ABSTRACT
Vibrio splendidus is one of the most opportunistic marine pathogens and infects many important marine animals, including the sea cucumber Apostichopus japonicus. In this study, two genes named DLD1 and DLD2, encoding dihydrolipoamide dehydrogenase (DLD) homologues in pathogenic V. splendidus, were cloned, and conditionally expressed in Escherichia coli BL21 (DE3). The enzymatic activities of DLD1 and DLD2 showed that they both belonged to the NADH oxidase family. Both DLD1 and DLD2 were located on the outer membrane of V. splendidus as detected by whole-cell ELISA. To study the adhesion function of DLD1 and DLD2, polyclonal antibodies were prepared, and antibody block assay was performed to detect the normal function of the two proteins. DLD1 and DLD2 were determined to play important roles in adhesion to different matrices and the adhesive ability of V. splendidus reduced more than 50% when DLD1 or DLD2 was defective.
Introduction
The sea cucumber Apostichopus japonicus (Echinodermata, Holothuroidea) is a marine animal with important economic values [Citation1] and is particularly important to the economy of North China [Citation2]. The bacterial infectious diseases of A. japonicus, such as skin ulcer syndrome (SUS) [Citation3], perioral swelling syndrome [Citation4] and rotting edge syndrome [Citation5], frequently occur in aquaculture. Vibrio sp. [Citation6], Pseudomonas sp. [Citation7] and spherical virus [Citation8] are the main pathogens of A. japonicus. In particular, Vibrio splendidus is considered to be the major pathogen that infects A. japonicus [Citation9]. However, until now, little is known about the pathogenic mechanism of V. splendidus, which has significantly hindered the development of specific prevention methods for A. japonicus.
The virulence factors of Vibrio sp. generally include adhesion factors, hemolysins, and extracellular products [Citation10]. The metalloproteinase Vsm is involved in the interaction between V. splendidus and A. japonicus and contributes to the cytotoxicity effects on the A. japonicus coelomocyte [Citation11–Citation13]. Hemolysin Vshppd not only is involved in the cytotoxicity to coelomocyte but also contributes to the stimulatory effect on the immune response [Citation14]. When expressed in the cytoplasm under the control of the CUP1 promoter, Vis was toxic to yeast, and catalytic variants lost the ability to kill the yeast host, indicating that the toxin exerts its lethality through its enzymatic activity [Citation15]. These studies on the pathogenicity of V. splendidus are far from enough.
In general, adhesion is the first step of bacterial infection and bacterial adherence is a complicated process of interaction between a pathogen and its host [Citation16,Citation17]. However, there has been no report on the adhesion factor of V. splendidus and its adhesive process until now. Flagellar assembly-associated proteins, such as flrA, flrB, and flrC, have been reported to be typical adhesion factors [Citation18]. In Vibrio mimicus, outer membrane protein U (OmpU) was shown to be involved in adhesion [Citation19]. V. splendidus possesses the characteristics of strong hydrophobicity and high biofilm formation ability [Citation20], which made us wonder whether it possesses adhesion factors or not, and what are the adhesion factors contributing to its pathogenicity. Till now, no adhesion factor has been reported in V. splendidus, so finding basic adhesion factors and exploring their ability is very important for the exploration of the pathogenicity of V. splendidus.
Dihydrolipoamide dehydrogenase (DLD) is present in a variety of organisms and is an oxidoreductase that is essential in energy metabolism [Citation21]. It is a part of three α-pyruvate dehydrogenase complex species, and belongs to the flavin protein oxidoreductase family [Citation22,Citation23]. DLD not only performs the function of oxidoreductase but also plays an important role in bacterial pathogenesis. DLD was determined to be one of the virulence determinants in Mycoplasma gallisepticum by signature sequence mutagenesis [Citation24]. DLD-deficient Streptococcus pneumoniae lost the ability to infect mice [Citation25]. In the present study, two DLD genes were cloned, and their enzymatic activities were characterized. The localization of DLDs was also determined using whole cell enzyme-linked immunosorbent assay (ELISA) and the adhesive ability of DLD was explored.
Materials and methods
Bacterial strains, culture conditions and chemicals
V. splendidus was isolated from A. japonicus suffering from SUS in an indoor farms in Jinzhou Hatchery in May 2013, and its identity was determined using 16S rDNA sequence. Its pathogenicity to A. japonicus was determined in our previous study [Citation26]. This bacterium was stored in glycerol at −80°C for further utilization. Unless otherwise stated, V. splendidus was cultured in modified Zobell’s 2216E medium at 28°C (tryptone, 5 g; yeast extract, 1 g; and FePO4, 0.01 g in 1 L aged seawater). Escherichia coli DH5α, S17λπ and BL21 (DE3) was cultured in Luria-Bertani (LB) medium at 37°C. Cell density was measured at 600 nm by a UV-Vis spectrophotometer (Beckman). Culture of V. splendidus or E. coli at an OD600 = 1.0 was corresponded to the cell density of 1.01 × 109 CFU mL−1. Ampicillin (Ap, 100 μg mL−1) and kanamycin (Kn, 50 μg mL−1) were used in this study. Plasmid pMD19-T, Taq and Pfu DNA polymerase was Clontech purchased from Takara (China). Restriction endonucleases were purchased from New England Biolabs. 5-([4,6-dichlorotriazin-2-yl] amino) fluorescein hydrochloride (5-DTAF) was purchased from Sigma (USA). All the other chemicals used in this study were purchased from Sangon (Shanghai, China) unless otherwise stated.
DNA manipulation and plasmid construction
The plasmid preparation, the extraction of DNA fragments from agarose gels and the purification of PCR products were performed using the respective kits from Omega Bio-Tek (GA) according to the manufacturer’s instructions. According to the genomic DNA of V. splendidus LGP32, we found two nucleotides sequences encoding DLD, and they were named DLD1 and DLD2. Four primers, DLD1F:5ʹ-CGCGGATCCATGAGCAAAGAAATTAAA-3ʹ, DLD1R: 5ʹ-ACGCTCGAGTTACTTCTTCTTTTTCACTG-3ʹ, DLD2F: 5ʹ-GGATCCATGAAACAAGTCAATGTAGATGTAGC-3ʹ, and DLD2R: 5ʹ-CTCGAGTTAACAACCAGGACCACAGTCTA-3ʹ were designed according to the two nucleotide sequences. Using the two pairs of primers, DLD1 and DLD2 were amplified and ligated to the pMD19-T. pET-28a-DLD1 or pET28a-DLD2 was constructed by ligating DLD1 or DLD2 between the BamH I and Xho I sites of pET28a.
Expression and purification of recombinant DLD
Overnight culture of E. coli BL21 (DE3)/pET28a-DLD1 or E. coli BL21 (DE3)/pET28a-DLD2 was inoculated into 100 mL LB medium with Kn and cultured at 37°C until the OD600 reached 0.5. Isopropyl-β-D -thiogalactopyranoside was added into the culture at a working concentration of 0.4 mM to induce the expression of DLD and the induction process lasted for 6 h at 28°C. Then, the cells were harvested by centrifuging at 8000 × g for 10 min. Cells were precipitated and used for the purification of recombinant protein. Recombinant His-tagged DLD1 or DLD2 was purified using Ni-NTA Sepharose column (Roche, Shanghai). The recombinant protein was determined by 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and the concentration of recombinant protein was measured using the BCA Protein Assay Kit (Sangon, China).
Antibody preparation
Polyclonal antibodies against DLD1 and DLD2 were generated by immunizing 4-week-old mice according to the method described in [Citation27]. Blood was taken from mice through the eyeball and polyclonal antibodies against DLD1 and DLD2 were stored at −80°C for further utilization.
Enzymatic assay
The DLD activity of DLD1 and DLD2 was determined based on the method of Serrano [Citation28] with slight modification. The DLD activity was calculated based on measurement of NADH oxidation using disulfide substrate as a substrate at 25°C. The reaction was monitored by the decrease in absorbance at 340 nm. Reaction mixtures contained 10 μM PBS (pH 7.0), 3 μM DL-lipoamide (ANPEL Laboratory Technologies, Shanghai), 2 μM NADH (Solarbio, China), and 100 μL of enzyme in a final volume of 3 mL.
Whole cell ELISA
To determine whether DLD was located on the cell surface, a whole cell ELISA was performed according to the protocol of Aquatic Diagnostic Ltd. (UK). Polyclonal antibodies against DLD1 and DLD2 were diluted 1:400 with PBS. A total of 100 μL of mid-logarithmic phase V. splendidus at approximately 1.0 × 107 CFU mL−1 was added separately to a 96-well ELISA plate coated with 0.05% (w/v) poly-L-lysine. The well without incubation with V. splendidus cells was used as a negative group. There were three replicates for each sample. Then, 100 μL of 2.5% (v/v) glutaraldehyde was added to each well to fix cells for 4 h, and each well was washed three times with 300 μL of PBS (the plate was inverted on absorbent paper to avoid cross-contamination). After fixation, 300 μL of 1% (w/v) bovine serum albumin was added to block the sample for overnight, followed by three times washes. The cells were treated with 1:400 diluted anti-DLD polyclonal antibody above. The wells treated without the polyclonal antibody were used as the control group. After washing, the wells were treated with horseradish peroxidase-conjugated goat anti-mouse IgG (Bios, China). Color development was performed using the TMB Kit (Solarbio, China) and then 50 μL of hydrochloric acid (1 M) was added to terminate the reaction. The absorbance of the developed color was read at 450 nm with a UV-Vis spectrophotometer (Beckman) [Citation29].
Fluorescence labeling of V. splendidus
To observe the adhesive ability of V. splendidus, V. splendidus cells were stained with 5-DTAF (Sigma, USA) as described by [Citation30] and visualized under a microscope. The working concentration of 5-DTAF was 0.005 mg mL−1 (dissolved in PBS). Five millilitres of mid-logarithmic phase V. splendidus (OD600 = 0.5) and 2 mL of the 5-DTAF solution were mixed in a dark environment at 28°C and incubated in an oscillating incubator for 1 h. Then, DTAF-labeled V. splendidus (Vs-DTAF) cells were collected by centrifugation at 8000 × g for 6 min and resuspended in PBS. The wells were repeatedly washed until the supernatant showed no color [Citation31,Citation32]. Finally, Vs-DTAF cells were observed under a fluorescence microscope (Nikon).
Adhesion of V. splendidus to polystyrene
In this study, the efficiency of V. splendidus to adhere to polystyrene was evaluated by cell number count (CP) and epifluorescence microscopy methods [Citation33]. Vs-DTAF and polyclonal antibodies (diluted 1:400 with PBS) were mixed at a ratio of 1:1 (v/v). Then, it was incubated in an oscillating incubator at 28°C for 2 h. In the control group, V. splendidus was mixed with an equal volume of PBS and incubated under the same conditions. Then, 400 μL of the incubated mixed solution was placed onto a polystyrene plate to adhere for 4 h in the dark. Then, the excess liquid was removed, and then the adhered cells were washed with PBS for 10 min. After washing, 2.5% glutaric dialdehyde (Solarbio, China) was added to the polystyrene plate for fixation. The polystyrene plate was observed under a fluorescence microscope and the elution was coated on 2216E medium after a 1.0 × 103-fold dilution. The adhesion rate was calculated based on the number of single colonies that emerged on the plate.
Coelomocyte culture
Healthy adult A. japonicus (weight 125 ± 15 g) were obtained from Zhangzi Island Aquaculture Company (Dalian, China) and acclimatized in 30 L of aerated natural seawater (salinity 28 psu, temperature 16°C) for 3 days. Primary coelomocytes were prepared according to our previous work [Citation34]. Briefly, the harvested cells were resuspended in L-15-S cell culture medium with sodium (Invitrogen, USA), at a final concentration of 1.0 × 106 cells mL−1. NaCl solution was utilized at a final concentration of 0.39 M to adjust the osmotic pressure. The cells were then dispensed into a 24-well culture microplate with 500 μL of L-15-S medium with sodium in each well. All the experiments were performed in 48 h to ensure that the cells were in a healthy adherent state.
Adhesion of V. splendidus to coelomocytes
According to the cell adhesion counting method of [Citation35], coelomocytes were cultured in a 24-well plate. A total of 0.5 mL of L-15-S cell culture medium with sodium (Invitrogen, USA) was added to each well until the monolayer cells were cultured. The wells were divided into three groups: one group was incubated with Vs-DTAF preincubated with polyclonal DLD1 antibody. The second group was incubated with Vs-DTAF preincubated with polyclonal DLD2 antibody. The control group was incubated with V. splendidus. The antibody was preincubated with Vs-DTAF at a ratio of 1:1 (v/v), with three replicates for each group. After incubation for 2 h, the spent culture medium was removed and 400 μL of the mixtures was added to each well. The 24-well plate was incubated at room temperature for 3 h. After that, the suspension was removed and the cells that adhered to the coelomocytes were resuspended in 1 mL of PBS. The resuspension was 1.0 × 101- to 1.0 × 104-fold diluted and plated on 2216E medium. The rest of the resuspension was stained with Dil (Beyotime, China) and visualized using a laser scanning spectral con-focal microscope (TCS SP2; Leica, Solms, Germany).
Adhesion of V. splendidus to tissues of A. japonicus
Ten individuals of healthy adult A. japonicus (weight 125 ± 15 g) were obtained from Zhangzi Island Aquaculture Company (Dalian, China) and acclimatized in 30 L of aerated natural seawater (salinity 28 psu, temperature 16°C) for 3 days. We divided the A. japonicus individuals into three groups in 10 L of aerated natural seawater, and there were three A. japonicus specimens for each group. A total of 0.1 L V. splendidus (OD600 = 1.0) and polyclonal antibodies (diluted 1:400 with PBS) were mixed at 28°C and incubated in an oscillating incubator for 2 h at a ratio of 1:1 (v/v) and then centrifuged at 8,000 g for 10 min. The control group was V. splendidus (0.1 L, OD600 = 1.0) mixed with the equivalent amount of PBS. After immersion infection for 24 h, the body wall, tentacle, muscle, respiratory tree and intestine of A. japonicus were collected and weighed. For each sample, three A. japonicus specimens were collected. Tissue samples were homogenized by a homogenizer with sterilized PBS and then the homogenate was 1.0 × 101- to 1.0 × 104-fold diluted and plated on 2216E medium. The plates were cultured at 28°C for 24 h [Citation36,Citation37].
Sequence analysis
The sequences of DLD1 and DLD2 were analyzed using the BLAST algorithm at the National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast). The deduced amino acid sequences were analyzed using the Expert Protein Analysis System (http://www.expasy.org/). The molecular mass (MM) and theoretical isoelectric point (pI) were calculated by the Protparam tool (http://www.expasy.ch/tools/protparam.html). The putative signal peptide cleavage site was identified using the SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/). Domain in these amino acid sequences was detected using the simple modular architecture research tool (SMART) program (http://www.smart.emblheidelbergde/) and multiple alignment analysis of proteins was performed using the Clustal Omega Multiple Alignment program (http://www.ebi.ac.uk/clustalw/) [Citation38]. The neighbor-joining phylogenetic tree was constructed using Mega 5.0 program with 1000 bootstraps [Citation39].
Results
Cloning of DLD and sequence analysis of DLD
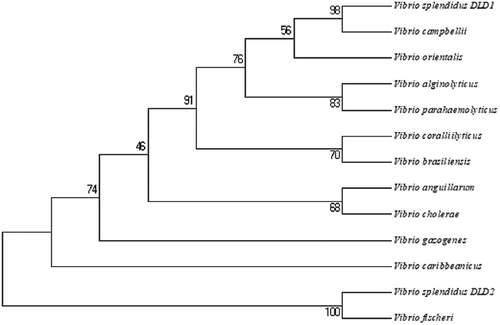
There were two proteins annotated as DLD, named DLD1 (WP_010435969.1) and DLD2 (WP_004729673.1) from the whole sequence of V. splendidus LGP32. DLD1 was amplified by PCR with the primers DLD1F and DLD1R. Nucleotide sequence analysis showed that the ORF of DLD1 was 1431 bp. DLD1 encoded a protein with an estimated molecular mass of 50.9 kDa, and its theoretical pI was 5.62. Likewise, DLD2 was amplified by PCR with the primers DLD2F and DLD2R. Nucleotide sequence analysis showed that the ORF of DLD2 was 1467 bp. DLD2 encoded a protein with an estimated molecular mass of 53.16 kDa, and its theoretical pI was 5.28. SingalP prediction revealed that both DLD1 and DLD2 had no typical signal peptide. BLAST showed that the amino acid sequences of DLD1 and DLD2 were similar to those of Vibrio alginolyticus, Vibrio crassostreae, and Vibrio fischeri, and the similarity reached 99%. Multiple sequence alignment showed a highly conserved amino acid sequence between DLD1 and other Vibrio spp., while DLD2 showed a slightly lower similarity ()). Smart analysis showed that both DLD1 and DLD2 contained a Pyr_redox domain and GIDA domain (,)). They both showed close evolutionary relationships with other Vibrio spp. in the phylogenetic tree analysis ().
Figure 1. (a): Multiple sequence alignment of DLDs from Vibrio spp. The GenBank accession numbers are as follows: DLDs of Vibrio fischeri WP_069594165.1, Vibrio splendidus (DLD1) WP_010435969.1, Vibrio splendidus (DLD2) WP_004729673.1, Vibrio campbellii WP_012128871.1, Vibrio coralliilyticus WP_006958123.1, Vibrio parahaemolyticus WP_021823119.1, Vibrio alginolyticus AGK62253.1, Vibrio mimicus WP_000031532.1, Vibrio cholerae WP_000031535.1, Vibrio anguillarum WP_013857673.1, Vibrio vulnificus RAH35251.1, Vibrio fluvialis WP_020430460.1. (b and c) showed the domains of DLD1 and DLD2.

Figure 2. Unrooted phylogenetic tree of DLDs from different Vibrio spp. constructed using MEGA 5.0 software. The scale bar represents a distance of 0.1 substitutions per site. The tree was obtained by bootstrap analysis with a neighbor-joining method, and numbers on branches represent bootstrap values for 1000 replications.
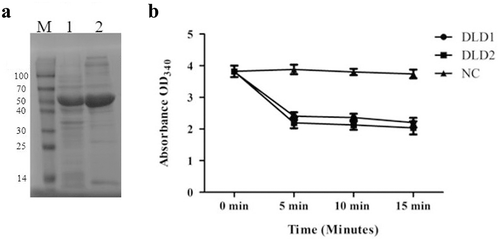
Purification of recombinant DLD and enzymatic activity
The recombinant DLD was expressed in E. coli BL21 cells, and purified using Ni-NTA Sepharose column. The recombinant proteins are shown in ). Before the addition of DLD1 or DLD2, the NADH solution was clear yellow. After the addition of the recombinant protein, the color of the mixture turned from yellow to colorless. The absorbance dropped rapidly within the first 5 min of the reaction and the solution tended to be colorless, with an OD340 decrease from 3.8 to 2.06 or 2.12 in the presence of DLD1 or DLD2, respectively. However, there were no obvious changes in the control group ()).
Figure 3. (a): the SDS-PAGE analysis of the recombinant proteins of DLD1 (lane 1) and DLD2 (lane 2), and M is the Marker (kDa). (b): the absorbance at OD340 after the reaction was catalyzed by DLD1 or DLD2. The reaction without DLD1 or DLD2 was used as a control (NC). Data are the means of three independent experiments, and are presented as means ± SD.
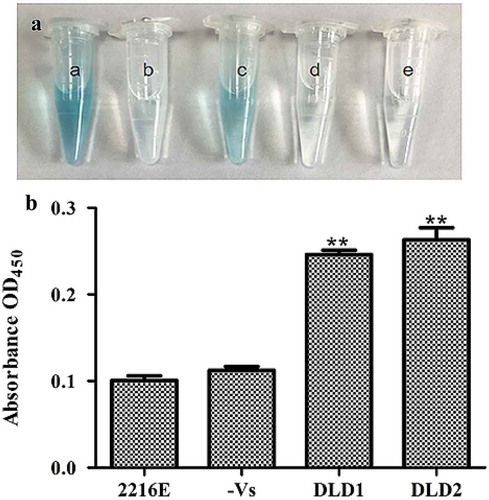
Localization of DLD in V. splendidus
We detected if DLDs were located on the cell surface of V. splendidus using whole cell ELISA. After the whole cell reacted with an antibody against DLD1 or DLD2, the color of the experimental group changed to blue, while there was no significant color change in the control group (no antibody) or negative group (no V. splendidus). The absorbance at OD450 of cells that reacted with DLD1 or DLD2 was 0.25 ()) or 0.27 ()), respectively, which was obviously higher compared with the control group. All the obtained results confirmed the fact that DLD1 and DLD2 are located on the cell surface of V. splendidus.
Figure 4. Determination of DLDs location using whole cell ELISA. The same amount of cells was used as antigen to react with DLD1 or DLD antibody 2. The developed color was measured at 450 nm. (a): a, the cells blocked by DLD1 antibody; b, the negative group; c, the cells blocked by DLD2 antibody; d, the negative group; e, the control group. (b): The absorbance at OD450 of the control group, negative group, V. splendidus reacted with DLD1 antibody and V. splendidus reacted with DLD2 antibody. Data are the means of three independent experiments, and are presented as means ± SD. *P< 0.05, **P< 0.01.
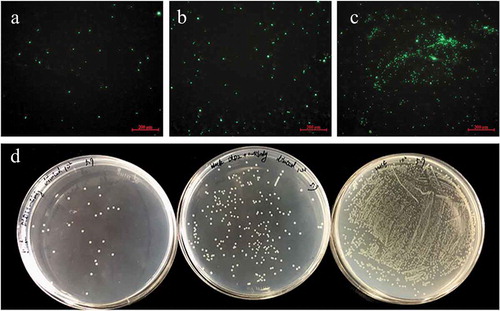
Adhesion of V. splendidus to polystyrene
For the adhesion assay, Vs-DTAF at a concentration of 8.0 × 107 CFU mL−1 was incubated with antibodies of either DLD1 or DLD2. The cells that adhered to the polystyrene were significantly reduced after incubation with the DLD1 or DLD2 antibody. Under the fluorescence microscope, the number of luminous V. splendidus cells preincubated with polyclonal DLD1 or DLD2 antibody was obviously less than that of the control group (–c)). Moreover, the cells blocked by the DLD1 antibody were slightly less adhesive than that of the cells blocked by the DLD2 antibody. Quantitative determination of the amount of the adhered bacteria was performed by the viable cell counting method ()). The adhesive rate to the polystyrene of V. splendidus without antibody blocking was approximately 1%, but after blocking with DLD1 or DLD2 antibody, the adhesive rate decreased to 0.149% or 0.168%, respectively.
Figure 5. The adhesive ability of V. splendidus to polystyrene demonstrated by fluorescence (a-c) and colony counting (d). The green fluorescence was generated by the labeled Vs-DTAF. a, the cells blocked by DLD1 antibody; b, the cells blocked by DLD2 antibody; c, the control group; d, quantity of the cells that adhered to polystyrene. From the left to right: V. splendidus treated with DLD1 antibody, V. splendidus treated with DLD2 antibody, and V. splendidus.
Adhesion of V. splendidus to coelomocytes
We further tested the adhesive ability of V. splendidus to coelomocytes after blocked with DLD1 or DLD2 antibody. Under the laser scanning spectral confocal microscope, the adhesive Vs-DTAF after antibody blocking (,)) was obviously less than that of the control group ()). Based on the number of bacteria that adhered to coelomocytes showed that after blocking with DLD1 or DLD2 antibody, the adhesion quantity was significantly reduced, and the number of cells blocked with the DLD1 antibody was reduced more than that of cells blocked with the DLD2 antibody ()). The adhesive rate of wide-type V. splendidus to the coelomocytes was 25% in the control group, while the adhesive rate of the cells blocked by the DLD1 antibody was 7.5% and the adhesive rate of the cells blocked by the DLD2 antibody was 12.5%. Thus, V. splendidus also showed adhesive ability to coelomocytes of A. japonicus, and DLD1 and DLD2 were also involved in the adhesion of V. splendidus to coelomocytes of A. japonicus.
Figure 6. The adhesive ability of V. splendidus to coelomocytes demonstrated by fluorescence (a-c) and colony counting (b). The green fluorescence was the labeled Vs-DTAF and the red fluorescence was the labeled coelomocyte. (a), the cells blocked by DLD1 antibody; (b), the cells blocked by DLD2 antibody; (c), the control group. (d), Colony counting of the cells that adhered to coelomocytes. After adhesion and washes, the resuspesion cell solution was 1.0 × 103-fold dilution, and 50 μL was spread on 2216E plate. Data were means of three independent experiments, and are presented as means ± SD. *P< 0.05, **P< 0.01.
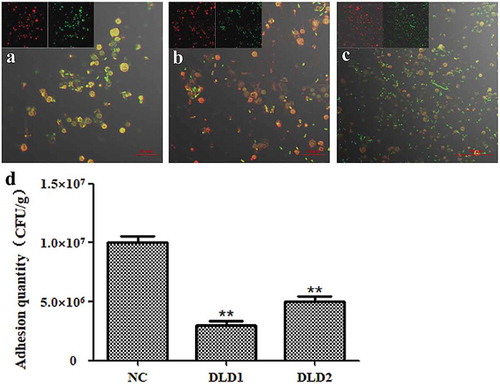
Adhesion of V. splendidus to tissues of A. japonicus
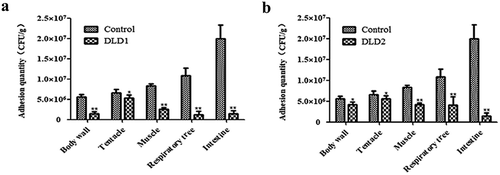
After immersion infection, the individual A. japonicus specimens started to shrink and become soft compared with the individuals in the control group. The color of the respiratory tree and intestine became noticeably darker. The quantitative detection of cell adhesion quantity is shown in . The adhesion quantities of the control group to the tissues of body wall, tentacle, muscle, respiratory tree and intestine were 5.57 × 106 CFU g−1, 6.54 × 106 CFU g−1, 8.39 × 106 CFU g−1, 1.07 × 107 CFU g−1 and 1.92 × 107 CFU g−1, respectively. Expect for the tentacle, the adhesion quantity of V. splendidus treated with antibodies to other tissues was obviously decreased. DLD1 and DLD2 showed different adhesive abilities to the body wall of A. japonicus, in which DLD1 showed a stronger adhesive ability than DLD2.
Figure 7. The adhesive ability of V. splendidus to different tissues of A. japonicus. The tissues of body wall, tentacle, muscle, respiratory tree, and intestine of A. japonicus specimens were collected and homogenized after 24 h immersion infection. The homogenate was 1.0 × 103-fold diluted and 50 μL was spread on 2216E plate. (a): the cells blocked by DLD1 antibody; (b): the cells blocked by DLD2 antibody; Data are means of three independent experiments, and are presented as means ± SD. *P < 0.05, **P< 0.01.
Discussion
In this study, full-length sequences of DLD1 and DLD2 in V. splendidus were amplified. It was found that DLD was widely present in Vibrio spp. with high conservation and the homology similarity of DLD1 and DLD2 to the DLDs from other Vibrio spp. reached 98%-100%. SingalP prediction revealed that both DLD1 and DLD2 had no typical signal peptide. Similar to glyceraldehyde-3-phosphate dehydrogenase in Lactobacillus reuteri [Citation40], DLDs are also located on the cell surface of V. splendidus without a typical signal peptide. Smart analysis showed that both DLD1 and DLD2 contained a Pyr_redox domain and GIDA domain. The GIDA domain is a tRNA modification enzyme that is found in bacteria and mitochondria and its precise molecular function is unknown [Citation41]. The Pyr_redox domain family includes both class I and class II oxidoreductases as well as NADH oxidases and peroxidases. This domain is actually a small NADH-binding domain within a larger FAD-binding domain [Citation42]. After analysis of the domain in DLDs from other bacteria, it was determined that the GIDA domain and Pyr_redox domain were ubiquitously present in DLDs. Thus, we found that the GIDA domain and Pyr_redox domain are probably the main functional domains of DLD. Unlike other bacteria, there were two genes coding DLDs in V. splendidus, and both DLD1 and DLD2 belonged to NADH oxidases.
DLD is a multiple enzyme that is ubiquitously present in microbes, plants, and animals and plays an important role in energy metabolism. Altered energy metabolism, including reductions in the activities of the key mitochondrial enzymes alpha-ketoglutarate dehydrogenase complex (KGDHC) and pyruvate dehydrogenase complex (PDHC), are characteristic of many neurodegenerative disorders including Alzheimer’s disease, Parkinson’s disease and Huntington’s disease. DLD is a critical subunit of KGDHC and PDHC [Citation43]. A previous study concluded that DLD is a multifunctional molecule. DLD plays a role not only in growth but also in biofilm formation. DLD is involved in drug resistance, and the expression of DLD differs among different wild type and drug-resistant strains of V. parahaemolyticus [Citation44]. DLD is also involved in biofilm formation in V. alginolyticus and the DLD-deficient strain was attenuated in swimming and significantly reduced in biofilm formation [Citation45]. Our study was the first to explore the function of DLD in V. splendidus. The two DLDs showed obvious adhesive abilities on various matrices, from polystyrene to host (coelomocytes and tissues). Additionally, the binding of adhesion factors to the host has strong tissue specificity in E. coli [Citation46]. In our present study, it seemed that V. splendidus adherence to the intestine was stronger than that in the other tissues, and both DLD and DLD2 showed the smallest ability to adhere to tentacles. Although DLD1 is similar to DLD2 both in enzymatic activity and adhesive ability, DLD2 showed weaker adhesive ability than DLD1, especially in the body wall.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen J. Present status and prospects of sea cucumber industry in China. In: Lovatelli A, Conand C, Purcell S, et al., editors. Advances in sea cucumber aquaculture and management. Rome: FAO; 2004. p. 25–38.
- Wang Y, Zhang C, Rong X, et al. Diseases of cultured sea cucumber, Apostichopus japonicus, in China. In: Lovatelli A, Conand C, Purcell S, et al., editors. Advances in sea cucumber aquaculture and management. Rome: FAO; 2004. p. 297–310.
- Wang YG, Rong XJ. Main problems and key points of integrated disease control technology in Apostichopus japonicus breeding in China. Shandong Fish. 2004;21(10):29–31. ( In Chinese with English abstract).
- Ma YX, Xu GR, Zhang EP. Bacterial pathogen of acute perioral swelling of young ginseng imitating Apostichopus japonicus. J Fish. 2006;30(2):377–382. ( In Chinese with English abstract).
- Wang YG, Rong XJ, Zhang CY. Main diseases and control techniques of cucumber culture. Mar Sci. 2005;29(3):1–7. ( In Chinese with English abstract).
- Ma Y, Xu G, Cang Y, et al. Bacterial pathogens of skin ulceration on disease in cultured sea cucumber Apostichopus japonicus (Selenka) juveniles. J Dalian Ocean Univ. 2006;21(1):13–18. (In Chinese with English abstract).
- Wang P, Chang Y, Yu J, et al. Acute peristome edema disease in juvenile and adult sea cucumbers Apostichopus japonicus (Selenka) reared in North China. J Invertebr Pathol. 2007;96:11–17.
- Deng H, He C, Zhou Z, et al. Isolation and pathogenicity of pathogens from skin ulceration disease and viscera ejection syndrome of the sea cucumber Apostichopus japonicus. Aquaculture. 2009;287:18–27.
- Liu H, Zheng F, Sun X, et al. Identification of the pathogens associated with skin ulceration and peristome tumescence in cultured sea cucumbers Apostichopus japonicus (Selenka). J Invertebr Pathol. 2010;105:236–242.
- Austin B, Austin DA. Bacterial fish pathogens: disease of farmed and wild fish. 3rd ed. Godalming: Springer-Praxis; 1999. (revised).
- Binesse J, Delsert C, Saulnier D, et al. Metalloprotease vsm is the major determinant of toxicity for extracellular products of Vibrio splendidus. Appl Environ Microbiol. 2008;74:7108–7117.
- Le Roux F, Binesse J, Saulnier D, et al. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl Environ Microbiol. 2007;73:777–784.
- Zhang C, Liang WK, Zhang WW, et al. Characterization of a metalloprotease involved in Vibrio splendidus infection in the sea cucumber, Apostichopus japonicus. Microb Pathog. 2016;101:96–103.
- Liang WK, Zhang C, Liu NN, et al. Cloning and characterization of Vshppd, a gene inducing haemolysis and immune response of Apostichopus japonicus. Aquaculture. 2016;464:246–252.
- Ravulapalli R, Lugo MR, Pfoh R. Characterization of Vis toxin, a novel ADP-Ribosyltransferase from Vibrio splendidus. J Biochem. 2015;54(38):5920–5936.
- Wilson M. Bacterial adhesion to host tissue: mechanisms and consequence. London: Cambridge University Press; 2002.
- Ofek I, Doyle RJ. Principles of bacterial adhesion. In: In bacterial adhesion to cells and tissues. Bacterial adhesion to cells and tissues. New York, NY: Springer; 1994. p. 1–15.
- Luo G, Huang L, Su Y, et al. FlrA, flrB and flrC regulate adhesion by controlling the expression of critical virulence genes in Vibrio alginolyticus. Emerging Microb Infect. 2016;5(8):2–11.
- Liu XQ. Adhesion function of V. mimimetica OmpU protein and determination of its receptor binding domain. Hefei, China: Anhui Agricultural University. 2014. ( In Chinese with English abstract).
- Li H, Wang Y, Li Q, et al. Hydrophobicity and biofilm formation ability of Vibrio brilliant. J Fish China. 2011;18(5):1084–1091. (In Chinese with English abstract).
- Moran JF, Sun Z, Sarath G, et al. Molecular cloning, functional characterization, and subcellular localization of soybean nodule dihydrolipoamide reductase. Plant Physiol. 2002;28(1):300–313.
- Williams CH, Zanetti G, Arscott LD, et al. Lipoamide dehydrogenase, glutathione reductase, thioredoxin reductase, and thioredoxin. J Biol Chem. 1967;242(22):5226–5231.
- Dason MJ, Conroy K, Mcquattie A, et al. Dihydrolipoamide dehydrogenase from Trypanosoma brucie. J Boil. 1987;243(3):661–665.
- Hudson P, Gorton TS, Papazisi L, et al. Identification of a virulence associated determinant, dihydrolipoamide dehydrogenase (lpd), in Mycoplasma gallisepticum through in vivo screening of transposon mutants. Infect Immun. 2006;74(2):931–939.
- Smith AW, Roche H, Trombe MC, et al. Characterization of the dihydrolipoamide dehydrogenase from Streptococcus pneumoniae and its role in pneumococcal infection. Mol Microbiol. 2002;44(2):431–448.
- Zhang WW, Liang WK, Li CH. Inhibition of marine Vibrio sp. by pyoverdine from Pseudomonas aeruginosa PA1. J Hazard Mater. 2016;302:217–224.
- Wang HH, Li CH, Wang ZH, et al. p44/42MAPK and p90RSK modulate thermal stressed physiology response in Apostichopus japonicus. Comp Biochem Physiol. 2016;196–197:57–66.
- Serrano A. Purification, characterization and function of dihydrolipoamide dehydrogenase from the cyanobacterium Anabaena sp. strain P.C.C. 7119. J Biochem. 1992;288:823–830.
- Zhang WW, Yin K, Li B, et al. A glutathione S-transferase from Proteus mirabilis involved in heavy metal resistance and its potential application in removal of Hg2+. J Hazard Mater. 2013;261:646–652.
- Sherr B, Sherr E, Fallon R. Use of monodispersed, fluorescently labeled bacteria to estimate in situ protozoan bacterivory. Appl Environ Microbiol. 1987;53:958–965.
- Cao HC, Wang HR, Wang MZ. Effects of yucca schidigera on determination of bacteria predation rate by human protozoa based on a fluorescence-labeled technique. Chinese J Anim Nutr. 2009;21(3):417–422.
- Rodrigo R, Claudio D, Miranda RO, et al. Characterization and pathogenicity of Vibrio splendidus strains associated with massive mortalities of commercial hatchery-reared larvae of scallop Argopecten purpuratus. J Invertebr Pathol. 2015;124:61–69.
- Silvânia QF, Nélio JA. Bacterial adherence to different inert surfaces evaluated by epifluorescence microscopy and plate count method. Braz Arch Bio Technol. 2004;47:77–83.
- Zhang PJ, Li CH, Zhang P, et al. iTRAQ-based proteomics reveals novel members involved in pathogen challenge in sea cucumber Apostichopus japonicus. PLoS One. 2014;9(6):e100492.
- Charland N, Nizet V, Rubens CE, et al. Streptococcus suisserotype 2 interactions with human brain microvascular endothelialcells. Infect Immun. 2000;68(2):637–643.
- Pathak S, Awuh JA, Leversen NA. Counting mycobacteria in infected human cells and mouse tissue: a comparison between qPCR and CFU. PLoS One. 2014;7(4):1–11.
- Ouwehand AC, Kirjavainen MM, Grok N. Adhesion of probiotic micro-organisms to intestinal mucus. Int Dairy J. 1999;9:623–630.
- Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948.
- Tamura J, Dudley M, Nei M, et al. MEGA4: molecular evolutionary genetics analysis (mega) software version 4.0. Mol Biol Evol. 2007;24:1596–1599.
- Zhang WM, Wang HF, Gao K. Lactobacillus reuteri glyceraldehyde-3-phosphatedehydrogenase functions in adhesion to intestinal epithelial cells. J Can Microbiol. 2015;61:373–380.
- Umeda N, Suzuk T, Yukawa M, et al. Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J Boilchem. 2005;280(2):1613–1624.
- Mande S, Sarfaty S, Allen M, et al. Protein-protein interactions in the pyruvate dehydrogenase multienzyme complex: dihydrolipoamide dehydrogenase complexed with the binding domain of dihydrolipoamide acetyltransferase. Structure. 1996;4(3):277–286.
- Klivenyi P, Starkov AA, Calingasan NY, et al. Mice deficient in dihydrolipoamide dehydrogenase show increased vulnerability to MPTP, malonate and 3-nitropropionic acid neurotoxicity. J Neurochem. 2004;88(6):1352–1360.
- Shen CJ, Kuo TY, Lin CC, et al. Proteomic identification of membrane proteins regulating antimicrobial peptide resistance in Vibrio parahaemolyticus. J Appl Microbiol. 2010;108(4):1398–1407.
- Pang HY, Chen LM, Huang YC. Construction and characterization of a mutant of Vibrio alginolyticus ZJ03 ?dldh Strain. J Biol Bull. 2014;10:161–167. ( In Chinese with English abstract).
- Willemsen P, Graaf F. Age and serotype dependent binding of K88 fimbriae to porcine intestinal receptors. Microb Pathog. 1992;12(5):367–375.
