ABSTRACT
Streptococcus suis is a major porcine bacterial pathogen and emerging zoonotic agent. S. suis 5ʹ-nucleotidase is able to convert adenosine monophosphate to adenosine, resulting in inhibiting neutrophil functions in vitro and it is an important virulence factor. Here, we show that S. suis 5ʹ-nucleotidase not only enables producing 2ʹ-deoxyadenosine from 2ʹ-deoxyadenosine monophosphate by the enzymatic assay and reversed-phase high performance liquid chromatography (RP-HPLC) analysis in vitro, but also synthesizes both 2ʹ-deoxyadenosine and adenosine in mouse blood in vivo by RP-HPLC and liquid chromatography with tandem mass spectrometry analyses. Cellular cytotoxicity assay and Western blot analysis indicated that the production of 2ʹ-deoxyadenosine by 5ʹ-nucleotidase triggered the death of mouse macrophages RAW 264.7 in a caspase-3-dependent way. The in vivo infection experiment showed that 2ʹ-deoxyadenosine synthesized by 5ʹ-nucleotidase caused monocytopenia in mouse blood. The in vivo transcriptome analysis in mouse blood showed the inhibitory effect of 5ʹ-nucleotidase on neutrophil functions and immune responses probably mediated through the generation of adenosine. Taken together, these findings indicate that S. suis synthesizes 2ʹ-deoxyadenosine and adenosine by 5ʹ-nucleotidase to dampen host immune responses, which represents a new mechanism of S. suis pathogenesis.
Introduction
Streptococcus suis is one of the leading causes of sepsis, meningitis, pneumonia, arthritis, and sudden death in pigs worldwide. Based on the antigenicity of the capsular polysaccharide (CPS), more than 30 serotypes have been identified [Citation1]. S. suis is also recognized as an important zoonotic pathogen for humans in close contact with infected pigs or contaminated pork. Human infection caused by S. suis has been reported in Asia, Europe, South America, North America, and Oceania [Citation2]. At least nine serotypes can cause infection in humans, including serotypes 2, 4, 5, 9, 14, 16, 21, 24, and 31 [Citation2–Citation4]. Moreover, in Vietnam and Thailand, S. suis has been identified as the first and second most common cause of adult meningitis, respectively [Citation5]. S. suis deploys a plethora of mechanisms to evade innate and adaptive immune responses: among the various virulence factors described for S. suis, the CPS plays essential roles in resistance to phagocytosis by several innate immune cells [Citation6,Citation7]; the suilysin and the cell wall also contribute to limiting complement-dependent clearance by dendritic cells [Citation7]; S. suis produces several immunoglobulin degrading enzymes contributing to immune escape during infection including IgA1 protease, porcine IgM-specific protease IdeSsuis, and porcine IgG-specific protease IgdE [Citation8–Citation10]; S. suis exploits several surface proteins and transcriptional regulators to resist killing by neutrophils, including MRP and Enolase [Citation11], NisKR [Citation12], CcpA [Citation13].
In our previous study, 5ʹ-nucleotidase (NT) of S. suis virulent strain GZ0565 was identified as a possible virulence factor through a comparative proteomics approach [Citation14]. Recently, Liu et al. reported that S. suis 5ʹ-nucleotidase (designated as Ssads in their work) is a surface protein and an important virulence factor. Ssads is able to convert adenosine monophosphate (AMP) to adenosine (Ado). They further demonstrated that S. suis could dampen neutrophil defenses through Ado synthesized by Ssads with reducing oxidative activity and degranulation of neutrophils in human blood in vitro [Citation15]. However, Ado production by S. suis Ssads has not been examined in vivo and the effect of Ssads through the generation of Ado on immune responses has not been investigated in vivo. Thammavongsa et al. reported that Staphylococcus aureus 5ʹ-nucleotidase (designated as AdsA in their work) could convert AMP to Ado, contributing to S. aureus escape from phagocytic clearance and formation of organ abscesses in vivo [Citation16]. Later, they showed that S. aureus AdsA could also convert deoxyadenosine monophosphate (dAMP) to 2ʹ-deoxyadenosine (dAdo), which triggered the caspase-3-mediated death of human macrophages [Citation17]. However, dAdo production by S. aureus AdsA has not been examined in vivo.
In the present study, we show that S. suis NT synthesizes both Ado and dAdo in mouse blood in vivo. The production of dAdo by NT triggers the death of mouse macrophages in a caspase-3-dependent way in vitro and causes monocytopenia in mouse blood in vivo. The in vivo transcriptome analysis in mouse blood shows the inhibitory effect of NT on neutrophil functions and immune responses probably mediated through the generation of Ado. These findings indicate that S. suis synthesizes dAdo and Ado by NT to dampen immune responses, which represents a new mechanism of S. suis pathogenesis.
Results
NT converts dAMP to dAdo
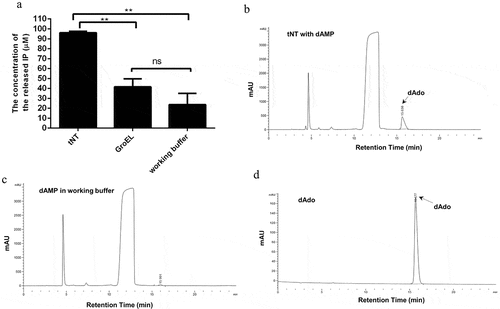
The previous studies showed that S. aureus AdsA could not only convert AMP to Ado, but also convert dAMP to dAdo [Citation16,Citation17]. S. suis NT shares high homology with S. aureus AdsA (identity 36.5% and similarity 51.5%) and also contains two 5ʹ-nucleotidase domains. To determine whether NT could function as a dAdo synthase, the ability of the recombinant truncated NT (tNT) to degrade dAMP was analyzed. Since the full length of NT could not be expressed water-soluble as described before [Citation15], the tNT (residues 36–377) containing two 5ʹ-nucleotidase domains was used. As shown in ), the concentration of inorganic phosphate (IP) in the tNT group (96.09 μM) was significantly higher than that of IP in GroEL group (41.61 μM, as a negative control) or working buffer group (23.69 μM), which suggested that tNT converted dAMP to IP and dAdo. Furthermore, the generation of dAdo by NT was directly detected by reversed-phase high performance liquid chromatography (RP-HPLC) analysis, as shown in . Taken together, these results indicate that S. suis tNT was also able to convert dAMP to dAdo.
Figure 1. S. suis NT is able to convert dAMP to dAdo. (a), The concentration of inorganic phosphate (IP) released from dAMP in the tNT group (96.09 μM) was significantly higher than that of IP in the GroEL group (41.61 μM) or working buffer group (23.69 μM). Statistical significance was examined by one-way ANOVA and Dunnett’s multiple comparisons test. ** P < 0.01; n = 3. The generation of dAdo by NT was detected by RP-HPLC analysis. The arrow indicates the dAdo produced. Samples are from tNT with dAMP group (b), the working buffer group (c), and the standard sample of dAdo (d).
To investigate whether NT can also degrade deoxyguanosine monophosphate (dGMP), deoxycytidine monophosphate (dCMP), and deoxythymidine monophosphate (dTMP), wild-type strain (WT) GZ0565, nt deletion mutant (Δnt), and the nt complemented strain C-nt were incubated with dGMP, dCMP, and dTMP, respectively. The substrate dAMP was used as a positive control. As shown in Supplementary Fig. S1, the concentration of IP in WT group or C-nt was significantly higher than that of IP in Δnt group, indicating that NT can not only convert dAMP to IP and dAdo, but also degrade dGMP, dCMP, and dTMP.
dAdo generated by NT triggers the death of mouse macrophages in a caspase-3-dependent way
dAdo has a cytotoxic effect on several human or animal immune cells [Citation18,Citation19]. The mouse macrophages RAW 264.7 were used to determine whether the dAdo generated by NT has the same effect. As shown in ), in all the groups in the presence of 100 μM adenosine deaminase inhibitor 2ʹ-deoxycoformycin (dCF), the viability of RAW 264.7 cells in the tNT+ dAMP (63 mM dAMP pretreated with 6 μM tNT at 37°C for 2 h) group was significantly lower than that of macrophages in the H2O, 6 μM tNT, 63 mM dAMP, or working buffer groups. There was no significant difference of cell viability between the tNT+ dAMP group and the 55 μM dAdo group (as a positive control) in the presence of dCF. A significant difference of cell viability was observed in tNT+ dAMP groups (with or without dCF) and dAdo groups (with or without dCF). The data suggest that dAdo generated by tNT has a cytotoxic effect on mouse macrophages RAW 264.7 in vitro. In addition, we have also examined the macrophage cytotoxicity in the presence of WT, Δnt, and C-nt using LDH (lactate dehydrogenase) release assay. As shown in the Supplementary Fig. S2, the cytotoxicity of macrophages in the presence of WT or C-nt was significantly higher than that of macrophages in the presence of Δnt. There was no significant difference of cytotoxicity of macrophages between WT group and C-nt group.
Figure 2. dAdo generated by tNT triggers caspase-3-dependent death of mouse macrophage RAW 264.7 cells. (a), In all the groups in the presence of 100 μM dCF (orange columns), the viability of RAW 264.7 cells in the tNT+ dAMP group (65.66%, 63 mM dAMP pretreated with 6 μM tNT at 37°C for 2 h) was significantly lower than that of cells in the H2O (95.33%), 6 μM tNT (86.00%), 63 mM dAMP (76.33%), or working buffer (81.33%) groups. There was no significant difference of cell viability between the tNT+ dAMP group and the 55 μM dAdo group (66.66%, as a positive control) in the presence of dCF. A significant difference of cell viability was observed in tNT+ dAMP groups (with or without dCF) and dAdo groups (with or without dCF). The viability of RAW 264.7 cells was measured by the trypan blue staining. Statistical significance was examined by one-way ANOVA and Dunnett’s multiple comparisons test. ns, not significant; *** P < 0.001; n = 3. (b), Western blot analysis showed that the band of active caspase-3 was detected in RAW 264.7 cells treated with 55 μM dAdo and 100 μM dCF for 12 h, 16 h, or 24 h. Jurkat cell extract treated with cytochrome c in vitro, obtained from Cell Signaling Technology (#9663), was used as a positive control. (c), The band of active caspase-3 was also detected in RAW 264.7 cells treated with dAMP that had been incubated with tNT in the presence of dCF for 12 h. β-actin was used as a loading control. Experiments were done in triplicate.
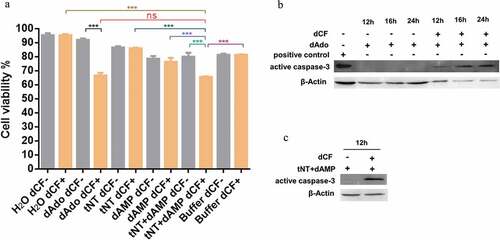
Western blot analysis was performed to test if the caspase-3 was involved in the death of mouse macrophages RAW 264.7. As shown in ), the band of active caspase-3 was detected in mouse macrophages treated with dAdo and dCF for 12 h, 16 h, or 24 h. The band of active caspase-3 was also detected in RAW 264.7 cells treated with tNT+ dAMP in the presence of dCF for 12 h ()). These results demonstrate that dAdo triggered the death of mouse macrophages in a caspase-3-dependent way.
S. suis NT produces both Ado and dAdo in mouse blood in vivo
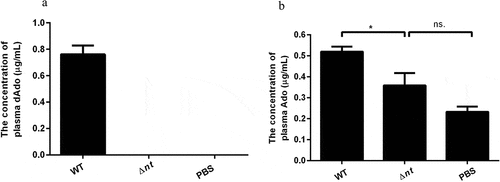
As shown in ), the concentration of plasma dAdo from mice infected with wild type strain was 0.761 μg/mL, while the concentration of plasma dAdo from mice inoculated with nt deletion mutant or PBS was below the method detection limit (0.019 μg/mL). The amount of plasma Ado from mice infected with WT strain (0.5194 μg/mL) was significantly higher than that of plasma Ado from mice injected with Δnt (0.3584 μg/mL) or PBS (0.2321 μg/mL). There was no significant difference of plasma Ado between Δnt infection group and mock-infected control group ()). The RP-HPLC chromatograms of above samples are shown in Supplementary Fig. S3. To further validate the results obtained by RP-HPLC analysis, we have reproduced the experiment and the dAdo and Ado in the plasma were detected by liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis. As shown in Supplementary Fig. S4, the concentration of plasma dAdo from mice infected with WT strain was significantly higher than that of plasma dAdo from mice injected with Δnt or PBS. The amount of plasma Ado from mice infected with WT strain was also significantly higher than that of plasma Ado from mice injected with Δnt or PBS.
Figure 3. The detection of Ado and dAdo in vivo. The abundance of Ado and dAdo in plasma was determined by RP-HPLC analysis. (a), The concentration of plasma dAdo from mice infected with the WT strain was 0.761 μg/mL, while the concentration of plasma dAdo from mice inoculated with Δnt or PBS was below the method detection limit (0.019 μg/mL). (b), The amount of plasma Ado from mice infected with the WT strain (0.5194 μg/mL) was significantly higher than that of plasma Ado from mice injected with Δnt (0.3584 μg/mL) or PBS (0.2321 μg/mL). The RP-HPLC chromatograms of above samples are shown in Supplementary Figure S3. Statistical significance was examined by one-way ANOVA and Dunnett’s multiple comparisons test. ns, not significant; *P < 0.05; n = 4.
NT causes monocytopenia in mouse blood in vivo
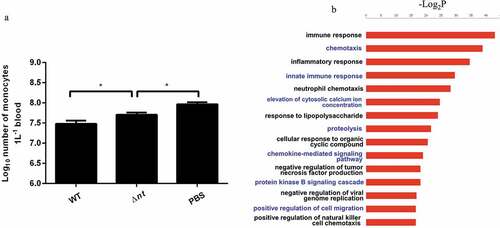
Since dAdo produced by NT has a cytotoxic effect on mouse macrophages in vitro, the number of monocytes from mouse blood was counted to evaluate the effect of dAdo in vivo. As shown in ), compared with the mock-infected group, the number of monocytes in blood from mice infected WT strain or Δnt was significantly less. The number of monocytes in blood from mice infected with the WT strain (3.2×107/L) was significantly less than that of monocytes in blood from mice infected with Δnt (5.2×107/L), verifying the significance of our in vitro findings.
Figure 4. S. suis NT contributes to dampening immune responses. (a), NT causes monocytopenia in mouse blood in vivo. The number of monocytes in blood from mice infected WT strain (3.2×107/L) was significantly less than that of monocytes in blood from mice infected with Δnt (5.2×107/L). Statistical significance was examined by one-way ANOVA and Dunnett’s multiple comparisons test. *P < 0.05; n = 5. (b), In vivo transcriptome analysis in mouse blood. Compared with blood from mice infected with the WT strain GZ0565, 320 genes were downregulated and 88 genes were upregulated in blood from mice infected with Δnt. The top five enrichment GO categories based on biological process for the total 408 differentially expressed genes belonged to immune response, chemotaxis, inflammatory response, innate immune response, and neutrophil chemotaxis.
To highlight the effect of dAdo in vivo, five mice per group were intraperitoneally injected with 0.5 mL of 121.3 μM dAdo or 0.5 mL of H2O per mouse. As shown in Supplementary Fig. S5, at 3 h after injection, the number of monocytes in blood from mice injected with dAdo was significantly less than that of monocytes in blood from mice injected with H2O.
In vivo transcriptome analysis in mouse blood
As shown above, NT synthesizes both Ado and dAdo in mouse blood in vivo. Next, we aimed at investigating the effect of NT through the generation of both Ado and dAdo on S. suis virulence. Compared with blood from mice infected with the WT strain GZ0565, 320 genes were downregulated and 88 genes were upregulated in blood from mice infected with Δnt (Supplementary Table S1). Interestingly, the top five enrichment GO categories based on biological process for the total 408 differentially expressed genes (DEGs) belonged to immune response, chemotaxis, inflammatory response, innate immune response, and neutrophil chemotaxis ()). Compared with the Δnt infection group, the concentration of dAdo in blood was significantly higher in the WT strain infection group. Nine genes involved in DNA damage response were downregulated in the Δnt infection group (i.e. upregulated in WT infection group). Seven genes involved in negative regulation of inflammatory response, four genes involved in negative regulation of cytokine production, and four genes involved in negative regulation of nitric oxide biosynthetic process were downregulated in the Δnt infection group (i.e. upregulated in the WT infection group). This effect was possibly mediated by Ado whose concentration was significantly higher in the WT infection group. The key DEGs involved in above GO categories or functions are listed in .
Table 1. The key DEGs in blood from mice between WT infection group and Δnt infection group.
To validate the results obtained by RNA sequencing (RNA-seq) analysis, five downregulated genes and five upregulated genes were analyzed by quantitative real-time PCR (qRT-PCR). As shown in Supplementary Fig. S6, the reliability of the transcriptional data obtained from the RNA-seq approach was confirmed by qRT-PCR analysis.
Discussion
This study demonstrates that S. suis NT is able to produce dAdo in vitro and in vivo. The production of dAdo by NT triggers cytotoxicity in mouse macrophages in vitro and causes monocytopenia in mouse blood in vivo. Thammavongsa et al. reported that production of dAdo by S. aureus AdsA induced caspase-3–mediated death of human macrophages [Citation17]. In the present study, we also show that the dAdo triggers caspase-3-dependent death of mouse macrophages. Carrera et al. showed that human monocytes were sensitive to dAdo and monocytes exposed to dAdo in vitro developed DNA strand breaks [Citation18]. DNA damage appears to be requisite for toxicity and dAdo causes the DNA damage through effects on enzymes implicated in DNA synthesis or repair [Citation18]. The in vivo transcriptome analysis in mouse blood indicated that the WT strain GZ0565 upregulated the expression of the host genes involved in DNA damage, which was possibly mediated by dAdo synthesized by NT. DNA from cells dying may provide sufficient dAdo to produce the effects on the function and survival of immune cells [Citation18]. Pathogenic bacteria may use the synthesized dAdo to evade host defenses. S. aureus produces two enzymes, nuclease and AdsA, to convert DNA from neutrophil extracellular traps to dAdo for the exclusion of macrophages from abscesses. Here, we show that S. suis produces dAdo by NT causing monocytopenia in mouse blood in vivo.
Ado also plays essential roles in regulating immune responses. It can inhibit pro-inflammatory and promote anti-inflammatory actions of immune cells [Citation20]. Recent studies highlight that pathogenic bacteria increase their virulence and survival in the host by synthesizing Ado. S. aureus and Bacillus anthracis escape from being killed by human neutrophils through producing Ado [Citation16]. Liu et al. reported that S. suis Ssads (designated as NT in our work) reduced oxidative activity and degranulation of human neutrophils by producing Ado. However, in the above studies, the effects of Ado produced by bacterial adenosine synthase on neutrophil functions were mainly investigated in vitro. In this study, we confirmed that S. suis NT synthesizes Ado in mouse blood in vivo. The in vivo transcriptome analysis in mouse blood indicated that the WT strain GZ0565 upregulated the expression of the genes involved in negative regulation of nitric oxide biosynthetic process (). In addition, we observed that two more interesting genes cd300ld and cxcr2 were downregulated in mouse blood in the WT infection group. The gene cd300ld encodes myeloid-associated immunoglobulin-like receptor (MAIR)-IV which regulates activation of neutrophils and plays an important role for innate immunity [Citation21]; the gene cxcr2 encodes CXC chemokine receptor 2, a major receptor regulating neutrophils recruitment, and binding of IL-8 to the receptor activates neutrophils; Herbold et al. showed that CXCR2 is important for regulating monocyte/macrophage recruitment in acute infections of the lungs caused by Streptococcus pneumoniae [Citation22]. These observations indicated that the inhibitory effect of NT on immune responses and neutrophil functions in vivo might be mediated through the generation of Ado. Moreover, a number of genes involved in negative regulation of immune responses were upregulated in mouse blood in the WT infection group, including the potent immunosuppressive cytokine IL-10 (). Ado increases IL-10 production by human monocytes [Citation23]. Additionally, the expression of gene cd300lb, encoding an activating immunoglobulin-like receptor (LMIR5), was decreased in mouse blood in the WT infection group. LMIR5 is a newly discovered inflammatory mediator and endogenous ligand of TLR4 involved in the positive regulation of inflammation [Citation24]. These findings may suggest the immunosuppressive activity of Ado on immune responses globally in vivo.
In conclusion, we have provided direct evidence that S. suis NT synthesizes dAdo in mouse blood, which causes monocytopenia in mouse blood in vivo. In addition, the in vivo transcriptome analysis in mouse blood shows that the inhibitory effect of NT on immune responses and neutrophil functions may be mediated through the generation of Ado ().
Figure 5. S. suis synthesizes dAdo and Ado by NT to dampen host immune responses. S. suis NT increases the concentration of dAdo and Ado in mouse blood in vivo. dAdo causes monocytopenia in mouse blood in vivo. In addition, the inhibitory effect of NT on immune responses and neutrophil functions in vivo may be mediated through the generation of Ado; the inhibitory effect of Ado on neutrophil functions through occupancy of A2a receptor was demonstrated by Liu et al. in vitro [Citation15]. The dot line means the directly inhibitory effect of Ado on immune responses and neutrophil functions in vivo needs to be further validated.
![Figure 5. S. suis synthesizes dAdo and Ado by NT to dampen host immune responses. S. suis NT increases the concentration of dAdo and Ado in mouse blood in vivo. dAdo causes monocytopenia in mouse blood in vivo. In addition, the inhibitory effect of NT on immune responses and neutrophil functions in vivo may be mediated through the generation of Ado; the inhibitory effect of Ado on neutrophil functions through occupancy of A2a receptor was demonstrated by Liu et al. in vitro [Citation15]. The dot line means the directly inhibitory effect of Ado on immune responses and neutrophil functions in vivo needs to be further validated.](/cms/asset/ed59d762-8e8a-4921-8bd8-0429a4457cd9/kvir_a_1520544_f0005_c.jpg)
Materials and methods
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids are listed in Supplementary Table S2. The S. suis serotype 9 virulent strain GZ0565 was isolated from a diseased pig with meningitis in Guangzhou, China, in 2005 [Citation14]. Escherichia coli BL21 (DE3) (Tiangen, China) was used for protein expression. All S. suis strains were grown in Todd-Hewitt broth (THB, Becton Dickinson, USA) and plated on Todd-Hewitt agar at 37°C, unless otherwise noted. E. coli strains were maintained in Luria-Bertani (LB, Becton Dickinson, USA) broth at 37°C. When necessary, appropriate antibiotics (Sigma-Aldrich, USA) were supplemented as follows: ampicillin, 100 μg/mL for E. coli; spectinomycin (SPC), 50 μg/mL for E. coli and 100 μg/mL for S. suis.
Ethics statement
All the animal experiments were approved by the Laboratory Animal Monitoring Committee of Jiangsu Province and performed in the Laboratory Animal Center of Nanjing Agricultural University (Permit number: SYXK (Su) 2017–0007).
Construction of nt deletion mutant and complemented strain
Deletion of nt in WT strain GZ0565 was achieved by allelic replacement as previously described [Citation25]. The primers used for constructing Δnt are listed in Supplementary Table S3. Upstream and downstream regions were amplified and fused by overlap-extension PCR. The fusion fragment was purified, digested with respective endonucleases, and then cloned into S. suis-E. coli shuttle vector pSET4s. Replication of pSET4s in E. coil is temperature independent, and it can replicate well at 37°C. However, replication of pSET4s in S. suis is temperature sensitive, and it can replicate on 28°C but not on 37°C. The recombinant plasmid was electrosporated into S. suis (Bio-Rad, Gene Pulser Xcell™) with the parameters: voltage 2300 V, capacitance 25 μF, resistance 200 Ω, and cuvette 1 mm. After electroporation, the cells were then diluted into five mL THB supplemented with 2% yeast extract and 10% sucrose without SPC and incubated at 28°C, 180 rpm for 4 h. The culture was taken and spread on THB agar plate with SPC. After incubation at 28°C for 24h, single colony was taken into five mL THB culture with SPC and incubated at 28°C, 180 rpm for 12–24 h. Then, 50 μL of above culture was taken into five mL THB culture with SPC at 37°C incubator for 12–24 h. The first single-crossover occurred. The culture was diluted into 10−5, and then spread on THB agar plate with SPC at 37°C. Single colony was taken into five mL THB culture without SPC at 28°C incubator for 12–24 h (without shaking). Since selection pressure was removed, S. suis is spontaneously loosing integrated plasmid by second single-crossover due to the homology sites between chromosome and plasmid. Five μL was taken into five mL THB culture without SPC at 28°C incubator for 12–24 h (without shaking). After repeating last step for 5–10 times, the culture was diluted into 10−5, and then spread on THB agar plates with SPC and without SPC at 37°C. After incubation at 37°C for 24h, select the colonies that only grew on plate without SPC. The deletion of nt was verified by PCR with primers NT-F/NT-R and L1/R2, and confirmed by DNA sequencing with primers NT-X/NT-Y.
The complementation of target gene into chromosomes has an obvious advantage over complementation by plasmids, preventing heterogeneity of gene expression because of the variation in the copy number of plasmids [Citation26]. From our RNA-seq data on S. suis strain GZ0565 (unpublished data), there is no transcript at the genomic intergenic region between BFP66_RS05970 and BFP66_RS05975 where the gene nt could be inserted into. The complemented strain for nt was constructed according to our previous report [Citation25]. A fragment of the genome between BFP66_RS05970 and BFP66_RS05975 was fused with nt and its promoter by overlap-extension PCR. The fusion product was cloned into pSET4s and then electroporated into competent cells of Δnt. The single-crossover mutant was selected as complemented strain at 37°C on THB agar under selection pressure with SPC. Complementation was verified by PCR and sequencing.
Expression of the recombinant truncated NT
Since the full length of NT could not be expressed water-soluble as described before [Citation15], the truncated NT (residues 36–377) containing two 5ʹ-nucleotidase domains was expressed. The primers for expressing tNT are listed in Supplementary Table S3. The detailed protocol for expressing and purifying tNT was described in our previous study [Citation27]. GroEL was expressed and purified in our previous study [Citation28].
The enzymatic activity of NT
The enzymatic activity of NT was carried out as described previously [Citation29], with some modifications. Briefly, six μM recombinant proteins tNT or GroEL were mixed with 63 mM dAMP in working buffer (240 mM Tris-HCl [pH 7.4], 2.4 mM MgCl2, 2.4 mM CaCl2, 2.5 mM MnCl2), resulting in the final volume of 500 μL. Then, the mixture was incubated at 37°C for 2 h. dAMP in working buffer was used as a control under the same reaction procedure. After incubation, 50 mM EDTA were used to stop the reaction. The reaction solution was centrifuged at 10,000 × g for 5 min to collect the supernatant. The IP released from dAMP was detected by Quanti-Chrom Phosphate Assay Kit DIPI-500 (Bioassay systems, USA) according to the manufacturer’s instructions. The dAdo released from dAMP was detected by RP-HPLC analysis. Samples were chromatographed on a 250 mm × 4.6 mm column (5μm particle size, Agilent C18). The mobile phase was 0.1% trifluoroacetic acid in deionized H2O. Samples were eluted for 25 min at a flow rate of 1.5 ml/min. Peaks were detected by UV absorbance at 260 nm. Commercial dAdo (Sigma-Aldrich, USA) was used as a standard sample.
To investigate whether NT can also degrade dGMP, dCMP, and dTMP, WT, Δnt, and C-nt were incubated with dGMP, dCMP, and dTMP, respectively. The substrate dAMP was used as a positive control. Briefly, log-phase cultures of bacteria were centrifuged, washed, and resuspended in working buffer. 0.5 mL of bacteria were mixed with 0.5 mL of 5mM dGMP, dCMP, dTMP, and dAMP in working buffer, respectively. The mixture was incubated at 37°C for 1.5 h. The IP resealed was detected as described above. The ability of NT to degrade these four substrates was calculated as: the concentration of IP resealed (μmol)/the number of bacteria (108 colony-forming units).
Detection of Ado and dAdo in mouse blood in vivo
Six-week-old female SPF CD1 mice (Beijing Vital River Laboratory Animal Co. Ltd) were used to determine the abundance of Ado and dAdo in mouse blood. Log-phase cultures of WT strain or Δnt were centrifuged, washed, and resuspended in PBS. The mice (12 mice per group) were inoculated by intraperitoneal injection of WT strain or Δnt at the dose of 7 × 107 colony-forming units (CFU) per mouse. The mice from the mock-infected group were injected with PBS using the same volume. At 12 hours post-infection, mouse plasma was collected in anticoagulant tubes. The extraction of Ado and dAdo from plasma was performed as previously described [Citation16]. Plasmas from three mice were pooled as one sample resulting in four samples in total for each group. One mL pooled plasma was taken for extracting Ado and dAdo. One hundred twenty-five μL perchloric acid (1.5 M) was added into one mL pooled plasma and then the sample was vortexed for 10 seconds. After centrifuging for 5 min at 16,000 × g, 850 μL supernatant was collected and neutralized with 49 μL 4 M KOH. After ice cooling for 10 min, the sample was centrifuged at 16,000 × g, 4°C for 5 min, adjusted to pH 6–7, diluted 1:3 with PBS, and filtered with a 0.22 μm syringe filter. The presence of Ado and dAdo in the sample was determined by RP-HPLC analysis. The samples underwent chromatography on a 250 mm × 4.6 mm column (5 μm particle size, Shiseido C18, Japan). The mobile phase consisted of solution A (0.1% trifluoroacetic acid in deionized H2O) and solution B (0.1% trifluoroacetic acid in acetonitrile). The sample was then eluted for 20 min at a flow rate of 0.5 mL/min. Peaks were detected by UV absorbance at 260 nm. Commercial Ado and dAdo (Sigma-Aldrich, USA) were used as standard samples.
To further validate the results of dAdo and Ado in the plasma of the mice infected with S. suis obtained by RP-HPLC analysis, we have reproduced the experiment and the dAdo and Ado in the plasma were detected by LC-MS/MS analysis (Thermo Scientific™ TSQ Quantum Ultra™ triple quadrupole mass spectrometer). The mice (9 mice per group) were inoculated by intraperitoneal injection of WT strain or Δnt at the dose of 7 × 107 CFU per mouse. The mice from the mock-infected group were injected with PBS using the same volume. At 12 hours post-infection, mouse plasma was collected in anticoagulant tubes. The extraction of Ado and dAdo from plasma was performed by a simple protein precipitation with acetonitrile [Citation30,Citation31]. Plasmas from three mice were pooled as one sample, resulting in three samples in total for each group. The chromatographic separation was performed on a 150 mm × 2.1 mm column (5 μm particle size, Shiseido C18, Japan) with a mobile phase of 10% acetonitrile in deionized H2O. The sample was then eluted for 8 min at a flow rate of 0.25 mL/min. Detection was carried out on a triple quadrupole tandem mass spectrometer equipped with electrospray ionization (ESI) in positive ion selective reaction monitoring (SRM) mode. The monitored transitions were 252.131→136.00 for dAdo and 268.300→136.11 for Ado, respectively.
Cellular cytotoxicity assay
The mouse macrophages RAW 264.7 (ATCC TIB-71) were purchased from ATCC. The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (Gibco, USA) at 37°C in 5% CO2. To measure cytotoxicity, RAW 264.7 cells (5 × 105 cells/well) were seeded into 24‑well plates and incubated overnight. The dCF (MedChem Express, USA) was added into the cells according to previous studies [Citation17,Citation32,Citation33]. The cells underwent 12 different treatments in the presence or absence of 100 μM dCF as indicated: sterile H2O; 55 μM dAdo (suspended in sterile H2O); 6 μM tNT; 63 mM dAMP (suspended in working buffer); tNT+ dAMP; working buffer only. For each treatment, the solution with the volume of 30 μL was added into cells with the volume of 500 μL culture medium. Then, the cells were incubated at 37°C in 5% CO2 for 24 hours. The cells were then stained with Trypan Blue Stain (0.4%) (Invitrogen, USA) and counted using Countess™ II FL Automated Cell Counter (Life Technologies, USA) with Countess™ Cell Counting Chamber Slides (Invitrogen, USA).
The mouse macrophages RAW 264.7 were interacted with mid-log growth phase bacteria at a MOI of 50 (bacteria: macrophage) for 2 h at 37°C in 5% CO2. The cytotoxicity assay was performed using LDH (lactate dehydrogenase) cytotoxicity assay kit (Beyotime Biotechnology, China) according to the manufacturer’s instructions. The percentage of cytotoxicity was calculated as: [(bacteria-treated LDH activity – spontaneous LDH activity)/(maximum LDH activity – spontaneous LDH activity)] ×100%. The experiments were repeated for four times.
Western blot
RAW 264.7 cells were treated with dAdo (in the presence or absence of dCF) for 12 h, 16 h, or 24 h, respectively. RAW 264.7 cells were also treated with tNT+ dAMP (in the presence or absence of dCF) for 12 h. Jurkat cell extract treated with cytochrome c in vitro, obtained from Cell Signaling Technology (#9663), was used as a positive control. The cells were then lysed, and cellular proteins were separated by 15% SDS-PAGE; the resolved proteins were transferred onto a PVDF membrane (GE Healthcare, USA). The detailed procedure for Western blotting was described in our previous study [Citation28]. The antibody against the cleaved caspase-3 or antibody against β-actin (1:1000 dilution, Cell Signaling Technology, USA) were used as the primary antibody. The anti-rabbit IgG–HRP or goat anti-mouse IgG–HRP (β-Actin) (1:2000 dilution, Boster, China) were used as the secondary antibody. The experiments were done in triplicate.
The number of monocytes in mouse blood
Five mice per group were intraperitoneally infected with the WT strain or Δnt at 7 × 107 CFU per mouse and the mock-infected mice were injected with PBS. The blood was collected in anticoagulant tubes at 12 h post-infection and the number of monocytes was counted by Auto Hematology Analyzer BC-5000 Vet (Mindary, China).
To highlight the effect of dAdo in vivo, five mice per group were intraperitoneally injected with 0.5 mL of 121.3 μM dAdo or 0.5 mL of H2O per mouse. The blood was collected in anticoagulant tubes at 3 h after injection and the number of monocytes was counted by Auto Hematology Analyzer BC-5000 Vet.
RNA preparation from mouse blood and in vivo transcriptome analysis
In vivo transcriptome analysis in mouse blood was performed according to our previous report [Citation34]. Bacteria were collected as described above. The mice were divided into two groups and intraperitoneally injected with the dose of 5 × 107 CFU of either the WT strain or Δnt. Blood was collected at 12 h after infection. Before transcriptome analysis, a preliminary experiment was preformed to show that the loads of bacteria in the blood between WT strain infection group and ∆nt infection group was comparable at 12 h after infection. RiboPure™-Blood Kit (Life Technologies, USA) was used to extract the blood total RNA according to the manufacturer’s instructions. Two biological replicates were prepared for each group. The RNA samples were then sent to BGI-Shenzhen, China, for RNA-seq. Sequencing library was carried out as previously described [Citation35]. The sequencing was performed by Illumina HiSeqTM 2000 according to the manufacturer’s protocol. The RNA-seq data were analyzed as previously described [Citation35]. NOISeq approach was used to calculate the probability value [Citation36]. NOISeq approach has obvious advantages over other most existing methodologies, such as Fisher’s exact test, edgeR, baySeq, and DESeq [Citation36]. The probability of 0.8 is chosen as a cutoff value for NOISeq [Citation36]. The differentially expressed genes (DEGs) were selected with probability ≥0.8 and the fold change ≥2. In total, 17,686 genes were detected and 408 genes of them (2.307%) were differentially expressed. Gene Ontology (GO) categories based on biological process were analyzed using the GO project (http://www.geneontology.org). This method firstly maps all DEGs to GO terms in the database (http://www.geneontology.org/), calculating gene numbers for every term, then hypergeometric test was used to find significantly enriched GO terms in DEGs comparing to the genome background. In addition, an online program, DAVID bioinformatics resources 6.8, was also used for GO analysis [Citation37].
Quantitative real-time PCR
The detailed procedure for qRT-PCR analysis was described in our previous study [Citation34]. For qRT-PCR analysis, three biological replicates were used for each group including two of them from transcriptome analysis. Specific primers for qRT-PCR are listed in Supplementary Table S3. qRT-PCR with SYBR Premix Ex TaqTM (Takara) was performed on QuantStudio 6 Flex (Thermo Fisher Scientific, China) according to the manufacturer’s protocol. The relative fold change was calculated based on the 2−ΔΔCT method.
Statistical analysis
Statistical significance was examined by one-way ANOVA and Dunnett’s multiple comparisons test using GraphPad Prism 6.
Abbreviations
| RP-HPLC | = | reversed-phase high performance liquid chromatography |
| NT | = | 5ʹ-nucleotidase |
| tNT | = | recombinant truncated 5ʹ-nucleotidase |
| AMP | = | adenosine monophosphate |
| Ado | = | adenosine |
| CPS | = | capsular polysaccharide |
| dAMP | = | deoxyadenosine monophosphate |
| dAdo | = | 2ʹ-deoxyadenosine |
| IP | = | inorganic phosphate |
| WT | = | wild type |
| Δnt | = | nt deletion mutant |
| C-nt | = | complemented strain |
| RNA-seq | = | RNA sequencing |
| qRT-PCR | = | quantitative real-time PCR |
| GroEL | = | belonging to the chaperonin family of molecular chaperones |
| GO | = | gene ontology |
| DEGs | = | differentially expressed genes |
| LC-MS/MS | = | liquid chromatography with tandem mass spectrometry |
Supplemental Material
Download Zip (4.2 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Segura M, Fittipaldi N, Calzas C, et al. Critical Streptococcus suis virulence factors: are they all really critical? PubMed PMID: 28274524 Trends Microbiol. 2017;257:585–599.
- Goyette-Desjardins G, Auger JP, Xu J, et al. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. PubMed PMID: 26038745 Emerg Microbes Infect. 2014;36:e45.
- Kerdsin A, Hatrongjit R, Gottschalk M, et al. Emergence of streptococcus suis serotype 9 infection in humans. PubMed PMID: 26362754 J Microbiol Immunol Infect. 2017;504:545–546.
- Hatrongjit R, Kerdsin A, Gottschalk M, et al. First human case report of sepsis due to infection with streptococcus suis serotype 31 in Thailand. BMC Infect Dis. 2015;15:392. PubMed PMID: 26420029.
- Fittipaldi N, Collis T, Prothero B, et al. Streptococcus suis meningitis, Hawaii. Emerg Infect Dis. 2009;15(12): 2067–2069. PubMed PMID: 19961708.
- Benga L, Fulde M, Neis C, et al. Polysaccharide capsule and suilysin contribute to extracellular survival of streptococcus suis co-cultivated with primary porcine phagocytes. PubMed PMID: 18565698 Vet Microbiol. 2008;1321–2:211–219.
- Lecours MP, Gottschalk M, Houde M, et al. Critical role for streptococcus suis cell wall modifications and suilysin in resistance to complement-dependent killing by dendritic cells. PubMed PMID: 21849289 J Infect Dis. 2011;2046:919–929.
- Seele J, Singpiel A, Spoerry C, et al. Identification of a novel host-specific IgM protease in streptococcus suis. J Bacteriol. 2013;195(5): 930–940. PubMed PMID: 23243300.
- Zhang A, Mu X, Chen B, et al. Identification and characterization of IgA1 protease from streptococcus suis. PubMed PMID: 19619964 Vet Microbiol. 2010;1401–2:171–175.
- Spoerry C, Seele J, Valentin-Weigand P, et al. Identification and characterization of IgdE, a Novel IgG-degrading protease of streptococcus suis with unique specificity for porcine IgG. PubMed PMID: 26861873 J Biol Chem. 2016;29115:7915–7925.
- Pian Y, Wang P, Liu P, et al. Proteomics identification of novel fibrinogen-binding proteins of Streptococcus suis contributing to antiphagocytosis. Front Cell Infect Microbiol. 2015;5:19. PubMed PMID: 25789245.
- Xu J, Fu S, Liu M, et al. The two-component system NisK/NisR contributes to the virulence of Streptococcus suis serotype 2. PubMed PMID: 24342108 Microbiol Res. 2014;1697–8:541–546.
- Willenborg J, Fulde M, de Greeff A, et al. Role of glucose and CcpA in capsule expression and virulence of Streptococcus suis. PubMed PMID: 21349980 Microbiology. 2011;157Pt 6:1823–1833.
- Wu Z, Zhang W, Lu C. Comparative proteome analysis of secreted proteins of Streptococcus suis serotype 9 isolates from diseased and healthy pigs. PubMed PMID: 18554861 Microb Pathog. 2008;453:159–166.
- Liu P, Pian Y, Li X, et al. Streptococcus suis adenosine synthase functions as an effector in evasion of PMN-mediated innate immunit. PubMed PMID: 24446521 J Infect Dis. 2014;2101:35–45.
- Thammavongsa V, Kern JW, Missiakas DM, et al. Staphylococcus aureus synthesizes adenosine to escape host immune responses. PubMed PMID: 19808256 J Exp Med. 2009;20611:2417–2427.
- Thammavongsa V, Missiakas DM, Schneewind O. Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. PubMed PMID: 24233725 Science. 2013;3426160:863–866.
- Carrera CJ, Terai C, Lotz M, et al. Potent toxicity of 2-chlorodeoxyadenosine toward human monocytes in vitro and in vivo. A novel approach to immunosuppressive therapy. PubMed PMID: 1700795 J Clin Invest. 1990;865:1480–1488.
- Blackburn MR, Kellems RE. Adenosine deaminase deficiency: metabolic basis of immune deficiency and pulmonary inflammation. Adv Immunol. 2005;86:1–41. PubMed PMID: 15705418.
- Hasko G, Pacher P. A2A receptors in inflammation and injury: lessons learned from transgenic animals. PubMed PMID: 18160539 J Leukoc Biol. 2008;833:447–455.
- Nakano T, Tahara-Hanaoka S, Nakahashi C, et al. Activation of neutrophils by a novel triggering immunoglobulin-like receptor MAIR-IV. PubMed PMID: 17543387 Mol Immunol. 2008;451:289–294.
- Herbold W, Maus R, Hahn I, et al. Importance of CXC chemokine receptor 2 in alveolar neutrophil and exudate macrophage recruitment in response to pneumococcal lung infection. Infect Immun. 2010;78(6):2620–2630. PubMed PMID: 20368349.
- Le Moine O, Stordeur P, Schandene L, et al. Adenosine enhances IL-10 secretion by human monocytes. J Immunol. 1996;156(11): 4408–4414. PubMed PMID: 8666814.
- Phongsisay V, Iizasa E, Hara H, et al. LMIR5 extracellular domain activates myeloid cells through toll-like receptor 4. PubMed PMID: 25004110 Mol Immunol. 2014;621:169–177.
- Wu Z, Wu C, Shao J, et al. The Streptococcus suis transcriptional landscape reveals adaptation mechanisms in pig blood and cerebrospinal fluid. RNA. 2014;20(6):882–898. PubMed PMID: 24759092.
- Dortet L, Mostowy S, Samba-Louaka A, et al. Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PubMed PMID: 21829365 PLoS Pathog. 2011;78:e1002168.
- Zhu H, Huang D, Zhang W, et al. The novel virulence-related gene stp of Streptococcus suis serotype 9 strain contributes to a significant reduction in mouse mortality. PubMed PMID: 21924346 Microb Pathog. 2011;516:442–453.
- Lai L, Dai J, Tang H, et al. Streptococcus suis serotype 9 strain GZ0565 contains a type VII secretion system putative substrate EsxA that contributes to bacterial virulence and a vanZ-like gene that confers resistance to teicoplanin and dalbavancin in Streptococcus agalactiae. Vet Microbiol. 2017;205:26–33. PubMed PMID: 28622857.
- Ma F, Guo X, Fan HExtracellular nucleases of streptococcus equi subsp. Zooepidemicus degrade neutrophil extracellular traps and impair macrophage activity of the hostAppl Environ Microbiol201783(2). doi: 10.1128/AEM.02468-16 PubMed PMID: 27815272.
- Sharma K, Singh R, Giri S, et al. Highly sensitive method for the determination of adenosine by LC-MS/MS-ESI: method validation and scope of application to a pharmacokinetic/pharmacodynamic study. PubMed PMID: 21491472 Biomed Chromatogr. 2012;261:81–88.
- Jia Y, Wang B, Wu X, et al. Simultaneous quantification of 2ʹ,3ʹ,5ʹ-tri-O-acetyl-N6-(3-hydroxylaniline)adenosine and its principal metabolites in hamster blood by LC-MS/MS and its application in pharmacokinetics study. J Chromatogr B Analyt Technol Biomed Life Sci. 2016;1022:46–53. PubMed PMID: 27082762.
- Dennis A, Jk CARSON, Matsumoto STEVEN, et al. Biochemical basis for the enhanced toxicity of deoxyribonucleosides toward malignant human T cell lines. Medical Sciences. 1979;76(5):2430–2433.
- Knudsen Rsw TB, Otey SK, Blackburn MR, et al. Effects of (R)-Deoxycoformycin (Pentostatin) on Intrauterine nucleoside catabolism and embryo viability in the pregnant mouse. WILEY-LISS INC. 1992;45:91–103.
- Wu Z, Shao J, Ren H, et al. A streptococcus suis LysM domain surface protein contributes to bacterial virulence. Vet Microbiol. 2016;187:64–69. PubMed PMID: 27066710.
- Chen HY, Shen H, Jia B, et al. Differential gene expression in ovaries of qira black sheep and hetian sheep using rNA-Seq technique. PubMed PMID: 25790350 PLoS One. 2015;103:e0120170.
- Tarazona S, Garcia-Alcalde F, Dopazo J, et al. Differential expression in RNA-seq: a matter of depth. PubMed PMID: 21903743 Genome Res. 2011;2112:2213–2223.
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. PubMed PMID: 19131956 Nat Protoc. 2009;41:44–57.
