The bacterial species Klebsiella pneumoniae is an increasingly important human pathogen. Dramatic increases in the levels of multidrug resistance (MDR) associated with this species [Citation1], particularly resistance to carbapenems and third-generation cephalosporins, pose an emerging global problem [Citation2]. The common drug-resistant strains of Klebsiella pneumoniae are ST11 or ST258, which were identified by multilocus sequence typing (MLST) [Citation3,Citation4]. Notably, hypervirulent variants of K. pneumoniae (hvKP), which cause severe infections such as liver abscesses, endophthalmitis, meningitis, osteomyelitis, necrotizing fasciitis in healthy and ambulatory individuals [Citation5], have been increasingly reported recently. hvKP often exhibit hypermucoviscosity and mainly belong to ST23 (by MLST) for the K1 capsular serotype and to ST86 and ST375 for the K2 serotype [Citation5,Citation6]. In contrast to classic K. pneumoniae (cKP), hvKP usually show a high level of susceptibility to the antimicrobial agents used for clinical treatment, except for ampicillin [Citation7]. However, in the last decade, hvKP with high degrees of drug resistance have increasingly emerged, even in the well-documented K1 and K2 serotype hvKP strains [Citation8–12]. So far, there are two papers reporting hypervirulent K. pneumoniae that produced DHA-1 beta-lactamase and ESBL in France [Citation8] and Korea [Citation12], respectively; however, the genetics investigationor genome backgrounds remains to be characterized. Recently the classic ST11 carbapenem-resistant K. pneumoniae strain has been reported to have acquired a virulence plasmid, resulting in a carbapenem-resistant hypervirulent variant of ST11 (hv-MDR-KP) [Citation13].
In cKP, the acquired resistance genes are found mainly embedded on plasmids, for example, the blaKPC-2 gene that confers carbapenems resistance [Citation14] and the mcr-1 gene that confers colistin resistance [Citation15]. Similarly, plasmids are also the major carriers for emerging drug-resistance genes in multidrug resistant hvKP (MDR hvKP). The blaCTX-M-3 gene was found on an IncL/M plasmid, in an ST86 and K2 hvKP isolated in France [Citation8]. A carbapenem-resistant ST23 hvKP with serotype K1 was reported in China, and its blaKPC-2 gene was located on a non-transferable plasmid [Citation10]. Plasmids carrying resistance genes with transferability to hvKP were also observed. The conjugation between typical carbapenemase-producing cKP (ST258) and K2 hvKP (ST65) showed that the plasmid carrying the blaKPC-2 or blaKPC-3 genes could be transferred to and retained in hvKP [Citation16]. Siu et al. reported a case of recurrent liver abscess caused by hvKP and successfully mimicked the blaCMY-2 gene transferred from E. coli to K2 hvKP in vitro [Citation17]. However, so far, there have been no reports of hvKP-borne resistance plasmids that are transferable to other hvKP strains.
Here, we present hypervirulent K. pneumoniae clinical strain RJA166 with resistance to all the third-generation cephalosporins and to the combination of β-lactam and β-lactamase inhibitors. RJA166 is a typical hvKP with a K1 capsular serotype and ST23 sequence type. The blaDHA-1 gene, as well as three other AMR genes, were identified as being on the natural plasmid pRJA166a and this resistance plasmid was subsequently determined to be transferable to the K1 and K2 hvKP strains and to the ST11 cKP.
First, we investigated the virulence of K. pneumoniae RJA166 in the G. mellonella larvae and mouse infection models. K. pneumoniae RJA166 was isolated from sputum specimen from a patient in Ruijin Hospital, Shanghai, China. It showed a viscous string longer than 40 mm (Figure S1a) by the string test. It was also determined to be ST23 and a capsular serotype K1 strain using PCR in our retrospective experiment. RJA166 was then determined to be as virulent as K. pneumoniae NTUH-K2044 in the killing assay on Galleria mellonella larvae, with the cKP HS11286 (ST11 and KL103 serotype [Citation18]) as control (Figure S1b, Table S1). For further confirmation of RJA166 hypervirulence, a mouse lethality assay was performed. RJA166 showed an LD50 of 2.3 × 102 CFU in BALB/c mice, which was comparable to hypervirulent NTUH-K2044 strain [Citation19,Citation20]. The data from the killing assays on both mice and G. mellonella show that RJA166 had the same virulence level as NTUH-K2044, which has been shown to be highly pathogenic in a previous study [Citation21], more so than the classic ST11 K. pneumoniae [Citation14]. In addition, the liver, spleen, and kidney of the mouse which was challenged by 103 CFU K. pneumoniae RJA166 and sacrificed 3 days after infection, were taken for the further histopathological examination. Compare to the control, the infected liver showed multiple microabscesses, with infiltration of inflammatory cells, including neutrophils and monocytes mainly in the part of the portal center infiltration mode, accompanied with vacuolization, focal hyperemia, and hemorrhage. Eosinophilic liver abscess exhibited pale stained nucleus, irregular shape, with the parenchyma becoming looser due to infiltration of inflammatory cells, whilst the normal liver cells have more obvious boundaries (Figure S2a and S2d). In comparison with the control kidney, there was local renal capsule inflammatory cell infiltration, glomerular lesions in varying degrees of atrophy, mild swelling of renal tubular epithelial cells, mild expansion of the lumen observed in the infected group (Figure S2b and S2e). The infected spleen showed white pulp atrophy, irregular structure, reduced number of lymphoid follicles. The remaining follicular morphology was also less regular, with large peripheral macrophage accumulation. Mild red pulp hyperemia, with slightly scattered thrombosis, were also observed (Figure S2c and S2f).
Secondly, we wanted to address the resistance of hvKP RJA166 to third-generation cephalosporins. The MIC data generated by the VITEK 2 Compact system showed that RJA166 was resistant to first-, second- and third-generation cephalosporins; aztreonam; and a combination of β-lactam and β-lactamase inhibitors (), suggesting that RJA166 is a ST23 MDR-hvKP. RJA166 also displayed low-level resistance to ertapenem but was susceptible to imipenem (). Additionally, this strain was determined to be susceptible to aminoglycosides and fluoroquinolones, such as amikacin, gentamycin, tobramycin and ciprofloxacin (). The resistance profile of RJA166 suggested the potential existence of a gene encoding the AmpC β-lactamase in its genome.
Table 1. Antimicrobial susceptibility test of the hypervirulent K. pneumoniae RJA166, RJF293 and pRJA166a/RJF293H transconjugants.
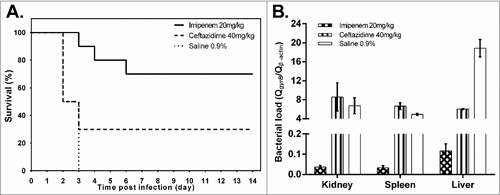
The resistance of hvKP RJA166 in a murine model was also determined by an antibiotic treatment assay. Ceftazidime (40 mg/kg) and imipenem (20 mg/kg) were administered to BALB/c mice intraperitoneally challenged by K. pneumoniae RJA166. The infected mice treated with ceftazidime displayed higher mortality (70%) than the mice in the imipenem group (30%). No survival was observed in the group treated with 0.9% saline () as a control. The bacterial loads analysis in murine tissues was determined to be consistent with the survival analysis, by quantitative real-time PCR with primers of bacterial gyrB gene and the murine β-actin gene (Table S2). The mice treated with ceftazidime showed higher bacterial loads in kidneys (p = 0.0383, by Student's t test), spleen (p < 0.0001) and liver (p<0.0001) () than the mice in the imipenem group. This demonstrates that the drug resistance of the ST23 MDR-hvKP RJA166 contributes to bacterial survival in the murine model under the treatment of third generation cephalosporins.
Figure 1. Effect of ceftazidime and imipenem on K. pneumoniae RJA166 infection in murine model. (a) The survival curve of the BALB/c mice infected with 1.8 × 103 CFU of Klebsiella pneumoniae RJA166 with different treatments. In the 14 days after bacteria challenge, mice treated by imipenem (20 mg/kg), ceftazidime (40 mg/kg) and saline (0.9%) showed mortalities of 30%, 70% and 100%, respectively. (b) The bacterial load of murine tissues in the mice treated with different antibiotic. The results were calculated by dividing the quantity of bacterial gyrase gene by the quantity of murine β-actin gene. The bacterial loads in the kidneys, spleens and livers from the mice treated with ceftazidime were significantly higher than those in the mice treated with imipenem, with the p value 0.0383, <0.0001 and <0.0001, respectively, as generated by Student's t test.
Thirdly, our genome analysis showed that the hvKP RJA166 carries a conjugative resistance plasmid pRJA166a. The whole genome sequencing showed that the ST23 MDR-hvKP RJA166 genome consists of a circular chromosome and three circular plasmids (Table S3; supplementary Materials and Methods), including the antibiotic resistance plasmid pRJA166a (230 Kb in size; ), the virulence plasmid pRJA166b (229 Kb; Figure S3a) and the plasmid pRJA166c with unknown function (111 Kb; Figure S3b and S3c). The RJA166 chromosome exhibits similar genetic background to the other sequenced K1 hvKP strains, as determined by the core chromosomal SNP-based phylogenetic analysis of 107 completely sequenced K. pneumoniae strains (Figure S4). The resistance plasmid pRJA166a exhibited a genetic background distinct from to the virulence plasmids frequently found in the sequenced K2 or K1 serotype hvKP strains. We thus presumed that the acquisition of a blaDHA-1-carrying plasmid by K1 hvKP resulted in a multidrug-resistant hypervirulent variant.
Figure 2. The genetic context of the blaDHA-1 gene in the resistance plasmid pRJA166a of K. pneumoniae RJA166. The plasmid shares the backbone of two previously reported plasmids pYNKP001-dfrA of Raoultella ornithinolytica YNKP001[ref. Citation22] and pKOX_R1 of Klebsiella michiganensis E718 [ref. Citation23]. The predicted antibiotic resistance gene is shown in red, the tra gene cluster is in dark green, and the IS element is shown in light green.
![Figure 2. The genetic context of the blaDHA-1 gene in the resistance plasmid pRJA166a of K. pneumoniae RJA166. The plasmid shares the backbone of two previously reported plasmids pYNKP001-dfrA of Raoultella ornithinolytica YNKP001[ref. Citation22] and pKOX_R1 of Klebsiella michiganensis E718 [ref. Citation23]. The predicted antibiotic resistance gene is shown in red, the tra gene cluster is in dark green, and the IS element is shown in light green.](/cms/asset/5b70583a-0bfb-40ce-b86e-b6ddd9767b33/kvir_a_1456229_f0002_oc.jpg)
We then typed the incompatible group of pRJA166a as IncHI5 and found that its backbone was highly homologous to the previously reported IncHI5-group plasmids, namely, pYNKP001-dfrA of Raoultella ornithinolytica YNKP001 [ref. Citation22] and pKOX_R1 of K. michiganensis E718 [ref. Citation23]. Notably, in addition to the region homologous to pKOX_R1 and pYNKP001-dfrA (Figure S5), pRJA166a has a 26-kb accessory region (RJA_27650..RJA_27825) associated with antibiotic resistance genes, integrons, transposons and insertion sequence-based mobile elements (). Across this mosaic-like, three-module region, the blaDHA-1 gene is present on Module II of the accessory resistance region, a relatively large portion of this 26-kb region. The AmpC-type β-lactamase gene blaDHA-1, first reported in 1997 [ref. Citation24], confers resistance to third-generation cephalosporins on the host. This gene is frequently carried by plasmids belonging to the IncH and IncR incompatibility groups. The genetic context of blaDHA-1 is organized as a conserved structure: sul1-qacEΔ1-ampR-blaDHA-1-pspABCDF-qnrB4. This structure is often associated with the class I integron and ISCR1; however, a complete integron was not found; there was no 5′-CS or int gene present in pRJA166a. This gene is highly conserved in the blaDHA-1-carrying plasmids of Klebsiella strains [Citation25,Citation26]. Module II is located upstream of ISCR1 in pRJA166a and in the IncF plasmid pKP048 [ref. Citation27], but downstream of ISCR1 in pYNKP001-dfrA. Additionally, the IS26 elements bracketed Module II-III are in opposite orientation, but no target site duplication was observed. Due to their opposite orientations, we propose that these IS26 elements and the Module II-III sandwiched between them might constitute a new composite transposon capable of being disseminated to other replicons [Citation28]. Thus, given the highly mosaic-like structure of the pRJA166a accessory region, we suggest that the present entity represents a relic of multiple past insertions, deletions and recombination events that may have since acquired multidrug resistance genes.
Figure 3. Organizational map of the 26-kb accessory resistance region (RJA_27650…RJA_27825) on pRJA166a. Two other blaDHA-1-carrying plasmids with the same incompatibility group (IncHI5), pKP048 and pYNKP001-dfrA, were compared. The three modules (I–III) referred to in the text are shown as red boxes while the module that contains the conserved blaDHA-1 context is highlighted in the black boxes with dashed borders. The protein-coding regions are shown as arrows with different colours, where green indicates an IS element, black indicates integrase gene, red indicates antibiotic resistance gene and grey indicates flanking gene. Genes identical to the 3′-CS of class 1 integron are shown with an orange background. Inverted repeats of IS elements are represented as small black triangle above the arrow. VR1 and VR2 are the variable regions of In469, as defined by Liang et al [Citation22].
![Figure 3. Organizational map of the 26-kb accessory resistance region (RJA_27650…RJA_27825) on pRJA166a. Two other blaDHA-1-carrying plasmids with the same incompatibility group (IncHI5), pKP048 and pYNKP001-dfrA, were compared. The three modules (I–III) referred to in the text are shown as red boxes while the module that contains the conserved blaDHA-1 context is highlighted in the black boxes with dashed borders. The protein-coding regions are shown as arrows with different colours, where green indicates an IS element, black indicates integrase gene, red indicates antibiotic resistance gene and grey indicates flanking gene. Genes identical to the 3′-CS of class 1 integron are shown with an orange background. Inverted repeats of IS elements are represented as small black triangle above the arrow. VR1 and VR2 are the variable regions of In469, as defined by Liang et al [Citation22].](/cms/asset/c40b8023-2e1d-4423-9371-72f1af429216/kvir_a_1456229_f0003_oc.jpg)
To date, third-generation cephalosporins have been the main antibiotics for the treatment for of infections caused by hypervirulent K. pneumoniae [Citation29]. Recently, the emergence of multidrug-resistant hypervirulent K. pneumoniae strains has been reported in different countries, which include a broad range of sequence types [Citation8,Citation11,Citation30]. The ST23 MDR-hvKP strain RJA166 presented here is the first hvKP strain with resistances to the third-generation cephalosporins collected in the ICU of our hospital. This type of K. pneumoniae strain has been reported in Korea [Citation12], but neither conjugation assay nor genetic background parsing was included in the previous publication. Thus, the approach of the emergence of multidrug resistant hypervirulent K. pneumoniae strains is still unclear. Our WGS data suggested that RJA166 carries three natural plasmids, including a resistance plasmid pRJA166a and a virulence plasmid pRJA166b. K. pneumoniae variants with both multidrug resistance and hypervirulence phenotypes potentially developed via two distinct approaches: (I) the multidrug-resistant hvKP (MDR-hvKP), origin from the hvKP; and (II) the hypervirulent multidrug-resistant K. pneumoniae (hv-MDR-KP), derived from the multidrug-resistant cKP. The development of the virulence and resistance traits of the ST23 MDR-hvKP presented here (approach I above) is distinct from that of the newly reported ST11 hv-MDR-KP [Citation13] (approach II above).
Finally, we investigated the conjugative transfer of the blaDHA-1-containing plasmid pRJA166a between diverse K. pneumoniae strains. By filter mating, the ceftazidime resistance was found to be successfully transferred from the ST23 MDR-hvKP RJA166 to RJF293H, which is a hygromycin-resistant mutant from the ST374 and K2 serotype hvKP clinical strain RJF293 [ref. Citation7]. The MIC tests of three randomly selected K. pneumoniae pRJA166a/RJF293H transconjugants (Tc1-3) revealed that the resistance to cephalosporin antibiotics was elevated (). PCR amplifications of fragments from the blaDHA-1, repA and traD genes of pRJA166a confirmed that the antibiotic resistance dissemination transfer was mediated by pRJA166a (Figure S6c). Then, the XbaI-PFGE and S1-nuclease PFGE suggested that the increase in antimicrobial resistance of these transconjugants was due to the acquisition of the plasmid pRJA166a (Figure S6a-b). Finally, the transconjugation frequency of pRJA166a from RJA166 to RJF293H was determined to be approximately 2.6 × 10−6 transconjugants per donor cell. We subsequently assessed the stability of pRJA166a in K. pneumoniae RJA166 and RJF293H transconjugants by a continuous passage. After 375 generations (15 days) on the LB agar without antibiotic selection, no loss of the plasmids in either host was observed. We further performed a conjugation assay to explore whether pRJA166a was able to disseminate across an expanded collection of K. pneumoniae. Similarly, the expected transfer of pRJA166a from RJA166 (ST23 and K1) to its closely related recipient NTUH-K2044IT (a hygromycin-resistant derivative of ST23 and serotype K1 K. pneumoniae NTUH-K2044; Table S1) was observed with a frequency of approximately 5.5 × 10−6 transconjugants per donor. Remarkably, pRJA166a can also be transferred into the carbapenem-resistant cKP strain HS11286 (ST11 and KL103), by using its hygromycin resistant derivative HS11286YZ6 (Table S1). The transfer frequencies to HS11286YZ6 were determined to be approximately 2.7 × 10−6 transconjugants per donor. However, the conjugative transfer of pRJA166a from RJA166 to the frequently used E. coli recipients either HB101 or J53 was not detectable using the same conditions described above.
The transfer of resistance plasmids has been previously reported to occur either from hvKP to E. coli [Citation10] or vice versa [Citation16,Citation17]. Our study showed the conjugative transfer of a resistance plasmid pRJA166a, which coded for the entire conjugation machinery, between hvKP. The entire plasmid pRJA166a transferred to both the K1 hvKP strain and the K2 hvKP strain, resulting in MDR-hvKP. Notably, the resistance plasmid pRJA166a kept its stability across the transconjugants. Although the pRJA166a acquisition comes at a fitness cost to the recipient (Figure S7a), the G. mellonella larvae infection assay confirmed that these obtained transconjugants are hypervirulent like the recipient (Figure S7b and S7c). Remarkably, pRJA166a was also transferable to the carbapenem-resistant ST11 cKP strain HS11286, which had already carried three resistance plasmids. These results indicated that the resistance plasmid pRJA166a carrying the blaDHA-1 gene could be disseminated among K. pneumoniae.
In conclusion, we investigated the genetic background and microbiological features of a hypervirulent clinical strain (RJA166) of K. pneumoniae with resistances to the third-generation cephalosporins (ST23 MDR-hvKP). The blaDHA-1-carrying natural plasmid from this K1 and ST23 hvKP that is transferable to hvKP and cKP might contribute to the rapid spread of drug resistance among K. pneumoniae and other bacterial pathogens.
Nucleotide sequence accession numbers
The sequences of K. pneumoniae RJA166 chromosome (accession number CP019047) and the three plasmids pRJA166a (CP019048), pRJA166b (CP019049) and pRJA166c (CP019050) were deposited in GenBank.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Virulence_MS_RJA166_supplementary_R1_letter_v3.pdf
Download PDF (4.6 MB)Additional information
Funding
References
- Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9:228–36. Available from: http://doi.org/10.1016/S1473-3099(09)70054-4.
- Counter resistance. Nat Microbiol. 2017;2:17071. Available from: http://www.nature.com/articles/nmicrobiol201771..
- Chen L, Mathema B, Pitout JDD, et al. Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. MBio. 2014;5:e01355-14–e01355-14. Available from: http://mbio.asm.org/cgi/doi/10.1128/mBio.01355-14.
- Maiden MC, Bygraves JA, Feil E, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95:3140–5. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid = 19708&tool = pmcentrez&rendertype = abstract.
- Shon AS, Bajwa RPS, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Virulence. 2013;4:107–18. Available from: http://www.tandfonline.com/doi/abs/10.4161/viru.22718.
- Holt KE, Wertheim H, Zadoks RN, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112:E3574–81. Available from: http://www.pnas.org/lookup/doi/10.1073/pnas.1501049112.
- Wang X, Xie Y, Li G, et al. Whole-genome-sequencing characterization of bloodstream infection-causing hypervirulent Klebsiella pneumoniae of capsular serotype K2 and ST374. Virulence. 2018;5594:00–00. Available from: https://www.tandfonline.com/doi/full/10.1080/21505594.2017.1421894.
- Surgers L, Boyd A, Girard PM, et al. ESBL-Producing Strain of hypervirulent Klebsiella pneumoniae K2, France. Emerg Infect Dis. 2016;22:1687–8. Available from: http://wwwnc.cdc.gov/eid/article/22/9/16-0681_article.htm.
- Zhang Y, Zeng J, Liu W, et al. Emergence of a hypervirulent carbapenem- resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect. 2015;71:553–60. Available from: 10.1016/j.jinf.2015.07.010.
- Zhang R, Lin D, Chan EW, et al. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother. 2016;60:709–11. Available from: http://aac.asm.org/lookup/doi/10.1128/AAC.02173-15.
- Shankar C, Nabarro LEB, Devanga K, et al. Draft genome sequences of three hypervirulent carbapenem-resistant Klebsiella pneumoniae isolates from bacteremia. Genome Announc. 2016;4:e01081–16. Available from: http://genomea.asm.org/lookup/doi/10.1128/genomeA.01081-16.
- Cheong HS, Chung DR, Lee C, et al. Emergence of serotype K1 Klebsiella pneumoniae ST23 strains co-producing the plasmid-mediated AmpC beta-lactamase DHA-1 and an extended-spectrum beta-lactamase in Korea. Antimicrob Resist Infect Control. 2016;5:50. Available from: http://aricjournal.biomedcentral.com/articles/10.1186/s13756-016-0151-2.
- Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2017;3099:1–10. Available from: http://linkinghub.elsevier.com/retrieve/pii/S1473309917304899.
- Liu P, Li P, Jiang X, et al. Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputum. J Bacteriol. 2012;194:1841–2. Available from: http://jb.asm.org/content/194/7/1841.
- Wong SCY, Tse H, Chen JHK, et al. Colistin-resistant Enterobacteriaceae carrying the mcr-1 gene among patients in Hong Kong. Emerg Infect Dis. 2016;22:1667–9. Available from: http://wwwnc.cdc.gov/eid/article/22/9/16-0091_article.htm.
- Siu L-KK, Huang DB, Chiang T. Plasmid transferability of KPC into a virulent K2 serotype Klebsiella pneumoniae. BMC Infect Dis. 2014;14:176. Available from: https://bmcinfectdis.biomedcentral.com/articles/10.1186/1471-2334-14-176.
- Lin Y-T, Pan Y-J, Lin T-L, et al. Transfer of CMY-2 cephalosporinase from Escherichia coli to virulent Klebsiella pneumoniae causing a recurrent liver abscess. Antimicrob Agents Chemother. 2015;59:5000–2. Available from: http://aac.asm.org/content/59/8/5000.
- Wyres KL, Wick RR, Gorrie C, et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom. 2016;2:e000102. Available from: http://mgen.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.000102.
- Fodah RA, Scott JB, Tam HH, et al. Correlation of Klebsiella pneumoniae comparative genetic analyses with virulence profiles in a murine respiratory disease model. PLoS One. 2014;9:1–11. Available from: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0107394.
- Lin T-L, Chuang Y-P, Huang Y-T, et al. Identification of an immuno-dominant protein from Klebsiella pneumoniae strains causing pyogenic liver abscess: implication in serodiagnosis. BMC Microbiol. 2014;14:321. Available from: https://bmcmicrobiol.biomedcentral.com/articles/10.1186/s12866-014-0321-4.
- Wu KM, Li NH, Yan JJ, et al. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol. 2009;191:4492–501. Available from: http://jb.asm.org/content/191/14/4492.
- Liang Q, Yin Z, Zhao Y, et al. Sequencing and comparative genomics analysis of the IncHI2 plasmids pT5282-mphA and p112298-catA and the IncHI5 plasmid pYNKP001-dfrA. Int J Antimicrob Agents. 2017;49:709–18. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0924857917301085.
- Huang TW, Wang JT, Lauderdale TL, et al. Complete sequences of two plasmids in a blaNDM-1-positive Klebsiella oxytoca isolate from Taiwan. Antimicrob Agents Chemother. 2013;57:4072–6. Available from: http://aac.asm.org/content/57/8/4072.
- Gaillot O, Clément C, Simonet M, et al. Novel transferable β-lactam resistance with cephalosporinase characteristics in Salmonella enteritidis. J Antimicrob Chemother. 1997;39:85–7. Available from: https://academic.oup.com/jac/article/39/1/85/734705.
- Verdet C, Benzerara Y, Gautier V, et al. Emergence of DHA-1-producing Klebsiella spp. in the Parisian region: Genetic organization of the ampC and ampR genes originating from Morganella morganii. Antimicrob Agents Chemother. 2006;50:607–17. Available from: http://aac.asm.org/content/50/2/607.
- Guo Q, Spychala CN, Mcelheny CL, et al. Comparative analysis of an IncR plasmid carrying armA, blaDHA-1 and qnrB4 from Klebsiella pneumoniae ST37 isolates. J Antimicrob Chemother. 2016;71(4):1–5.
- Jiang Y, Yu D, Wei Z, et al. Complete nucleotide sequence of Klebsiella pneumoniae multidrug resistance plasmid pKP048, carrying blaKPC-2, blaDHA-1, qnrB4, and armA. Antimicrob Agents Chemother. 2010;54:3967–9. Available from: http://aac.asm.org/cgi/doi/10.1128/AAC.00137-10.
- He S, Hickman AB, Varani AM, et al. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. MBio. 2015;6:e00762. Available from: http://mbio.asm.org/content/6/3/e00762-15.
- Siu LK, Yeh KM, Lin JC, et al. Klebsiella pneumoniae liver abscess: A new invasive syndrome. Lancet Infect Dis. 2012;12:881–5. Available from: https://www.sciencedirect.com/science/article/pii/S1473309912702050.
- Robin F, Hennequin C, Gniadkowski M, et al. Virulence factors and TEM-type β-lactamases produced by two isolates of an epidemic Klebsiella pneumoniae strain. Antimicrob Agents Chemother. 2012;56:1101–4. Available from: http://aac.asm.org/content/56/2/1101.
