82 cells
In order to determine if protein A in S. pseudintermedius can function as an immune evasion factor, phagocytosis assays were performed using canine whole blood, neutrophils and DH82 canine macrophage-like cells. S. pseudintermedius isolate 08–1661 was labeled with a pH-sensitive dye, pHrodo™ Red which enables easy visualization of phagocytosis. The labeled bacteria fluoresce in the acidic environment of phagosomes, allowing the differentiation and visualization of phagocytosed bacteria from those that are merely adhered to the cell surface.
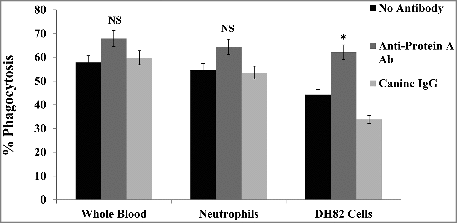
The percentage of phagocytosis after 30 min at 37°C in whole blood, neutrophils and DH82 macrophage-like cells was 57.8%, 54.6% and 44.3% respectively compared to the control which was incubated on ice for the same duration (). To test the involvement of protein A in phagocytosis, pHrodo™ Red labeled bacteria were pre-incubated with 100 μg/ml of either chicken anti-protein A antibody or canine IgG prior to the start of phagocytosis. With both canine blood and neutrophils, pre-incubation of the bacteria with anti-protein A antibody enhanced phagocytosis by at least 10%, although this difference was not significant (). In DH82 cells however, phagocytosis was significantly enhanced by 17.8% in the presence of anti-protein A antibody compared to the control without antibody strongly suggesting that blocking cell wall-associated protein A makes S. pseudintermedius more susceptible to phagocytosis. Canine IgG did not have any effect on phagocytosis (). The duration for phagocytosis was determined by a time course experiment and 30 min was found to be optimal as with previous studies involving S. aureus.Citation43,Citation44 Taken together, these results suggest that protein A in S. pseudintermedius could function as an immune evasion factor similar to protein A in S. aureus, and prevent bacteria from being opsonized and phagocytosed.
Figure 8. Phagocytosis of S. pseudintermedius. The involvement of protein A in immune evasion was demonstrated by phagocytosis using pHrodo™ Red-labeled 08–1661 in whole blood, neutrophils and DH82 cells. Pre-incubation of bacteria with anti-protein A antibody enhanced phagocytosis (red). The values represent average from three independent experiments. (#P < 0.05 was considered significant). NS – not significant.
Discussion
Protein A in S. aureus is an important virulence and immune evasion factor. Nearly all S. aureus isolates tested have been found to produce protein A.Citation23 It is expressed on the peptidoglycan cell wall and is also secreted into the bacterial culture supernatant during exponential growth phase.Citation24,Citation26-28,Citation45 One of the most important features of protein A is its ability to bind IgG via its Fc region, thereby preventing opsonization and phagocytosis.Citation29,Citation30,Citation46 This helps the bacteria evade the host immune response, leading to persistent infections.Citation20-22,Citation31 S. pseudintermedius is an important opportunistic pathogen in dogs. It causes skin and soft tissue infections and is implicated as the main causative agent of canine pyoderma.Citation2,Citation4,Citation15 S. pseudintermedius contains several genes predicted to encode virulence factors similar to S. aureus including genes predicted to encode protein A.Citation2,Citation15,Citation47 Protein A in S. pseudintermedius has not been previously studied or characterized.
In this study, 18 S. pseudintermedius clinical isolates representing the major clonal populations (sequence types) in the United States and Europe were selected and studied. First, the isolates were screened for the presence of the spsQ gene using gene-specific primers and conventional PCR. The mRNA expression profile of spsQ was determined by quantitative real-time PCR. Cell-wall anchored protein A was measured by flow cytometry and the spsQ mRNA levels were correlated to the amount of protein A on the surface. It was found that isolates that had low spsQ mRNA levels also had correspondingly low amounts of cell wall-associated Protein A ( and ). It was found that the gene sequence of protein A in S. pseudintermedius was similar within sequence types and isolates belonging to the same sequence type had similar reactivity with anti-protein A antibody as demonstrated experimentally.
It was also determined that canine IgG reacts with protein A on the surface of S. pseudintermedius via its Fc fragment suggesting that it may help the bacteria escape phagocytosis and serve as an immune evasion factor. Canine IgG used in this study shows both Fc and Fab-mediated binding to protein A, although it predominantly binds via the Fc region. The dominant clonal populations in the United States and Europe, ST68 and ST71, bound more canine IgG and had a higher density of protein A on their surface than isolates of other STs. This property could help explain why ST68 and ST71 are so successful.Citation19 It should be noted that the anti-protein A antibody used in this study was raised against S. aureus protein A. Since this antibody raised against protein A in S. aureus recognizes and binds to protein A on S. pseudintermedius, protein A in these two species apparently share epitopes, however, differences between the two species may affect relative antibody reactivity. Also, there is a possibility that this antibody reacts with epitopes shared on other surface proteins, particularly immunoglobulin binding proteins like Sbi. This is true in the case of S. aureus, where spa deletion mutants have been shown to bind anti-Protein A antibod.Citation48-50 In fact, isolate 57395 which was negative for the spsQ gene by conventional PCR showed some binding to anti-protein A antibody. The presence of extracellular protein A in S. pseudintermedius suggests that it could also function as a superantigen. The toxic effects, if any, of S. pseudintermedius protein A on B-cells are yet to be determined. Phagocytosis experiments with whole blood, neutrophils and DH82 macrophage-like cells indicate that blocking cell wall-associated protein A makes the bacteria more susceptible to phagocytosis. A non-toxigenic protein A-based vaccine has shown promise in animal model of S. aureus infection.Citation38 Phagocytosis and B-cell apoptosis assays with a non-toxigenic version of S. pseudintermedius protein A similar to that in S. aureus protein A are yet to be performed to determine if protein A is a potential vaccine candidate for S. pseudintermedius infection. Taken together, the results suggest that protein A in S. pseudintermedius shares common features with its S. aureus counterpart, and could serve as a potent virulence factor.
Vaccine studies with S. aureus have indicated that a multivalent vaccine with different epitopes would probably work best in preventing infections.Citation36,Citation51,Citation52 Keeping the same factors in mind, the successful development of a vaccine against S. pseudintermedius would involve identifying antigenic targets representative of a wide variety of strains and sequence types. Understanding the nature of surface proteins and their role in virulence and pathogenesis is critical for vaccine development against S. pseudintermedius infection.
Materials and methods
Bacterial strains and growth conditions
S. pseudintermedius clinical isolates (Table 1) were obtained as stock cultures from the Clinical Bacteriology Laboratory (College of Veterinary Medicine, University of Tennessee, Knoxville, Tennessee). Eighteen strains were selected based on their sequence types. Representatives from the most prevalent sequence types in the United States and Europe were chosen for this study. The genetic background of the isolates was determined by multilocus sequence typing (MLST) as described previously.Citation16 The antibiotic susceptibility profile was determined by disk diffusion and interpreted as recommended by the Clinical and Laboratory Standards Institute (CLSI), 2016 guidelines.Citation53 The detection of the mecA gene was performed as described previously.Citation19 S. aureus strain Cowan 1 and the spsP and spsQ negative S. pseudintermedius strain 57395 (a kind gift from Dr. Vincent Perreten, Institute of Veterinary Bacteriology, Vetsuisse Faculty, University of Bern, Bern, Switzerland) were used as the positive control and negative control strains for spa respectively. Bacteria from blood agar plates were grown overnight in 5 ml of Trypticase Soy Broth (TSB, Becton Dickinson, BD 257486) at 37°C in a shaking incubator at 225 rpm. Fifty microliters of overnight culture were inoculated into 5 ml of fresh, sterile TSB and incubated for 3 hours at 37°C with shaking to obtain log phase cultures (OD600 = 0.4–0.6) which were used for all the experiments unless mentioned otherwise.
DNA extraction and conventional PCR
Bacterial DNA was extracted from overnight cultures using a commercial kit (MoBio Ultra Clean DNA Isolation Kit, # 12224–50) according to the manufacturer's protocol. PCR for the spsQ gene was performed using the following primers: spsQ forward 5′-ATCTCAACCTGCTCCTGATTAC-3′ and spsQ reverse 5′-GCATCTTTCGCTTTGTCCATAC-3′. The following cycling conditions were performed: initial denaturation at 95°C for 90 seconds, 30 cycles of annealing at 55°C for 30 seconds and extension at 72°C for 1 minute followed by a final extension at 72°C for 5 minutes. PCR products were sequenced at The University of Tennessee Genomics Core facility, aligned and compared using Geneious 9.1.0 (Biomatters).
RNA extraction and quantitative real time RT-PCR
Total RNA was extracted from log-phase bacterial cultures using a commercial kit (MoBio Ultra Clean RNA Isolation kit, # 15800–50) according to the manufacturer's protocol. Quantitative reverse-transcriptase PCR (RTqPCR) for spsQ was performed using the TaqMan RNA-to-CT 1-Step kit on an ABI StepOne PCR System (Applied Biosystems) using the primer-probe set: 5′-/56-FAM/CGCCAAGTT/ZEN/TCGATGAAGCGCAA/3IABkFQ/-3′, 5′-CCGTTACGTTGCTCTTCAGTA-3′, 5′-TAACCAAACACCTACACAACCT-3′. The following cycling conditions were performed: 48°C for 30 minutes, 95°C for 10 minutes, 95°C for 15 seconds and 40 cycles of 60°C for 1 minute. qPCR data were analyzed by the 2−ΔΔCT method and normalized against 16S rRNA as the endogenous control as previously described.Citation54
Preparation of canine fab and fc fractions from canine IgG and biotin conjugation
Lyophilized canine IgG (IR-DG2-GF, Innovative Research) was dissolved in sterile PBS (pH 7.2). Canine Fab and Fc fractions were prepared using a commercial kit according to the manufacturer's instructions (Pierce Fab preparation kit, Thermo Scientific, # 44985). Briefly, 1 mg/ml of canine IgG equilibrated in digestion buffer was added to papain (immobilized on beads) and incubated for six hours at 37°C with shaking. Enzyme treated-IgG was then added to a protein A column to bind Fc fragments. Unbound Fab fragments were collected and Fc fragments were eluted and immediately adjusted to neutral pH. All fragments were equilibrated in PBS.
Fc and Fab fragments prepared above and whole canine IgG were conjugated to biotin (EZ-Link™ Sulfo-NHS-LC-Biotin, No-Weigh™ Format, ThermoFisher Scientific, # 21327) according to the manufacturer's protocol. Two milligrams each of whole IgG, Fab and Fc were biotinylated. Unbound biotin was removed and the immunoglobulin fractions were equilibrated in PBS by ultrafiltration. The concentration of the fragments was measured using UV280 and the fragments were stored at 4°C until used.
Canine IgG binding assay by flow cytometry
One milliliter of log phase bacterial culture was centrifuged at 10,000xg for 1 minute. The bacteria were washed and suspended in 1 ml of PBS and incubated with 100 μl of canine IgG at concentrations of 10 μg/ml, 100 μg/ml or 1000 μg/ml for 30 minutes at room temperature. The cells were washed twice with PBS and incubated with 100 μl of 1:100 dilution of goat anti-canine IgG (H+L)-FITC conjugate (Southern Biotech, # 6070-02) for 30 minutes at room temperature in the dark. For the binding assay with canine Fab and Fc fractions, log phase bacteria were incubated with 2 μg/ml biotin-conjugated whole IgG, Fab or Fc at room temperature for 30 minutes. Bacteria were washed as before and incubated with 1:500 dilution of avidin-FITC conjugate (Sigma-Aldrich, A2050) at room temperature for 30 minutes in the dark. Unbound conjugate was removed by washing and the amount of binding was determined by flow cytometry.
Protein A binding assay by flow cytometry
One milliliter of log phase bacterial culture was centrifuged at 10,000xg for 1 minute. The pellets were washed twice with PBS and suspended in 1 ml PBS and incubated with 100 μl of 1:100 dilution of chicken anti-protein A-FITC conjugate (Gallus Immunotech, APA-F) at room temperature for 30 minutes in the dark. Unbound antibody was removed by washing and the amount of binding was determined by flow cytometry.
Protein A competitive binding assay
Bacteria were prepared and washed as previously described. One hundred microliters of 1:100 dilution of chicken anti-protein A antibody (Gallus Immunotech, APA) was added to the bacterial cells and incubated for 30 minutes at room temperature. The bacteria were washed as before and 100 μl of 2 μg/ml of biotin-conjugated canine Fc was added and incubated at room temperature for 30 minutes. The bacteria were washed and a 1:500 dilution of avidin-FITC conjugate was added and incubated for an additional 30 minutes in the dark. After washing to remove unbound conjugate, the amount of Fc binding was determined by flow cytometry.
Measurement of secreted protein A by ELISA
Secreted protein A in log-phase culture supernatants was measured using an antigen capture ELISA. Briefly, 96-well flat bottom, costar plates (Corning Inc., # 3590) were coated overnight at 4°C with 1 μg/ml of mouse monoclonal anti-protein A antibody clone SPA-27 (Mouse IgG1 isotype, Sigma-Aldrich, B3150) in PBS. Bacterial supernatant containing secreted protein A was added and incubated at 37°C for 1 hour. Bound protein was detected using a 1:500 dilution of HRP-conjugated chicken anti-protein A antibody (Gallus Immunotech, APA-HRP) followed by addition of 100 μl/well of tetramethylbenzidine (TMB) substrate (Thermo Scientific, N301). Plates were washed in between each step with PBS containing 0.05% polysorbate-20 using an ELX405 auto plate washer (BioTek Instruments Inc.). The reaction was stopped by adding 50 μl/well of 0.18M sulfuric acid. The optical density was measured at 450 nm using an ELX800 Universal Microplate Reader (BioTech Instruments Inc.).
Labeling of bacteria with pHrodo™ Red Amine-reactive Dye
Bacterial isolate 08–1661 was labeled with pHrodo Red, succinimidyl ester (pHrodo™ Red, SE, ThermoFisher Scientific, P36600) according to the manufacturer's instructions with minor variations. Briefly, 1 mg of the pHrodo™ Red SE was dissolved in 150 μl of dimethyl sulfoxide (DMSO) to make a stock solution of 10.2 mM. Five milliliters of overnight bacterial culture was centrifuged at 10,000xg for 5 minutes and the pellet was washed twice with PBS. One milligram of the pellet was suspended in 750 μl of fresh sodium bicarbonate buffer, pH 8.5. The prepared dye was diluted into the bacterial suspension at a final concentration of 1 mM. The tubes were incubated at room temperature for 1 hour in the dark with gentle rocking. After incubation, the cells were washed with 750 μl of Hank's balanced salt solution by centrifugation at 15,000xg for 1 minute. The bacterial pellet was suspended in 1 ml of 100% methanol. The bacteria were centrifuged again at 15,000xg for 1 minute to remove methanol. Finally, the labeled bacteria were resuspended in 500 μl of HBSS and stored in aliquots at −80°C until use.
Phagocytosis in canine blood
Fresh blood was collected in heparinized tubes from healthy dogs following an approved IACUC protocol (UTK IACUC protocol #2474-0716). Briefly, 100 μl of blood was mixed with an equal volume of pHrodo™ Red-labeled bacteria (containing 5 × 105 bacteria) and incubated at 37°C for 30 minutes to facilitate phagocytosis. Control tubes containing all components were incubated on ice for the same duration. Phagocytosis was stopped by addition of ice-cold PBS containing 5 mM EDTA. The cells were washed by centrifugation at 5000xg for 10 minutes, suspended in 2 ml of RBC lysis buffer (Sigma-Aldrich, R7757) and incubated at 37°C for 20 minutes. The lysed RBCs were removed by centrifugation. The pellet was washed twice with PBS and the amount of phagocytosis was determined by flow cytometry. The same assay was performed after isolating neutrophils from whole blood using a method described previously.Citation55
Phagocytosis in DH82 cells
The canine macrophage-like cell line, DH82, was obtained from the American Type Culture Collection (ATCC, CLR-10389), propagated and maintained in Eagle's Minimum Essential Medium (EMEM, Quality Biological Inc., # 112-018-101) containing 15% heat-inactivated fetal bovine serum (Sigma Aldrich, F4135) and 1% penicillin-streptomycin-amphotericin-B mixture (Lonza, # 17–602E). Cells were grown to confluency, trypsinized and seeded into 24-well Costar tissue culture plates (Corning Inc., CLS3527) at a density of 1 × 106 cells per well and allowed to adhere. pHrodo™ Red-labeled bacteria were added at a ratio of 1:100 (macrophage: bacteria) and incubated for 30 minutes at 37°C to facilitate phagocytosis. A control plate with the same components was incubated on ice for the same duration. After incubation, phagocytosis was stopped by the addition of ice cold PBS containing 5 mM EDTA. The plate was allowed to sit for 5 minutes at room temperature. The PBS-EDTA was gently aspirated and the adherent cells were harvested by standard trypsin treatment for 5 minutes at 37°C. The contents from each well were transferred into a microcentrifuge tube and centrifuged at 5000xg for 10 minutes. Cells were washed twice and suspended in 1 ml of PBS and analyzed by flow cytometry. Each sample was run in triplicate and analyzed individually. Non-infected cells served as negative control to set the cut-off for the discrimination of pHrodo™ Red-negative and -positive cells, and the cells incubated on ice served as control to determine the percentage of phagocytosis.
Flow cytometry analysis
Binding experiments with canine IgG, anti-protein A antibody and phagocytosis assay using labeled bacteria were performed by flow cytometry. The amount of binding or phagocytosis was determined by measuring the fluorescence using an Attune acoustic focusing cytometer (Applied Biosystems) at excitation/emission wavelengths of 488/519 nm for green fluorescence and 560/585 nm for red fluorescence. Data were acquired in linear mode for forward and side scatter and logarithmic mode for fluorescence. For each sample, 10,000 events were measured.
Statistical analysis
A one-way ANOVA and multiple comparisons using Tukey's honest significant difference were performed to test if there was a significant difference in the expression of spsQ among bacterial isolates, and also to determine if there was a significant difference in the amount of surface-expressed protein A, secreted protein A and percentage of phagocytosed bacteria among isolates of various sequence types. Each experiment was repeated at least three times and a p-value of <0.05 was considered significant. All analyses were conducted using the GraphPad Prism software (Version 7, GraphPad Software Inc.).
Disclosure of interest
The authors report no conflict of interest.
Acknowledgements
The authors would like to thank Dr. Vincent Perreten (Institute of Veterinary Bacteriology, Vetsuisse Faculty, University of Bern, Bern, Switzerland) for the spa negative isolate 57395. This research was supported in part by a grant from the University of Tennessee, Center of Excellence in Livestock Diseases and Human Health.
Additional information
Funding
References
- Ross Fitzgerald J. The Staphylococcus Intermedius Group of Bacterial Pathogens: Species Re‐Classification, Pathogenesis and the Emergence of Meticillin Resistance. Vet Dermatol. 2009;20(5–6):490–495.
- Van Duijkeren E, Catry B, Greko C, et al. Review on Methicillin-Resistant Staphylococcus Pseudintermedius. J Antimicrob Chemotherx. 2011;66(12):2705–2714. doi:10.1093/jac/dkr367.
- Couto N, Martins J, Lourenço AM, et al. Identification of Vaccine Candidate Antigens of Staphylococcus Pseudintermedius by Whole Proteome Characterization and Serological Proteomic Analyses. J Proteomics. 2016;133:113–124. doi:10.1016/j.jprot.2015.12.017.
- Bannoehr J, Guardabassi L. Staphylococcus Pseudintermedius in the Dog: Taxonomy, Diagnostics, Ecology, Epidemiology and Pathogenicity. Vet Dermatol. 2012;23(4):253–e52. doi:10.1111/j.1365-3164.2012.01046.x.
- Beck KM, Waisglass SE, Dick HL, et al. Prevalence of Meticillin‐Resistant Staphylococcus Pseudintermedius (Mrsp) from Skin and Carriage Sites of Dogs after Treatment of Their Meticillin‐Resistant or Meticillin‐Sensitive Staphylococcal Pyoderma. Veterinary Dermatology. 2012;23(4):369–e67. doi:10.1111/j.1365-3164.2012.01035.x.
- Beever L, Bond R, Graham PA, et al. Increasing Antimicrobial Resistance in Clinical Isolates of Staphylococcus Intermedius Group Bacteria and Emergence of Mrsp in the Uk. The Veterinary Record. 2015;176(7):172–172. doi:10.1136/vr.102651.
- Feng Y, Tian W, Lin D, et al. Prevalence and Characterization of Methicillin-Resistant Staphylococcus Pseudintermedius in Pets from South China. Veterinary Microbiology. 2012;160(3):517–524.
- Haenni M, de Moraes NA, Châtre P, et al. Characterisation of Clinical Canine Meticillin-Resistant and Meticillin-Susceptible Staphylococcus Pseudintermedius in France. Journal of Global Antimicrobial Resistance. 2014;2(2):119–123. doi:10.1016/j.jgar.2014.02.002.
- Nienhoff U, Kadlec K, Chaberny IF, et al. Methicillin-Resistant Staphylococcus Pseudintermedius among Dogs Admitted to a Small Animal Hospital. Veterinary Microbiology. 2011;150(1):191–197.
- Wang Y, Yang J, Logue CM, et al. Methicillin‐Resistant Staphylococcus Pseudintermedius Isolated from Canine Pyoderma in North China. Journal of Applied Microbiology. 2012;112(4):623–630. doi:10.1111/j.1365-2672.2012.05233.x.
- Ruscher C, Lübke-Becker A, Wleklinski CG, et al. Prevalence of Methicillin-Resistant Staphylococcus Pseudintermedius Isolated from Clinical Samples of Companion Animals and Equidaes. Veterinary Microbiology. 2009;136(1):197–201.
- Moodley A, Damborg P, Nielsen SS. Antimicrobial Resistance in Methicillin Susceptible and Methicillin Resistant Staphylococcus Pseudintermedius of Canine Origin: Literature Review from 1980 to 2013. Veterinary Microbiology. 2014;171(3):337–341.
- Sasaki T, Kikuchi K, Tanaka Y, et al. Methicillin-Resistant Staphylococcus Pseudintermedius in a Veterinary Teaching Hospital. Journal of Clinical Microbiology. 2007;45(4):1118–1125. doi:10.1128/JCM.02193-06.
- Dos Santos TP, Damborg P, Moodley A, et al. Systematic Review on Global Epidemiology of Methicillin-Resistant Staphylococcus Pseudintermedius: Inference of Population Structure from Multilocus Sequence Typing Data. Front Microbiol. 2016;7.
- Bannoehr J, Ben Zakour NL, Reglinski M, et al. Genomic and Surface Proteomic Analysis of the Canine Pathogen Staphylococcus Pseudintermedius Reveals Proteins That Mediate Adherence to the Extracellular Matrix. Infection and Immunity. 2011;79(8):3074–3086. doi:10.1128/IAI.00137-11.
- Solyman S, Black CC, Duim B, et al. Multilocus Sequence Typing for Characterization of Staphylococcus Pseudintermedius. Journal of Clinical Microbiology. 2013;51(1):306–310. doi:10.1128/JCM.02421-12.
- Perreten V, Kadlec K, Schwarz S, et al. Clonal Spread of Methicillin-Resistant Staphylococcus Pseudintermedius in Europe and North America: An International Multicentre Study. J Antimicrob Chemother. 2010;dkq078.
- Black C, Solyman SM, Eberlein LC, et al. Identification of a Predominant Multilocus Sequence Type, Pulsed-Field Gel Electrophoresis Cluster, and Novel Staphylococcal Chromosomal Cassette in Clinical Isolates of Meca-Containing, Methicillin-Resistant Staphylococcus Pseudintermedius. Veterinary Microbiology. 2009;139(3):333–338.
- Videla R, Solyman SM, Brahmbhatt A, et al. Clonal Complexes and Antimicrobial Susceptibility Profiles of Staphylococcus Pseudintermedius Isolates from Dogs in the United States. Microbial Drug Resistance. 2017. [Epub ahead of print]. doi:10.1089/mdr.2016.0250.
- Foster TJ. Immune Evasion by Staphylococci. Nature Reviews Microbiology. 2005;3(12):948–958.
- Kim HK, Thammavongsa V, Schneewind O, et al. Recurrent Infections and Immune Evasion Strategies of Staphylococcus Aureus. Current Opinion in Microbiology. 2012;15(1):92–99. doi:10.1016/j.mib.2011.10.012.
- Rooijakkers, SH, Van Kessel KP, Van Strijp JA. Staphylococcal Innate Immune Evasion. Trends in Microbiology. 2005;13(12):596–601. doi:10.1016/j.tim.2005.10.002.
- Forsgren A. Significance of Protein a Production by Staphylococci. Infection and Immunity. 1970;2(5):672.
- Sjöquist J, Movitz J, Johansson IB, et al. Localization of Protein a in the Bacteria. European Journal of Biochemistry. 1972;30(1):190–194. doi:10.1111/j.1432-1033.1972.tb02086.x.
- Björk, I, Petersson BÅ, Sjöquist J. Some Physicochemical Properties of Protein a from Staphylococcus Aureus. European Journal of Biochemistry. 1972;29(3):579–584. doi:10.1111/j.1432-1033.1972.tb02024.x.
- Movitz J. Formation of Extracellular Protein a by Staphylococcus Aureus. European Journal of Biochemistry. 1976;68(1):291–299. doi:10.1111/j.1432-1033.1976.tb10788.x.
- Lindmark R, Movitz J, Sjöquist J. Extracellular Protein a from a Methicillin‐Resistant Strain of Staphylococcus Aureus. European Journal of Biochemistry. 1977;74(3):623–628. doi:10.1111/j.1432-1033.1977.tb11431.x.
- Becker S, Frankel MB, Schneewind O, et al. Release of Protein a from the Cell Wall of Staphylococcus Aureus. Proceedings of the National Academy of Sciences. 2014;111(4):1574–1579. doi:10.1073/pnas.1317181111.
- Forsgren A, Sjöquist J. “Protein a” from S. Aureus I. Pseudo-Immune Reaction with Human Γ-Globulin. The Journal of Immunology. 1966;97(6):822–827.
- Kronvall G, Frommel D. Definition of Staphylococcal Protein a Reactivity for Human Immunoglobulin G Fragments. Immunochemistry. 1970;7(1):124IN19127–126IN20. doi:10.1016/0019-2791(70)90036-4.
- Dossett JH, Kronvall G, Williams RC Jr, et al. Antiphagocytic Effects of Staphylococcal Protein A. J Immunol. 1969;103(6):1405–1410.
- Silverman GJ, Goodyear CS. A Model B-Cell Superantigen and the Immunobiology of B Lymphocytes. Clinical Immunology. 2002;102(2):117–134. doi:10.1006/clim.2001.5143.
- Kristiansen SV, Pascual V, Lipsky PE. Staphylococcal Protein a Induces Biased Production of Ig by Vh3-Expressing B Lymphocytes. The Journal of Immunology. 1994;153(7):2974–2982.
- Silverman GJ, Goodyear CS. Confounding B-Cell Defences: Lessons from a Staphylococcal Superantigen. Nature Reviews Immunology. 2006;6(6):465–75. doi:10.1038/nri1853.
- Palmqvist N, Silverman GJ, Josefsson E, et al. Bacterial Cell Wall-Expressed Protein a Triggers Supraclonal B-Cell Responses Upon in Vivo Infection with Staphylococcus Aureus. Microbes and Infection. 2005;7(15):1501–1511. doi:10.1016/j.micinf.2005.05.008.
- Daum RS, Spellberg B. Progress toward a Staphylococcus Aureus Vaccine. Clinical Infectious Diseases. 2012;54(4):560–7. doi:10.1093/cid/cir828.
- Kim HK, Emolo C, DeDent AC, et al. Protein a-Specific Monoclonal Antibodies and Prevention of Staphylococcus Aureus Disease in Mice. Infection and Immunity. 2012;80(10):3460–3470. doi:10.1128/IAI.00230-12.
- Kim HK, Cheng AG, Kim HY, et al. Nontoxigenic Protein a Vaccine for Methicillin-Resistant Staphylococcus Aureus Infections in Mice. Journal of Experimental Medicine. 2010;207(9):1863–1870. doi:10.1084/jem.20092514.
- Kim HK, Emolo C, Missiakas D, et al. A Monoclonal Antibody That Recognizes the E Domain of Staphylococcal Protein A. Vaccine. 2014;32(4):464–469. doi:10.1016/j.vaccine.2013.11.054.
- Garbacz K, Zarnowska S, Piechowicz L, et al. Pathogenicity Potential of Staphylococcus Pseudintermedius Strains Isolated from Canine Carriers and from Dogs with Infection Signs. Virulence. 2013;4(3):255–259. doi:10.4161/viru.23526.
- Moodley A, Stegger M, Ben Zakour NL, et al. Tandem Repeat Sequence Analysis of Staphylococcal Protein a (Spa) Gene in Methicillin-Resistant Staphylococcus Pseudintermedius. Veterinary Microbiology. 2009;135(3):320–326.
- Riley MC, Perreten V, Bemis DA, et al. Complete Genome Sequences of Three Important Methicillin-Resistant Clinical Isolates of Staphylococcus Pseudintermedius. Genome announcements. 2016;4(5):e01194–16. doi:10.1128/genomeA.01194-16.
- Lu T, Porter AR, Kennedy AD, et al. Phagocytosis and Killing of Staphylococcus Aureus by Human Neutrophils. Journal of Innate Immunity. 2014;6(5):639–649. doi:10.1159/000360478.
- Salgado, M, Pignatari ACC, Bellinati-Pires R. Phagocytosis and Killing of Epidemic Methicillin-Resistant Staphylococcus Aureus by Human Neutrophils and Monocytes. Brazilian Journal of Infectious Diseases. 2004;8(1):80–89.
- Schneewind O, Model P, Fischetti VA. Sorting of Protein a to the Staphylococcal Cell Wall. Cell. 1992;70(2):267–281. doi:10.1016/0092-8674(92)90101-H.
- Forsgren A, Sjöquist J. “Protein a” from Staphylococcus Aureus Iii. Reaction with Rabbit Γ-Globulin. The Journal of Immunology. 1967;99(1):19–24.
- Geoghegan JA, Smith EJ, Speziale P, et al. Staphylococcus Pseudintermedius Expresses Surface Proteins That Closely Resemble Those from Staphylococcus Aureus. Veterinary Microbiology. 2009;138(3):345–352.
- Smith EJ, Visai L, Kerrigan SW, et al. The Sbi Protein Is a Multifunctional Immune Evasion Factor of Staphylococcus Aureus. Infection and Immunity. 2011;79(9):3801–3809. doi:10.1128/IAI.05075-11.
- Zhang L, Jacobsson K, Vasi J, et al. A Second Igg-Binding Protein in Staphylococcus Aureus. Microbiology. 1998;144(4):985–991. doi:10.1099/00221287-144-4-985.
- Atkins KL, Burman JD, Chamberlain ES, et al. S. Aureus Igg-Binding Proteins Spa and Sbi: Host Specificity and Mechanisms of Immune Complex Formation. Molecular Immunology. 2008;45(6):1600–1611. doi:10.1016/j.molimm.2007.10.021.
- Begier E, Seiden DJ, Patton M, et al. Sa4ag, a 4-Antigen Staphylococcus Aureus Vaccine, Rapidly Induces High Levels of Bacteria-Killing Antibodies. Vaccine. 2017;35(8):1132–1139. doi:10.1016/j.vaccine.2017.01.024.
- Bagnoli F, Bertholet S, Grandi G. Inferring Reasons for the Failure of Staphylococcus Aureus Vaccines in Clinical Trials. The Staphylococci and Staphylococcal Pathogenesis. 2012;7.
- Institute CaLS. Clsi: Performance Standards for Antimicrobial Susceptibility Testing 26 ed. Wayne, PA; Clsi Supplement M100s. Vol. 36: 2016.
- Black CC, Eberlein LC, Solyman SM et al. The Role of Meca and Blaz Regulatory Elements in Meca Expression by Regional Clones of Methicillin-Resistant Staphylococcus Pseudintermedius. Vet Microbiol. 2011;151(3–4):345–53. doi:10.1016/j.vetmic.2011.03.026.
- Oh H, Siano B, Diamond S. Neutrophil Isolation Protocol. JoVE (Journal of Visualized Experiments). 2008. (17):e745–e745.
