ABSTRACT
Parasitic fungi are the only pathogens that can infect insect hosts directly through their proteinaceous exoskeleton. Penetration of the cuticle requires the release of fungal enzymes, including proteinases, which act as virulence factors. Insects can sense fungal infections and activate innate immune responses, including the synthesis of antifungal peptides and proteinase inhibitors that neutralize the incoming proteinases. This well-studied host response is epigenetically regulated by histone acetylation/deacetylation. Here we show that entomopathogenic fungi can in turn sense the presence of insect-derived antifungal peptides and proteinase inhibitors, and respond by inducing the synthesis of chymotrypsin-like proteinases and metalloproteinases that degrade the host-derived defense molecules. The rapidity of this response is dependent on the virulence of the fungal strain. We confirmed the specificity of the pathogen response to host-derived defense molecules by LC/MS and RT-PCR analysis, and correlated this process with the epigenetic regulation of histone acetylation/deacetylation. This cascade of responses reveals that the coevolution of pathogens and hosts can involve a complex series of attacks and counterattacks based on communication between the invading fungal pathogen and its insect host. The resolution of this process determines whether or not pathogenesis is successful.
Introduction
The insect integument provides an efficient physical barrier against infection, and most pathogens therefore infect their insect hosts via the oral route when ingested with food. Only pathogenic fungi such as Metarhizium robertsii (Hypocreales, Clavicipitaceae), formerly known as Metarhizium anisopliae, can invade insect hosts directly through the cuticle. The latter is a structurally and chemically complex tissue, comprising a waxy layer of fatty acids, lipids and sterols, covering an epicuticle that predominantly consists of sclerotized proteins and smaller amounts of chitin.Citation1–3
The binding of fungal conidia to the insect cuticle is mediated by hydrophobic interactions and results in germination, accompanied by the release of enzymes such as lipases and proteinases.Citation4 Entomopathogenic fungi enter their host by a combination of physical force and enzymatic degradation.Citation5 The conidia have limited resources to produce enzymes and the host cuticle contains proteinase inhibitors. Theory suggests that only fungal proteinases that are uninhibited or incompletely inhibited by host-derived proteinase inhibitors can operate as virulence factors because they alone would fulfill the functions necessary for successful infection, such as penetration of the exoskeleton, utilization of host proteins for nutrition, suppression of host cellular immune responses, and degradation of host defense molecules.Citation6 The demand for proteolytic enzymes that are not susceptible to host proteinase inhibitors has imposed diversifying selection during host–pathogen coevolution, resulting in the broad spectrum of proteolytic enzymes produced by entomopathogenic fungi such as M. robertsii. This spectrum encompasses subtilisin-like and chymotrypsin-like proteinases as well as metalloproteinases.Citation7
The selective pressure on fungal virulence-associated proteinases is enhanced by the ability of insect hosts to sense fungal infections and respond by activating immune responses, including the synthesis of antifungal peptides and proteinase inhibitors. The response to M. robertsii involves transcriptomic reprogramming regulated by epigenetic mechanisms such as microRNA expression, which controls protein synthesis at the post-transcriptional level, and the acetylation and deacetylation of histones mediated by histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively.Citation8 The opposing activities of these enzymes are tightly regulated. The acetylation of histones by HATs promotes access to DNA and thus facilitates gene expression, whereas the removal of acetyl groups by HDACs has the opposite effect.Citation9–11
The use of next-generation sequencing to study M. robertsii and its insect hosts at the transcriptomic level has identified a large number of genes that are regulated in either the pathogen or the host during pathogenesis.Citation7 However, these approaches cannot distinguish the causes and consequences of mutually-induced transcriptional responses. Such insights require studies focusing on the interacting molecules. For example, M. robertsii is known to produce a thermolysin-like metalloproteinase from the highly toxic M4 family, which includes a number of prominent virulence factors associated with human pathogens.Citation12 Interestingly, the thermolysin-encoding genes have diversified in the genus Metarhizium.Citation13
The larvae of the greater wax moth Galleria mellonella (Lepidoptera, Pyralidae) can be used as a model host to decipher molecular and cellular interactions between entomopathogenic fungi and their host insects. The presence of even minute amounts of the M. robertsii thermolysin-like metalloproteinase is sufficient to induce immune responses that are qualitatively (spectrum of induced immunity-related proteins) and quantitatively (expression levels of these induced proteins) comparable with those observed following the injection of microbial elicitors.Citation14,15 The thermolysin-mediated hydrolysis of hemolymph proteins and extracellular matrix proteins such as type IV collagen results in the formation of specific peptides which function as danger signals.Citation16,17 The latter indicate the presence of the microbial metalloproteinases and in turn induce the synthesis of the insect metalloproteinase inhibitor (IMPI), which is currently the only known peptide-based and specific inhibitor of thermolysin-like metalloproteinases in animals.Citation18,19 Interestingly, the immunity-related transcriptome of G. mellonella contains at least eight homologous IMPI genes.Citation20,21 The diversification of thermolysin-like metalloproteases in M. robertsii and IMPI genes in G. mellonella may reflect reciprocal adaptations that have occurred during host–pathogen coevolution.Citation20
Transcriptional reprogramming of the host during infection therefore appears to be triggered by microbial enzymes that activate immunity-related genes, which in turn cause feedback regulation in the pathogen.Citation15,17 As proof of principle, we infected G. mellonella larvae with virulent and virulence-attenuated strains of M. robertsii to investigate the strain-dependent expression of selected IMPI homologs. It is unclear how the fungi cope with the destructive anti-fungal molecules and remain viable as a pathogen in the hostile host environment. We therefore designed a reductionist approach to determine whether entomopathogenic fungi can sense the presence of host defense molecules and respond by inducing the synthesis of proteolytic enzymes against these new targets. We used gelatin as a simple model substrate to represent insect cuticle proteins. Metarhizium robertsii conidia placed on gelatin-containing agar or in liquid culture medium can only grow if they produce proteinases to utilize gelatin as a nutritional substrate, and the expression of such proteinases is easy to measure. The addition of insect-derived antifungal peptides such as metchnikowin, or class-specific proteinase inhibitors, allowed us to experimentally mimic a host immune response.Citation22 Our experiments were designed to determine whether parasitic fungi can sense the induction of immune responses in the infected host and counteract the response in a specific manner. We investigated whether host defense molecules such as antimicrobial peptides and proteinase inhibitors can in turn affect conidial germination thereby causing epigenetically regulated transcriptional reprogramming in the entomopathogenic fungi, to induce the synthesis of further enzymes as a counterattack (reciprocal feedback regulation).
Results
Manipulation of immunity-related gene expression in G. mellonella larvae by M. robertsii strains
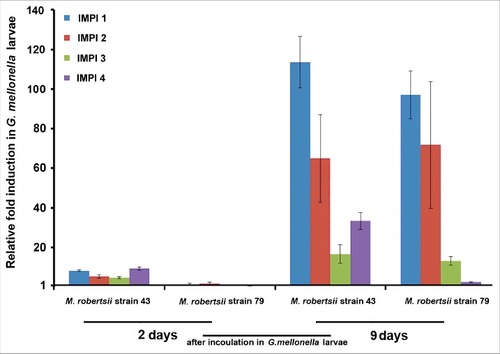
We measured immunity-related gene expression in G. mellonella larvae by inoculating them separately with two strains of M. robertsii. We selected fungal strains showing contrasting virulence in G. mellonella following infection by injection or through the natural route.Citation9 For example, the mortality of the larvae was 100% on day 5 after the injection of strain 43 conidia at 102 cfu/ml, whereas no larval mortality was observed with the same dose of strain 79 at the same time point (Supplementary Fig. 1). When these two fungal strains infected the G. mellonella larvae separately by penetrating the cuticle (natural route), we observed the induction of different homologs of IMPI 2 d after inoculation (). In larvae exposed to strain 79, the expression of all IMPI homologs except IMPI4 was similar to the levels in larvae inoculated with strain 43 after 9 d ().Citation20 We compared the expression levels of individual IMPI homologs in larvae infected with the different fungal strains at each specific time point, between time points in larvae infected with a particular fungal strain, and between treated and untreated last-instar larvae. There was no statistically significant difference in the fold change of IMPI expression when we compared the larvae injected with different strains at any specific time point. There was an overall increase in the expression of IMPI 1 and IMPI 2 between 2 and 9 d in larvae infected with either strain (IMPI 1, p = 0.0002, IMPI 2, p = 0.0003). However, only in larvae infected with strain 43 was IMPI 1 significantly upregulated after 9 d compared to the untreated control (p = 0.01667). In larvae infected with strain 79, the expression of IMPI 3 was also upregulated after 2 d (p = 0.02362).
Figure 1. Transcriptional activation of G. mellonella IMPI homologs in response to pathogenic strains of M. robertsii. Expression levels were determined by RT-PCR relative to the same genes in untreated last-instar larvae and were normalized against the 18S rRNA gene. Data are means of three independent measurements ± standard deviations.
Recognition of antifungal compounds by M. robertsii strains
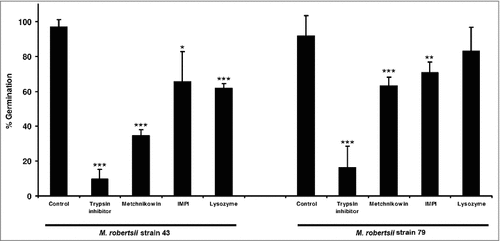
We prepared 1% gelatin substrates individually supplemented with trypsin inhibitor, metchnikowin, IMPI or lysozyme, and then monitored conidial germination (). Conidia grown on 1% gelatin without supplements were used as a control. The control germination frequencies of strains 43 and 79 were 97% and 92%, respectively. After 26 h, the strongest suppression of conidial germination in strains 43 and 79 was achieved by trypsin inhibitor (germination frequencies of 9.8% and 17%, respectively), followed by metchnikowin (germination frequencies of 34% and 63%, respectively), and then IMPI (germination frequencies of 65% and 70%, respectively) (). Lysozyme also inhibited the germination of strains 43 and 79 (germination frequencies of 61% and 84%, respectively). We observed significant differences in the germination frequency of conidia in the presence and absence of antimicrobial peptides for both strain 43 (p = 5.045 × 10−7) and strain 79 (p = 8.598 × 10−8). Compared to untreated controls, the germination frequency of strain 43 conidia was significantly repressed in the presence of trypsin inhibitor (p < 0.001), metchnikowin (p < 0.001), IMPI (p = 0.0298) and lysozyme (p < 0.001). Similarly significant differences were observed for strain 79 in the presence of trypsin inhibitor (p < 0.001), metchnikowin (p < 0.001) and IMPI (p = 0.00157), but not in the presence of lysozyme. We did not compare differences in germination frequencies between strains 43 and 79 in the presence and absence of antimicrobial peptides because this was beyond the scope of the current study. In , the p-values (###p < 0.001, ##p < 0.01, #p < 0.05) represent significant differences in germination frequencies when comparing conidia in the presence and absence (untreated control) of the treatments described above.
Figure 2. Germination of M. robertsii conidia in the presence of antimicrobial peptides and proteinase inhibitors. Conidia of M. robertsii strains 43 and 79 were inoculated in gelatin supplemented with trypsin inhibitor, metchnikowin, IMPI or lysozyme. Germination frequencies were measured 26 h post-inoculation. Data are means of three independent experiments ± standard errors (###p<0.001, ##p<0.01, #p<0.05; treatments compared to untreated controls).
Antifungal peptides induce M. robertsii extracellular proteinase activity
The extracellular proteinase activity of M. robertsii culture supernatants exposed to different antimicrobial compounds was determined using fluorescein isothiocyanate (FITC)-labeled casein as a substrate (). The p-values in (##p < 0.01, #p < 0.05) represent proteinase activities in strains 43 and 79 after treatment, compared to the corresponding untreated control. In the presence of antimicrobial peptides, the proteinase activity differed significantly from the control (black bar) after 24 h in strain 43 (p = 0.009285) and strain 79 (p = 0.009285). Compared to the untreated control, the proteinase activity of strain 43 was significantly enhanced only in response to metchnikowin (p = 0.00402). In contrast, the proteinase activity in strain 79 was significantly reduced in response to metchnikowin (p < 0.04), but it increased significantly in response to trypsin inhibitor, IMPI and lysozyme (in all cases, p < 0.01). We did not detect any significant differences in proteinase activity after 72 h in either strain (data not shown).
Figure 3. Extracellular proteinase activity of M. robertsii in the presence of antimicrobial peptides and proteinase inhibitors. (A) Nonspecific extracellular proteinase activity in M. robertsii strains 43 and 79 following the application of trypsin inhibitor, metchnikowin, IMPI or lysozyme was determined by fluorimetry using casein as a substrate. (B) Metalloproteinase activity was determined using the fluorigenic peptide Mca-RPPGFSAFK (Dnp)-OH as a substrate. Data are means of three independent experiments ± standard errors (###p<0.001, ##p<0.01, #p<0.05; treatments compared to untreated controls).
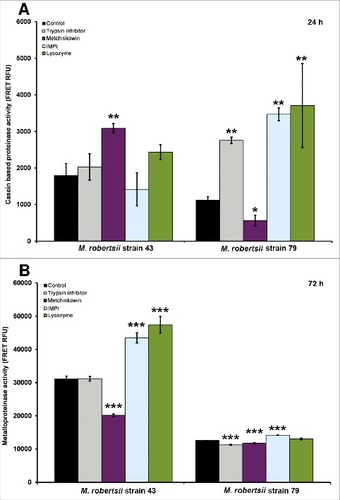
We also used the fluorigenic peptide Mca-RPPGFSAFK (Dnp)-OH as a substrate to measure metalloproteinase-specific activity (). The p-values in (###p < 0.001) represent metalloproteinase activities in strains 43 and 79 after treatment compared to the corresponding untreated control. We did not detect any significant differences in metalloproteinase activity after 24 h in either strain (data not shown). However, in the presence of antimicrobial peptides, the metalloproteinase activity differed significantly from the control (black bar) after 72 h in strain 43 (p = 8.98 × 10−8) and strain 79 (p = 1.458 × 10−7). Compared to the control, the metalloproteinase activity of strain 43 was significantly enhanced only in response to IMPI (p < 1 × 10−6) and lysozyme (p = 1.6 × 10−6) but was significantly reduced in response to metchnikowin (p < 1 × 10−6). Metalloproteinase activity in strain 79 declined in response to trypsin inhibitor (p < 1 × 10−4) and metchnikowin (p = 0.000203), and increased slightly in response to IMPI (p < 1 × 10−4).
Metarhizium robertsii can selectively regulate the expression of virulence genes to circumvent insect antifungal responses
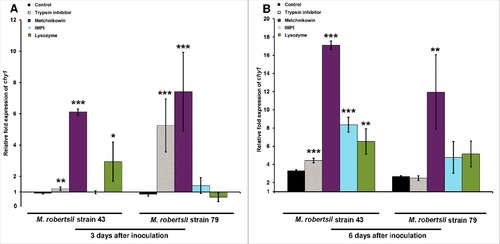
The ability of M. robertsii to counteract insect antifungal responses was investigated by monitoring chymotrypsin and metalloproteinase gene expression by RT-PCR. We found that the inoculation of M. robertsii onto a gelatin substrate supplemented individually with trypsin inhibitor, IMPI, metchnikowin or lysozyme resulted in the differential expression of chy1, mep1 and a gene encoding an M4 metallopeptidase-like protein. We observed the differential expression of chy1 in strain 43 in the presence and absence of antimicrobial peptides 3 days (p = 5.064 × 10−5) and 6 days (p = 1.711 × 10−7) after inoculation, and similarly in strain 79 after 3 days (p = 1.711 × 10−7) and 6 days (p = 0.0208). The chy1 gene was selectively and strongly induced in response to gelatin supplemented with metchnikowin in strain 43 (p < 0.001) and in strain 79 (p<1 × 10−4) 3 days after inoculation (), whereas in response to trypsin inhibitor chy1 was induced to a limited extent in strain 43 (p = 0.00765) and more robustly in strain 79 (p = 0.0112). Lysozyme triggered the upregulation of chy1 only in strain 43 (p = 0.04352). The expression of chy1 increased further in strain 43 (p < 0.001) and strain 79 (p = 0.00209) 6 days after inoculation in response to metchnikowin (). We also observed the selective upregulation of chy1 in response to trypsin inhibitor (p < 0.001), lysozyme (p = 0.00515), and IMPI (p < 0.001) in strain 43, but not in strain 79, 6 days after inoculation (). The p-values in (###p < 0.001, ##p < 0.01, #p < 0.05) represent gene expression levels in strains 43 and 79 after treatment compared to the corresponding untreated controls.
Figure 4. Differential expression of M. robertsii chy1 in response to proteinase inhibitors and antimicrobial peptides. The expression of M. robertsii chy1 after (A) 3 d and (B) 6 d exposure to gelatin and gelatin supplemented with trypsin inhibitor, metchnikowin, IMPI or lysozyme was determined by RT-PCR relative to expression levels in PDA without antimicrobial compounds or proteinase inhibitor. Metarhizium robertsii strains 43 and 79 inoculated in gelatin were used as controls. Values were normalized against the housekeeping gene gpd. Data are means of three independent experiments ± standard errors (###p<0.001, ##p<0.01, #p<0.05; treatments compared to untreated controls).
Similarly, we observed the differential expression of mep1 in strain 43 (p = 1.137 × 10−7) and strain 79 (p = 1.318 × 10−5) in the presence and absence of antimicrobial peptides 3 days after inoculation, but no differences in expression 6 days after inoculation. The expression of mep1 was induced in strain 79 in response to metchnikowin (p < 0.001), trypsin inhibitor (p < 0.001), IMPI (p < 0.001) and lysozyme (p = 0.0026) 3 days after inoculation, but in strain 43 mep1 was only induced in response to lysozyme (p < 0.001) at the same time point (). After 6 days, the opposite mep1 expression profile was observed in strain 43 in response to each of the antimicrobial peptides, although the differences were statistically insignificant (). The p-values in (###p < 0.001, ##p < 0.01) represent gene expression levels in strains 43 and 79 after treatment compared to the corresponding untreated controls. Initially the expression of the M4 metallopeptidase was induced in both strains, particularly in response to metchnikowin, but the expression levels only increased in strain 43 at a later time point (Supplementary Fig. 2).
Figure 5. Differential expression of M. robertsii mep1 in response to proteinase inhibitors and antimicrobial peptides. The expression of M. robertsii mep1 after (A) 3 d and (B) 6 d exposure to gelatin and gelatin supplemented with trypsin inhibitor, metchnikowin, IMPI or lysozyme was determined by RT-PCR relative to expression levels in PDA without antimicrobial compounds and proteinase inhibitors. Metarhizium robertsii strains 43 and 79 inoculated in gelatin were used as controls. Values were normalized against the housekeeping gene gpd. Data are means of three independent experiments ± standard errors (###p<0.001, ##p<0.01, #p<0.05; treatments compared to untreated controls).
To determine whether the selective reprogramming of virulence genes in response to specific antimicrobial challenges was regulated by histone acetylation, we selected HDAC and HAT genes based on their gene ontology (GO), and monitored their expression at the same time as the virulence genes (Supplementary Tables 1, 2 and 3). We found that the combined expression of HDACs and HATs complemented the expression of chymotrypsin and metalloproteinases, particularly in response to metchnikowin and IMPI, 3 and 6 days after inoculating strain 43 in gelatin (Supplementary Figures 3A and 4A). Similarly, the expression of HDACs and HATs complemented the expression of proteinases following exposure to metchnikowin, trypsin inhibitor and IMPI, 3 and 6 days after inoculating strain 79 in gelatin (Supplementary Figure 3B and 4B).
Degradation of metchnikowin and trypsin inhibitor by M. robertsii
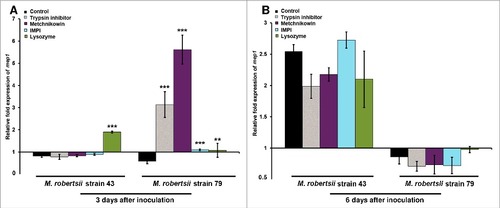
We used LC/MS to determine whether the observed fungal responses to metchnikowin ultimately led to the degradation of the antifungal peptide and proteinase inhibitor. We therefore inoculated M. robertsii conidia into potato dextrose broth (PDB), supplemented the broth with either metchnikowin or trypsin inhibitor, and monitored their degradation for 3 days by screening for their multiply-charged pseudomolecular ions. The antifungal peptide metchnikowin () and trypsin inhibitor () were represented by strong and well-defined pseudomolecular ions at time points 0 and 72 h, similar to their purified state (Supplementary Figures 5–8). However, the metchnikowin signal declined substantially when exposed to strains 79 () and 43 (), and a similar profile was observed for trypsin inhibitor following exposure to strains 79 () and 43 (). These data confirm that both strains can degrade these insect-derived antimicrobial compounds albeit at different rates, with the more aggressive strain 43 achieving more rapid degradation than the less aggressive strain 79.
Figure 6. Detection of multiply-charged pseudomolecular ions of metchnikowin. (A-B) Extracted ion current of the 5+ (m/z = 606.0) and 4+ (m/z = 757.5) charged pseudomolecular ions of metchnikowin following inoculation in PDB containing M. robertsii strains 43 or 79 at time point 0. (C-D) Corresponding data 72 h after inoculation for strain 79. (E-F) Corresponding data 72 h after inoculation for strain 43. The close-up shows the ESI+ mass spectrum in the m/z range 500–1100 (tR 19.1–19.7 min).
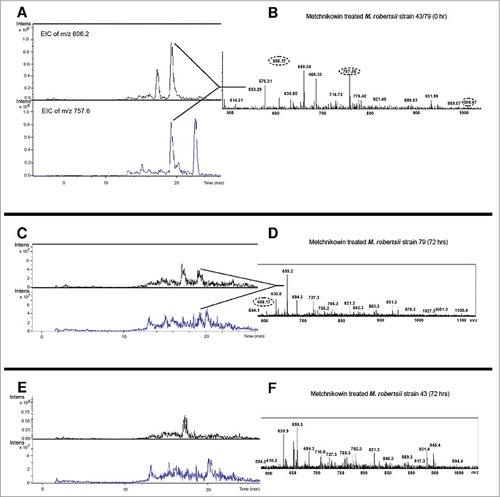
Figure 7. Detection of multiply-charged pseudomolecular ions of trypsin inhibitor. (A-B) Extracted ion current of the 5+ (m/z = 1052.4) and 4+ (m/z = 1110.8) charged pseudomolecular ions of trypsin inhibitor following inoculation in PDB containing M. robertsii strains 43 or 79 at time point 0. (C-D) Corresponding data 72 h after inoculation for strain 79. (E-F) Corresponding data 72 h after inoculation for strain 43. The close-up shows the ESI+ mass spectrum in the m/z range 800–1400 (tR 28.9–29.7 min).
Discussion
The pathogenicity of fungi relies on their ability to overcome innate immunity, and in the case of entomopathogenic fungi this involves the secretion of proteinases that breach the cuticle and allow colonization of the insect host.Citation1,20,23 Insects can detect these proteinases and release peptide-based proteinase inhibitors and antimicrobial peptides as defense molecules.Citation24 However, this antagonistic communication is not achieved by the expression of a static panel of inhibitors, as evident from the rapid emergence of insect resistance against fungi and other entomopathogens.Citation20 For example, insects have evolved resistance against harmful stimuli by increasing the number, expression level and/or functional diversity of genes encoding antimicrobial peptides, enzymes and detoxifying chemicals, albeit with an associated fitness penalty.Citation25,26
Metarhizium spp. are known to respond to insect gene diversification by modulating the release of subtilisin-like proteinases, trypsin-like serine proteinases and metalloproteinases, which in turn determine their host range.Citation27,28 It is unclear whether such responses improve the ability of fungi to destroy insect defense molecules, but the interaction between fungal toxins and insect defense molecules is likely to exert strong positive selection pressure on proteinases and proteinase inhibitors that cannot be inhibited.Citation6 For example, the IMPI produced by G. mellonella appears to have undergone a functional shift from a serine proteinase inhibitor to a M4 metalloproteinase inhibitor.Citation29 Thus, fungal pathogens coevolve infection strategies to withstand specialized host antimicrobials through reciprocal adaptation. Our data show that M. robertsii can not only sense the presence of proteinase inhibitors and antifungal peptides produced by insects, but can counterattack by selectively expressing chymotrypsin and metalloproteinases that target the insect defense molecules for degradation. This implies that the fungus has evolved a strategy that reduces fitness costs and avoids the unnecessary waste of stored insecticidal resources during cuticle penetration, when external resources are unavailable.
Pathogenic fungi survive inside their insect hosts by releasing secondary metabolites.Citation30 For example, insecticidal destruxin and cytochalasin produced by M. robertsii affect the morphology and cytoskeletal arrangements of insect immune-system cells. Destruxin causes cytoskeleton remodeling in hemocytes and tetanic paralysis in insects by opening Ca2+ channels and triggering membrane depolarization.Citation31,32 Cytochalasin inhibits the formation of bundled actin filaments and microtubules, but has a lower impact than destruxin on G. mellonella larvae.Citation31 M. robertsii also evades the insect immune response by expressing the collagenous protein Mcl1 to prevent phagocytosis and encapsulation by hemocytes.Citation33 The exposure of G. mellonella larvae to destruxin and cytochalasin can selectively regulate the expression of antimicrobial proteins such as lysozyme and IMPI, mimicking infection by pathogenic strains of M. robertsii.Citation32 This indicates the existence of an adapted fungal strategy to release secondary metabolites that modulate the insect immune system and thus ensure its activity remains below harmful levels.Citation34 However, these inferences do not explain how two pathogenic strains (43 and 79) of M. robertsii differ in their ability to kill G. mellonella larvae upon exposure. The high-level expression of IMPI homologs was achieved earlier in larvae infected with the more aggressive strain 43 than the less aggressive strain 79. Even purified IMPI inhibited the germination of M. robertsii conidia similarly to lysozyme, trypsin inhibitor and the antifungal peptide metchnikowin. This suggests that pathogenicity in M. robertsii is determined by the timely release of selective proteinases which destroy the antimicrobials released by the host to prevent hyphal penetration of the insect cuticle. Moreover, the growth of non-virulent strains of M. robertsii can be arrested by increasing the lysozyme concentration, but this is not effective against virulent strains, suggesting the importance of proteinases as a strategy to disrupt antimicrobial resistance.Citation6 This was confirmed by the induction of proteinase activity in M. robertsii conidia following the switch from potato dextrose agar (PDA) to gelatin as a source of nutrients, because the latter mimics the sclerotic layer of the insect cuticle.
We observed higher metalloproteinase activity in the two different M. robertsii strains when they encountered alternative insect-derived antimicrobial compounds in the gelatin substrate, indicating their ability to sense and induce specific counterattacks against the antifungal components metchnikowin, trypsin inhibitor and lysozyme. Similarly, bacterial pathogens activate specific virulence genes such as phoQ when they recognize antimicrobial peptides.Citation35 The presence of metchnikowin specifically induced the expression of M. robertsii genes encoding chymotrypsin, M4 and M43 metallopeptidases. The expression levels were epigenetically regulated by histone acetylation, which is known to regulate the life cycles of human pathogens and innate immune response in insects following infection by the entomopathogenic bacterium Bacillus thuringiensis (Bacillales, Bacillaceae).Citation10,36–41 For example, acetylation of histone H3 lysine 56 maintains the genome integrity and virulence of Candida albicans (Saccharomycetales, Saccharomycetaceae).Citation39,40 The induction of HDAC and HAT genes in M. robertsii particularly in response to metchnikowin is an indicator of epigenetically regulated transcriptional activation, which has been customized to degrade this specific antifungal peptide. Metchnikowin specifically inhibits the growth of filamentous fungi and affects conidial germination in M. robertsii.Citation42
Serine proteinases and metalloproteinases targeting metchnikowin were abundant and active in strains 43 and 79. We observed the complete degradation of metchnikowin by strain 43, as shown by the rapid disappearance of pseudomolecular ions representing this compound, whereas degradation took longer and was less efficient in the presence of strain 79. A similar profile was observed for the degradation of trypsin inhibitor, suggesting that the aggressiveness of fungal strains may depend on their ability to recognize and counteract diverse insect defense molecules. Strain-dependent differences in virulence affect the host range of M. robertsii. Pathogens with a broad host range are not specifically adapted to subvert host immune responses, and thus aggressively kill insects with toxins. Specialized pathogenic strains kill their host more slowly, achieving the maximum utilization of host nutrients by prolonging the infection process.Citation27,43
We show that the entomopathogenic fungus M. robertsii can sense antimicrobial peptides and proteinase inhibitors produced by insects and then counterattack by expressing selected proteinases that cause the specific degradation of these defense compounds. The rapidity of the response is strain dependent and epigenetically regulated to induce insect mortality after infection. In conclusion, our results indicate that fungal pathogenesis is the culmination of attacks and counterattacks between pathogens and insects that has required the coevolution of reciprocal communication involving specific interacting molecules. The experimental manipulation of virulence factors and the expression of antifungal molecules will further improve our understanding of the evolutionary arms race between entomopathogenic fungi and their insect hosts.
Materials and methods
Insect rearing and infection
Galleria mellonella larvae were reared on an artificial diet (TropicShop, Germany, 20329) at 32°C in darkness. Last-instar larvae, each weighing 250–350 mg, were used for all experiments. Approximately, 104 conidia/ml from strains 43 and 79 were serially diluted and separately injected into larvae to determine mortality at 27°C. Simultaneously, natural infection was mimicked by exposing larvae to culture plates containing M. robertsii conidia for 5 min. Conidia that had adhered to the larval cuticle following exposure to the fungus were counted using a Neubauer Chamber by dipping individual larvae (at least five larvae per strain) into 1 ml PBS containing Triton X-100. The number of M. robertsii conidia was 2902/larva (standard error = 100.1). Inoculated larvae were maintained at 27°C, relative humidity 80%, and fed on an artificial diet for RNA isolation on the second and ninth days.Citation9 All experiments were repeated with a new batch of insects and fungal inoculum.
Fungal strains, culture with antifungal compounds and isolation of conidia
Strains 43 and 79 of the parasitic fungus M. robertsii were obtained from the Julius-Kühn-Institute (Darmstadt, Germany) and were maintained on PDA (Carl Roth) at 27°C for 10 days to initiate conidiogenesis. The conidia were washed with 0.02% Triton X-100, filtered through miracloth to remove mycelia and sonicated before inoculation. We inoculated 100 μl of conidia (∼7 × 107 conidia/ml) from each strain into 1% gelatin (Merck-Millipore, 9000-70-8) or PDB (Carl Roth) supplemented with insect-derived antimicrobial peptides and proteinase inhibitors, and maintained the cultures aerobically at 27°C. The precise response of M. robertsii to insect antimicrobials was determined by growing the fungal strains on gelatin rather than a standard growth medium such as PDA. The germination of conidia in 1% gelatin with or without insect-derived antimicrobial peptides and proteinase inhibitors was measured 26 h after inoculation using a Zeiss Axioplan microscope. Trypsin inhibitor (Carl Roth, 5279.1), lysozyme (Carl Roth, 8259.1), IMPI1 (prepared as previously describedCitation19) and synthetic metchnikowin in dimethylformamide (JPT Peptide Technologies) were dissolved in sterile water before mixing with 1% gelatin or PDB. The final concentrations were prepared as follows: trypsin inhibitor (6 μl of a 2.5 mg/ml stock dissolved in 1.5 ml medium to a final concentration of 10 μg/ml), metchnikowin (7.5 μl of a 10 mg/ml stock dissolved in 1.5 ml medium to a final concentration of 50 μg/ml), IMPI1 (55 μl of a 1.354 mg/ml stock dissolved in 1.5 ml medium to a final concentration of 50 μg/ml), and lysozyme (7.5 μl of a 10 mg/ml stock dissolved in 1.5 ml medium to a final concentration of 50 μg/ml).
Metarhizium robertsii proteinase assay
Metarhizium robertsii extracellular proteinases including metalloproteinases were detected in culture supernatants by inoculating 100 μl M. robertsii conidia (∼7 × 107 conidia/ml) into PDB supplemented with trypsin inhibitor, metchnikowin, IMPI1 or lysozyme (prepared as described above) and incubating the cultures at 27°C. The supernatants were collected after 24 and 72 h and centrifuged at 3000 × g for 5 min in order to remove fungal debris. Extracellular proteinase activity was measured in 100 µl culture supernatant using 100 µl FITC-labeled casein (Elastin Products Company, JB890) or the metalloproteinase-specific substrate Mca-RPPGFSAFK (Dnp)-OH fluorigenic peptide (R & d Systems, ES005). Cleavage of the substrate by fungal proteinases resulted in free reporter molecules which were quantified using a fluorescence microtiter plate reader (BioTek) at 27°C.
RNA isolation and RT-PCR
RNA from naturally infected larvae, and from fungal strains exposed to antifungal peptides and proteinase inhibitors, was isolated using the RNeasy Plant Mini Kit (Qiagen, 74903) following the manufacturer's recommendations. The quality of RNA was determined by ethidium bromide gel staining and the quantity was determined by spectrophotometry. Complementary DNA was synthesized using the First Stand cDNA synthesis kit (Thermo Fisher Scientific, K1621). Quantitative real-time PCR was carried out using the CFX 96 real-time PCR system (BioRad, Mx3000P) and the SensiMix-Sybr No-Rox Kit (Bioline, BIO-98005) according to the manufacturers' protocols, with the gene-specific primers listed in Supplementary Table 4. Gene expression in M. robertsii and G. mellonella was measured using CFX Manager software (BioRad) relative to the reference genes gpd (MAA_07675) and 18S rRNA (AF286298), respectively.
Liquid chromatography–mass spectrometry
The degradation of antimicrobial compounds was monitored using a high-resolution micrOTOF Q-II mass spectrometer (Bruker Daltonics) equipped with an orthogonal electrospray ionization source and coupled to an UltiMate 3000 HPLC instrument (Dionex) controlled by HyStar v3.2. Metarhizium robertsii conidia (∼7 × 107 conidia/ml) were inoculated into PDB with or without metchnikowin or trypsin inhibitor. Fungal culture supernatants were sampled after 72 h, and were screened in positive ion mode for multiply-charged pseudomolecular ions of metchnikowin and trypsin inhibitor in the m/z range 500–1500. The capillary voltage was set to −4500 V, the end plate offset to −500 V, and the nebulizer pressure was adjusted to 1.60 bar. Dry N2 gas (99.9999%) was provided at a flow rate of 8 l/min and a temperature of 250°C. The target mass was set to m/z 1000 at a trap drive level of 100% using the smart parameter settings option. The instrument was operated in the maximum resolution scan mode.
Samples (10 µl) were injected into an Acclaim 120 C18, 3 µm, 120 Å, 2.1 × 150 mm column (Dionex) at a flow rate of 0.25 ml/min at 35°C. UV spectra were recorded at λ = 190–400 nm. Eluent A was water plus 0.1% formic acid, and eluent B was 80% acetonitrile in water plus 0.1% formic acid. LC-MS grade acetonitrile and formic acid were sourced from Sigma-Aldrich and water was purified using a Merck-Millipore Milli-Q system. The column was held at 95% A 5% B for 5 min, then a linear gradient from 5% to 100% B was applied over 40 min. Thereafter, the column was held at 100% B for 10 min, and returned to the initial conditions in 2 min before equilibration for 8 min.
Data analysis
Statistical analysis () was carried out using R v3.3.3 (2017-03-06) (https://www.R-project.org). For (showing the expression of four IMPI homologs) exploratory data analysis using normal q-q-plots did not reveal any evidence against the assumption of normality for the untransformed expression values. Based on this, comparisons with the untreated control larvae (relative expression was set to 1, and hence was constant) were performed using two-sided one-sample t-tests. Comparisons between strains at each time point were performed using Welch's two-sided two-sample t-tests (which allow different variances in the underlying normal populations). The p-values were Holm-adjusted for multiple testing within each family of tests per gene. For (showing the germination frequency of conidia), 3 (proteinase and metalloproteinase activities), 4 (chy1 expression levels) and 5 (mep1 expression levels), exploratory data analysis using q-q-plots for the residuals of one-factorial analysis of variance (ANOVA) models for both the untransformed and log-transformed expression values was performed to determine whether normality could be assumed in either case. Depending on this, pairwise comparisons with the control were carried out using Dunnett's procedure with the untransformed or log-transformed expression values (R-package “multicomp”) together with a heteroscedasticity-consistent estimator of the covariance matrix (R-package “sandwich”)0Citation44,45
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
2016VIRULENCE0433R2-s02.docx
Download MS Word (18.5 MB)Acknowledgments
The authors thank Jennifer Schwarz and Rene Röhrich for excellent technical support, and Dr. Richard M. Twyman for professional editing of the manuscript and Dr. Gerrit Eichner (Mathematical Institute, Justus-Liebig University), for providing excellent support in the statistical analysis of the data. The authors acknowledge the constructive recommendations provided by anonymous reviewers.
Additional information
Funding
References
- Vilcinskas A, Götz P. Parasitic Fungi and their Interactions with the Insect Immune System. In: Stothard JR, Baker RM, Rollinson D, editors. Advances in Parasitology USA: Academic Press. 1999. p. 267–313. doi:10.1016/S0065-308X(08)60244-4
- St Leger RJ, Joshi L, Bidochka MJ, et al. Biochemical characterization and ultrastructural localization of two extracellular trypsins produced by Metarhizium anisopliae in infected insect cuticles. Appl Environ Microbiol. 1996;62(4):1257–64
- St Leger RJ, Charnley AK, Cooper RM. Characterization of cuticle-degrading proteases produced by the entomopathogen Metarhizium anisopliae. Arch Biochem Biophys. 1987;253(1):221–32
- Freimoser FM, Hu G, St Leger RJ. Variation in gene expression patterns as the insect pathogen Metarhizium anisopliae adapts to different host cuticles or nutrient deprivation in vitro. Microbiology. 2005;151(Pt 2):361–71. doi:10.1099/mic.0.27560-0
- Gillespie JP, Bailey AM, Cobb B, et al. Fungi as elicitors of insect immune responses. Arch Insect Biochem Physiol. 2000;44(2):49–68. doi:10.1002/15206327(200006)44:2<49::AID-ARCH1>3.0.CO;2-F
- Vilcinskas A. Coevolution between pathogen-derived proteinases and proteinase inhibitors of host insects. Virulence. 2010;1(3):206–14. doi:10.4161/viru.1.3.12072
- Staats CC, Junges A, Guedes RL, et al. Comparative genome analysis of entomopathogenic fungi reveals a complex set of secreted proteins. BMC Genomics. 2014;15:822. doi:10.1186/1471-2164-15-822
- Mukherjee K, Vilcinskas A. Development and immunity-related microRNAs of the lepidopteran model host Galleria mellonella. BMC Genomics. 2014;15:705. doi:10.1186/1471-2164-15-705
- Mukherjee K, Fischer R, Vilcinskas A. Histone acetylation mediates epigenetic regulation of transcriptional reprogramming in insects during metamorphosis, wounding and infection. Front Zool. 2012;9(1):25. doi:10.1186/1742-9994-9-25
- Vilcinskas A. The role of epigenetics in host-parasite coevolution: lessons from the model host insects Galleria mellonella and Tribolium castaneum. Zoology (Jena, Germany). 2016;119(4):273–80. doi:10.1016/j.zool.2016.05.004
- Mukherjee K, Twyman RM, Vilcinskas A. Insects as models to study the epigenetic basis of disease. Prog Biophys Mol Biol. 2015;118(1–2):69–78. doi:10.1016/j.pbiomolbio.2015.02.009
- Adekoya OA, Sylte I. The thermolysin family (M4) of enzymes: therapeutic and biotechnological potential. Chem Biol Drug Des. 2009;73(1):7–16. doi:10.1111/j.1747-0285.2008.00757.x
- Qazi SS, Khachatourians GG. Hydrated conidia of Metarhizium anisopliae release a family of metalloproteases. J Invertebr Pathol. 2007;95(1):48–59. doi:10.1016/j.jip.2006.12.002
- Altincicek B, Linder M, Linder D, et al. Microbial metalloproteinases mediate sensing of invading pathogens and activate innate immune responses in the lepidopteran model host Galleria mellonella. Infect Immun. 2007;75(1):175–83. doi:10.1128/iai.01385-06
- Griesch J, Wedde M, Vilcinskas A. Recognition and regulation of metalloproteinase activity in the haemolymph of Galleria mellonella: a new pathway mediating induction of humoral immune responses. Insect Biochem Mol Biol. 2000;30(6):461–72
- Altincicek B, Berisha A, Mukherjee K, et al. Identification of collagen IV derived danger/alarm signals in insect immunity by nanoLC-FTICR MS. Biol Chem. 2009;390(12):1303–11. doi:10.1515/bc.2009.128
- Berisha A, Mukherjee K, Vilcinskas A, et al. High-resolution mass spectrometry driven discovery of peptidic danger signals in insect immunity. PLoS One. 2013;8(11):e80406. doi:10.1371/journal.pone.0080406
- Clermont A, Wedde M, Seitz V, et al. Cloning and expression of an inhibitor of microbial metalloproteinases from insects contributing to innate immunity. Biochem J. 2004;382(Pt 1):315–22. doi:10.1042/bj20031923
- Wedde M, Weise C, Nuck C, et al. The insect metalloproteinase inhibitor gene of the lepidopteran Galleria mellonella encodes two distinct inhibitors. Biol Chem. 2007;388(1):119–27. doi:10.1515/BC.2007.013
- Joop G, Vilcinskas A. Coevolution of parasitic fungi and insect hosts. Zoology (Jena, Germany). 2016;119(4):350–8. doi:10.1016/j.zool.2016.06.005
- Vogel H, Altincicek B, Glockner G, et al. A comprehensive transcriptome and immune-gene repertoire of the lepidopteran model host Galleria mellonella. BMC Genomics. 2011;12:308. doi:10.1186/1471-2164-12-308
- Rahnamaeian M, Langen G, Imani J, et al. Insect peptide metchnikowin confers on barley a selective capacity for resistance to fungal ascomycetes pathogens. J Exp Bot. 2009;60(14):4105–14. doi:10.1093/jxb/erp240
- Griesch J, Vilcinskas A. Proteases released by entomopathogenic fungi impair phagocytic activity, attachment and spreading of plasmatocytes isolated from haemolymph of the greater wax moth Galleria mellonella. Biocontrol Sci Technol. 1998;8(4):517–31. doi:10.1080/09583159830036
- Vilcinskas A, Wedde M. Inhibition of Beauveria bassiana Proteases and Fungal Development by Inducible Protease Inhibitors in the Haemolymph of Galleria mellonella larvae. Biocontrol Sci Technol. 1997;7(4):591–602. doi:10.1080/09583159730640
- Dubovskiy IM, Whitten MM, Kryukov VY, et al. More than a colour change: insect melanism, disease resistance and fecundity. Proc Biol Sci. 2013;280(1763):20130584. doi:10.1098/rspb.2013.0584
- Vilcinskas A. Evolutionary plasticity of insect immunity. J Insect Physiol. 2013;59(2):123–9
- Wang S, Leclerque A, Pava-Ripoll M, et al. Comparative genomics using microarrays reveals divergence and loss of virulence-associated genes in host-specific strains of the insect pathogen Metarhizium anisopliae. Eukaryot Cell. 2009;8(6):888–98. doi:10.1128/ec.00058-09
- Wang S, Fang W, Wang C, et al. Insertion of an esterase gene into a specific locust pathogen (Metarhizium acridum) enables it to infect caterpillars. PLoS Pathog. 2011;7(6):e1002097. doi:10.1371/journal.ppat.1002097
- Arolas JL, Botelho TO, Vilcinskas A, et al. Structural evidence for standard-mechanism inhibition in metallopeptidases from a complex poised to resynthesize a peptide bond. Angew Chem Int Ed Engl. 2011;50(44):10357–60. doi:10.1002/anie.201103262
- Schrank A, Vainstein MH. Metarhizium anisopliae enzymes and toxins. Toxicon. 2010;56(7):1267–74. doi:10.1016/j.toxicon.2010.03.008
- Vilcinskas A, Matha V, Götz P. Effects of the entomopathogenic fungus Metarhizium anisopliae and its secondary metabolites on morphology and cytoskeleton of plasmatocytes isolated from the greater wax moth, Galleria mellonella. J Insect Physiol. 1997;43(12):1149–59. http://doi.org/10.1016/S0022-1910(97)00066-8
- Pal S, St Leger RJ, Wu LP. Fungal peptide Destruxin A plays a specific role in suppressing the innate immune response in Drosophila melanogaster. J Biol Chem. 2007;282(12):8969–77. doi:10.1074/jbc.M605927200
- Wang C, St Leger RJ. A collagenous protective coat enables Metarhizium anisopliae to evade insect immune responses. Proc Natl Acad Sci USA. 2006;103(17):6647–52. doi:10.1073/pnas.0601951103
- de Bekker C, Quevillon LE, Smith PB, et al. Species-specific ant brain manipulation by a specialized fungal parasite. BMC Evol Biol. 2014;14:166. doi:10.1186/s12862-014-0166-3
- Bader MW, Sanowar S, Daley ME, et al. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122(3):461–72. doi:10.1016/j.cell.2005.05.030
- Yamauchi Y, Boukari H, Banerjee I, et al. Histone deacetylase 8 is required for centrosome cohesion and influenza A virus entry. PLoS Pathog. 2011;7(10):e1002316. doi:10.1371/journal.ppat.1002316
- Chaal BK, Gupta AP, Wastuwidyaningtyas BD, et al. Histone deacetylases play a major role in the transcriptional regulation of the Plasmodium falciparum life cycle. PLoS Pathog. 2010;6(1):e1000737. doi:10.1371/journal.ppat.1000737
- Bougdour A, Maubon D, Baldacci P, et al. Drug inhibition of HDAC3 and epigenetic control of differentiation in Apicomplexa parasites. J Exp Med. 2009;206(4):953–66. doi:10.1084/jem.20082826
- Lopes da Rosa J, Boyartchuk VL, Zhu LJ, et al. Histone acetyltransferase Rtt109 is required for Candida albicans pathogenesis. Proc Natl Acad Sci USA. 2010;107(4):1594–9. doi:10.1073/pnas.0912427107
- Wurtele H, Tsao S, Lepine G, et al. Modulation of histone H3 lysine 56 acetylation as an antifungal therapeutic strategy. Nat Med. 2010;16(7):774–80. doi:10.1038/nm.2175
- Mukherjee K, Grizanova E, Chertkova E, et al. Experimental evolution of resistance against Bacillus thuringiensis in the insect model host Galleria mellonella results in epigenetic modifications. Virulence. 2017:1–13. doi: 10.1080/21505594.2017.1325975. ( in press).
- Bolouri Moghaddam MR, Vilcinskas A, Rahnamaeian M. The insect-derived antimicrobial peptide metchnikowin targets Fusarium graminearum beta(1,3)glucanosyltransferase Gel1, which is required for the maintenance of cell wall integrity. Biol Chem. 2016;398(4):491–8. doi:10.1515/hsz-2016-0295
- Baumler A, Fang FC. Host specificity of bacterial pathogens. Cold Spring Harb Perspect Med. 2013;3(12):a010041. doi:10.1101/cshperspect.a010041
- Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models. Biom J. 2008;50(3):346–63. doi:10.1002/bimj.200810425
- Zeileis A. Econometric computing with HC and HAC Covariance matrix estimators. J Statistical Software. 2004;11(10):1–17. doi: 10.18637/jss.v011.i10
