ABSTRACT
To investigate the virulence of capsular polysaccharide export protein (Wza) in carbapenem-resistant Acinetobacter baumannii and its effect on capsule formation.
wza gene knockout and complementation strains were constructed, and changes in bacterial virulence were observed using in vitro adhesion, antiserum complement killing, anti-oxidation experiments, and infections in Galleria mellonella and mice. The effect of wza knockout on the genes wzb and wzc and wzi were assessed by RT-PCR.
We successfully constructed wza knockout and complementation strains. Compared with wild-type (WT) strains, wza knockout strains displayed lower adhesion to A549 cells (p = 0.044), lower antiserum complement killing ability (p = 0.001), and lower mortality of G. mellonella (p = 0.010) and mice (p = 0.033). Expression levels of wzb, wzc and wzi were decreased in wza knockout strains. The antioxidant capacity of Wza knockout bacteria was only slightly decreased. Complementation of the wza gene returned the adhesion ability, antiserum complement killing ability, and mortality of G. mellonella and mice to WT levels. Expression of wzb, wzc and wzi was also returned to WT levels following wza complementation.
The results clearly demonstrate that Wza is toxic. Wza affects the expression of other proteins of the Wzy capsule polysaccharide synthesis pathway, which affects the assembly, export, and extracellular fixation of capsular polysaccharide, resulting in synergistic effects that decrease bacterial virulence.
Introduction
Acinetobacter baumannii (AB) is a non-fermentative, oxidase-negative, non-flagellate Gram-negative bacterium that is widely distributed in nature and considered a conditional pathogen [Citation1]. It can cause a variety of clinical infections including pneumonia, meningitis, bacteremia, skin and soft tissue infections, peritonitis, and catheter-related and urinary tract infections [Citation2]. Due to increases in the number of invasive surgical operations, and the wide use of antibiotics and hormones, multi-drug-resistant AB (MDRAB) is increasing in prevalence, as is the mortality rate of patients. Worryingly, there are few novel antibacterial drugs for treating these so-called superbugs, hence understanding the mechanisms of virulence of MDRAB could be important for developing effective new antibacterial drugs.
In recent years, reports on highly virulent AB have increased. Jacobs et al. (2014) reported the highly virulent AB5075 strain. The survival rate of Galleria mellonella and mice following AB5075 infection is consistently lower than 25%, and much lower than with other infectious strains [Citation3]. Harris et al. (2013) also reported the highly virulent LAC-4 strain, and infection with this strain resulted in all C57BL/6 and BAMH/C mice dying within 48 h at a dose of 108 colony-forming units (CFU) [Citation4]. Jones et al (2015) also reported some highly virulent strains that were 10 to 100 times more concentrated in the lungs of infected mice than normal strains [Citation5]. In China, there have also been reports of highly virulent strains. Liu et al. (2016) isolated the highly virulent AB strain CCGGD201101 from diseased chickens, and virulence experiments using healthy chicks confirmed that it was more toxic than AB ATCC17978 [Citation6].
Studies on the virulence of AB have identified various virulence factors including outer membrane protein A (OmpA) [Citation7,Citation8], phospholipase D [Citation9], capsular polysaccharide [Citation10], biofilm formation-related protein (Bap) [Citation11], O-glycosylation system protein [Citation12], Acinetobacter trimeric transporter (Ata) [Citation13], the Csu chaperone usher-type pilus [Citation14], the iron acquisition system [Citation15], and secretion of serine proteases [Citation16]. The main virulence mechanisms include iron uptake systems, biofilm formation-related proteins, and damage repair systems [Citation17]. Although these findings have expanded our understanding of the virulence of AB, it remains necessary to identify key virulence factors that promote bacterial homeostasis and cause disease.
The capsular polysaccharide on the surface of AB is considered an important virulence factor because bacteria evade or counteract host immune responses through capsular polysaccharides. Capsular polysaccharides play a key role in biofilm formation and surface colonization [Citation18]. The synthesis of capsular polysaccharide is dependent on the Wzy capsule polysaccharide synthesis pathway [Citation19,Citation20]. Wza is an outer membrane protein encoded by the wza gene. Wza is responsible for transporting capsular polysaccharides from the periplasmic space to the surface of the bacterium, and it participates in the formation of bacterial capsules [Citation21]. Homologues of Wza are found widely in Gram-negative bacteria, and Wza is reported to be a toxic factor in Escherichia coli [Citation22], Klebsiella pneumoniae [Citation23], Riemerella pestis [Citation24], and Vibrio alginolyticus [Citation25]. Other studies have reported the properties of Wza molecules in AB. Russo et al. constructed the wza mutant with Tn mutagenesis and its complementary derivative, and showed that wza mutants were capsule deficient by Western analysis. The wza mutants showed significantly decreased survival in human ascites fluid, human serum and in a rat soft tissue model compared to that of WT strains [Citation18]. Similarly, Skerniškytė J et al constructed a wza mutant and showed that the wza mutant displayed an increase in serum-sensitivity and an impairment in CPS synthesis compared with the parent strain [Citation26]. A study from our group reported finding the wza gene in the highly virulent strain Lac-4 and other highly virulent strains, and that the wza gene is an important toxic factor of AB [Citation27]. To understand further the role of Wza in capsule formation in highly virulent AB strains from clinical isolates, we constructed wza knockout and complementation strains by homologous recombination and gene complementation, respectively, and assessed changes in capsular growth, biofilm formation, and infection in vivo and in vitro. We also measured the expression levels of the RNA wzb, wzc and wzi. The results provide a basis for understanding the role of Wza in the virulence of AB.
Materials and methods
Strains, plasmids, reagents, and culture media
Bacterial strains and plasmids used in this study are listed in . The control strain AB ATCC17978 was purchased from the American Type Culture Collection, USA. The virulent AB strain SKLX024256 was isolated from the First Affiliated Hospital of Zhejiang University School of Medicine, and the multilocus sequence type (Oxford MLST scheme) was 195. Bacterial culturing was performed using Mueller-Hinton Agar (MHA) medium (Cat. No. CM0337B; OXOID) or Mueller-Hinton Broth (MHB) medium (Cat. No. CM0405; OXOID). A bacterial genome extraction kit (Cat. No. 51,306; QIAGEN), a plasmid DNA extraction kit (Cat. No. 12,143; QIAGEN), a gel extraction DNA purification kit (Cat. No. 9762; TaKaRa), a reverse transcription kit (Code No. RR037A; TaKaRa), and an RT-PCR SYBR Green Supermix kit (Cat. No. 170–8894; Bio-Rad) were employed. PCR amplification was achieved using DNA polymerase (TaKaRa, Code. No. R045A). The amplified gene fragment was ligated with the vector using recombinant enzyme Exnase II (ClonExpress II, C112-01/02; Vazyme). Bacterial RNA was extracted using a PureLink RNA Mini Kit (Cat. No. 12183018A; Invitrogen). All strains and plasmids are listed in . All primers were designed and synthesized by Biosune company, and are listed in Supplementary Table S1 and Table S2.
Table 1. List of bacterial strains and plasmids used in this study
Wza gene knockout
Gene knockout was performed as previously described [Citation28,Citation29]. Primers wza-MF1/wza-MR1 (Table S1) were used to amplify the homologous arm A fragment upstream of the AB SKLX024256 wza gene. Primers wza-MF2/wza-MR2 (Table S1) were used to obtain the homologous arm B fragment downstream of the AB SKLX024256 wza gene. The A and B fragments were used as templates to carry out overlapping PCR amplification. The AB fragment was purified and ligated with the suicide vector pLP12 using the recombinant enzyme Exnase II. The resulting recombinant plasmid was transformed into competent E. coli DH5α cells and transformed cells were spread on MH plates (12 μg/mL TC, 0.3% D-Glucose) . Positive recombinant clones containing the AB fragment were screened using primers pLP-UF/pLP-UR. After screening, the positive recombinant clone was cultured, and plasmid pLP12-wza was extracted and purified, and transformed into competent E. coli β 2163 cells. Cells were spread on an MH plate (12 μg/mL TC, 0.3 mM DAP, and 0.3% D-Glucose), and positive clones were picked.
E. coli β 2163 cells carrying pLP12-wza and AB SKLX024256 cells were cultured overnight. Then, 100 μL culture was centrifuged and the supernatant was removed. The cell pellet was resuspended in 10 μL MH medium, spread on an MH plate (0.3 mM DAP + 0.3% D-Glucose), and incubated at 37°C for 6 h; and then the cells were resuspended in 1 mL MH medium. A 100 μL sample was then spread on an MH plate (12 μg/mL TC + 0.3% D-Glucose). Only AB cells containing the insertion in the designated site of the chromosome were able to survive. Clones were picked and streaked on MH plates (12 μg/mL TC + 0.3% D-Glucose) to obtain a pure clone. The pure clone was cultured and the insertion mutation was detected using primers wza-TF/wza-TR.
One insertion mutant was spread on an MH plate (0.3% D-Glucose) and incubated overnight, and then one clone was streaked on an MH plate (0.4% L-arabinose), incubated overnight, and checked using primers wza-TF/wza-TR, and the PCR product was submitted for sequencing.
Wza gene complementation
Gene complementation was performed as previously described [Citation30,Citation31]. The wza gene fragment was amplified using primers wza-RF/wza-RR. Using the pBAD33-TCR plasmid as template, primers pBAD33-TCR-ZF/pBAD33-TCR-ZR were used to amplify the pBAD33-TCR vector fragment, and the product was ligated with the fragment amplified from the pBAD33-TCR vector using ligation recombinase Exnase II . The recombinant plasmid was transformed into competent E. coil DH-5α cells, and transformed cells were spread on an MH plate (12 μg/mL TC) and cultured. After screening of recombinant clones, the pBAD33-TCR-wza vector was successfully obtained. The pBAD33-TCR-wza plasmid was used to transform competent cells of E. coli β 2163 cells, and transformed cells were spread on an MH plate (TC = 12 μg/mL, DAP = 0.3 mM) and incubated overnight. Positive clones were screened and identified as E. coli β 2163-[pBAD33TCR-wza]. AB wza knockout strains and E. coli β 2163 -[pBAD33TCR -wza] were cultured overnight, and 100 μL culture was centrifuged, resuspended in 30 μL MH medium, spread on an MH plate (0.3 mM DAP), and incubated at 37°C for 6 h. Cells were resuspended in 1 mL MH medium, spread on an MH plate (12 μg/mL TC), and incubated overnight; the recombinant clone was screened, and the PCR product was submitted for sequencing.
Growth assay
The growth assay was performed as previously described [Citation6]. AB strains cultured overnight in MH broth were normalized to an OD600 of 0.02 in the same medium and grown at 37°C for 18 h with vigorous aeration (220 rpm). The culture cell density was determined every hour by measuring OD600. The growth rates were estimated using Graphpad software.
Transmission electron microscopy (TEM)
After centrifugation of a 3 mL overnight culture was centrifuged. The supernatants were again discarded, and the pellet was resuspended in PBS (0.1 mol/L, pH 7.4) containing 2.5% glutaraldehyde and incubated at 4°C overnight. After three washes with 0.1 M PBS and fixing with 1% citrate fixative for 1 h, samples were rinsed three times with water and dehydrated using a gradient of alcohol (50%, 70%, 90%, and then 100%). Embedding agent and pure acetone (1:1, 3:1) were added for gradient embedding, and samples were finally mixed with pure embedding agent and incubated at 37°C for 12 h, 45°C for 12 h, and 60°C for 48 h. Samples were sliced using an ultra-thin trimming machine and observed under a Philips TECNAI-10 operated at 80 kV.
Bacterial biofilm test
Bacteria were cultured in LB medium for 14 h and diluted with MH medium to an absorbance at 600 nm (OD600) of 0.5, and 1 mL was placed on a NEST glass plate, incubated for 48 h at 37°C, and rinsed three times with PBS. A 6 μM sample of FITC-conA dye (Invitrogen, L13152) was then added and incubated for 30 min, and samples were rinsed three more times with PBS and observed under a confocal microscope. AB ATCC17978 served as the control strain.
Antioxidant test
An overnight bacterial culture was diluted to OD600 = 0.5 and spread evenly on an MH plate, and a 6 mm circular piece of filter paper was placed on top. A 20% H2O2 solution was then dripped on to the filter paper and plates were incubated at 37°C overnight. The inhibition zone was measured using Vernier calipers. AB ATCC17978 served as the control strain.
Anti-complement killing test
Serum was collected from healthy mice and centrifuged to obtain normal mouse serum. This was placed in a water bath at 56°C for 30 min to inactivate complement, generating inactive serum. An overnight bacterial culture was diluted to a cell density of 2 × 106 CFU/mL, and normal and inactivated sera (180 μL) were separately mixed with 20 μL bacterial suspension and incubated at 37°C for 1 h. Samples were diluted 100-fold, spread onto plates, and incubated overnight, and colonies on plates were counted. The bacterial survival rate was calculated using the following formula:
Bacterial survival rate = (number of colonies with normal serum/number of colonies with inactivated serum) × 100%.
AB ATCC17978 served as the control strain.
Cell adhesion test
An overnight bacterial culture was diluted to OD600 = 0.5, and 100 μL was placed in a 24-well cell culture plate containing A549 lung epithelial cells at 80% density and incubated at 37°C for 5 h. Non-adhered cells were washed with sterile PBS, incubated with 1% Triton X-100 for 5 min, and diluted 10-fold, and the number of viable bacteria were counted. AB ATCC17978 served as the control strain.
G. mellonella infection experiment
An overnight bacterial culture was diluted to a cell density of 1 × 106 CFU/mL, and G. mellonella individuals weighing ~250 mg were randomly divided into groups with 15 individuals in each group. Each individual was injected with 20uL of a 1 × 106 CFU/mL bacterial suspension, incubated at 37°C, and assessed once every 8 h for 7 days. The acupuncture method was used to judge survival status, and death was defined as no response in the test. The Kaplan-Meier estimator method was used to plot a survival curve for G. mellonella. PBS served as the negative control, and AB ATCC17978 was served as the control strain.
Mouse infection experiment
An overnight bacterial culture was diluted to a cell density of 1 × 108 CFU/mL and used to infect 6-week-old BAMH/C immunocompromised mice by supernatant atomization [Citation32]. Mice were then observed for 7 days after infection, and a survival curve was plotted using the Kaplan-Meier estimator method. The control strain was AB ATCC17978.
RT-qPCR measurement of wzb, wzc, and wzi expression
A 3 mL overnight bacterial culture was centrifuged, and bacterial RNA was extracted using a PureLink RNA Mini Kit (Invitrogen) following the manufacturer’s instructions. RNA was reverse-transcribed using a reverse transcription kit (TaKaRa) to obtain cDNA and RT-PCR was performed using an RT-PCR kit (Bio-Rad). Expression levels of wzb,wzc and wzi were determined based on the results of RT-qPCR. 16sRNA was used as an internal reference . The control strain was AB ATCC17978. The primers are listed in Supplementary Table S2.
Statistical analysis
For statistical analysis, values are presented as means±SD (standard deviation). Data are given as mean±standard deviations from three independent experiments. Rank-sum test was performed for pair-wise comparisons of groups. A two-tailed P < 0.05 was considered significant. The Kaplan-Meier method was used to estimate the survival distribution function.
Results
Construction of wza knockout and complementation strains
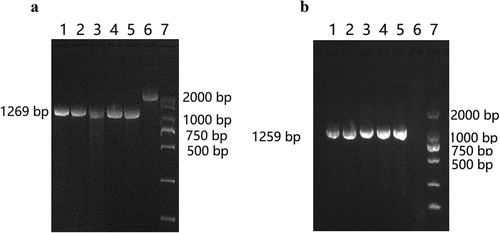
As shown in ) (lane 6), upstream and downstream regions of the wza gene were amplified from the WT strain. The entire 2373 bp fragment comprises 622 bp upstream of the wza gene, the 1104 bp wza gene itself, and 647 bp downstream of the wza gene. The wza knockout strain was successfully engineered, as shown in lanes 1–4; the amplified fragment is 1269 bp, comprising the 622 bp upstream fragment and the 647 bp downstream fragment. As shown in ) (lanes 1–4), the complementation strain was also successfully obtained; the amplified 1259 bp fragment consists of the 1104 bp wza gene and 155 bp from the pBAD33-TCR vector. The DNA sequencing results further confirmed that both wza knockout and complementation strains were successfully constructed.
Figure 1. Detection of Wza knockout and complementation strains
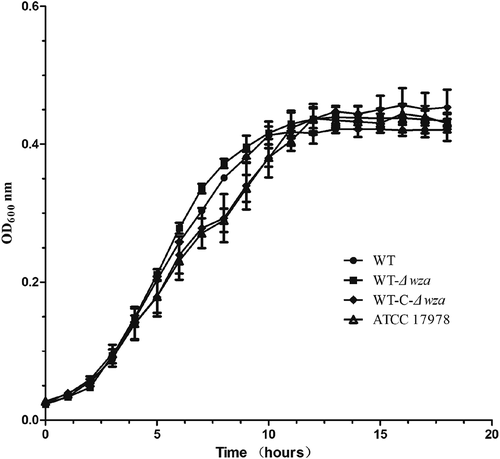
Growth kinetics
To compare the in vitro growth rates in different AB strains, growth kinetics were determined for each strain at 37°C in MH broth (). In general, the growth rates of the wza knockout strain and wza complementation strain were similar to those of the wild type strains.
Bacterial morphology and biofilm formation
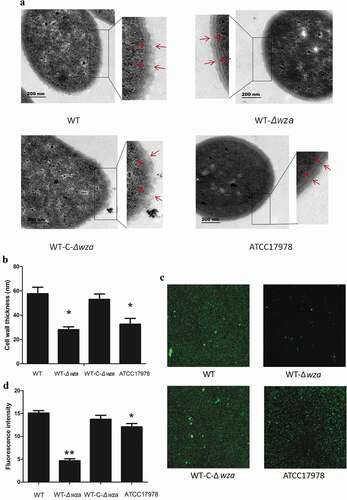
TEM was employed to visualize the cell wall of wza knockout and complementation strains. As shown in ), the cell wall of the wza knockout strain was smoother than that of the WT strain. Additionally, the thickness of the cell wall of the wza knockout strain was 32.26 nm, significantly lower than that of the WT strain (61.48 nm; p = 0.003; ). We found the wza gene knockout to be deficient in bacterial capsule. Furthermore, the thickness of the cell wall of the wza complementation strain was returned to that of the WT strain, and the cell surface recovered its rough appearance. Moreover, the cell wall thickness of the WT strain was thicker than that of the AB ATCC17178 control strain (37 nm; p = 0.011). The confocal fluorescence microscopy results also showed that the fluorescence intensity of the wza knockout strain was weaker than that of the WT strain (p < 0.01; ), while the fluorescence intensity of the wza complementation strain was restored to the level of the WT strain. Biofilm growth was also reduced in the AB ATCC17978 control strain compared with the WT strain. These results demonstrate that the wza gene affected the growth of bacterial biofilms.
Figure 3. Bacterial morphology and biofilm formation
Cell adhesion, anti-complement killing, and anti-oxidation in vitro
The A549 non-small cell lung cancer cell line was used as an in vitro bacterial adhesion model, and the results showed that knockout of the wza gene resulted in significantly lower adhesion than observed with the WT strain (3.69 × 108 vs. 2.47 × 109; p = 0.005). Furthermore, cell adhesion of the wza complementation strain covered to the level of the WT strain. Similarly, the adhesion ability of the AB ATCC17178 strain was significantly lower than that of the WT strain (p < 0.01; ). This indicates that the wza gene is involved in the bacterial adhesion process.
Figure 4. Cell adhesion, anti-complement killing, and anti-oxidation in vitro
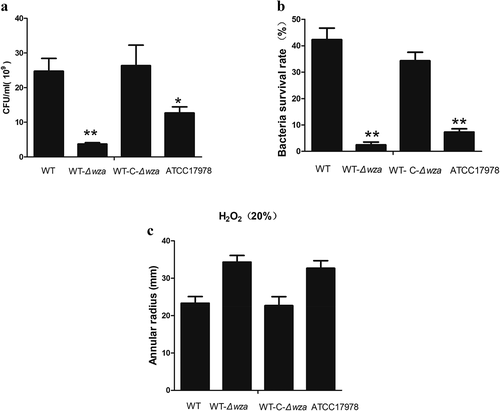
As shown in ), the survival rate of the WT strain was 42.3% at 90% normal serum concentration, while the survival rate of the wza knockout strain was only 2.5% under the same conditions (p = 0.001). These results indicate that the wza gene is important for resistance to complement-related killing. In another experiment, we used 20% H2O2 to test the tolerance of bacteria to oxidants. The result showed that the diameter of the inhibition zone of the WT strain was lower than that of the wza knockout strain (23.3 mm vs. 34.3 mm; p = 0.070), but the difference was not significant. This indicates that tolerance to 20% H2O2 was decreased following loss of the wza gene. The inhibition zone diameter of the complementation strain was 22.7 mm. Thus, the wza gene appears to be involved in the antioxidant process, at least in vitro. Additionally, the WT strain was more tolerant to 20% H2O2 than the AB ATCC17978 control strain (p = 0.108).
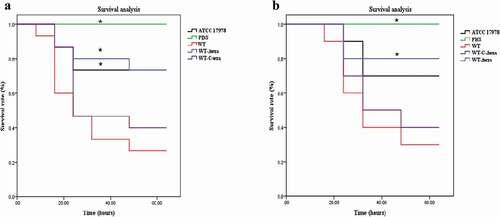
Bacterial infection of G. mellonella and mice
As shown in ), G. mellonella died within 48 h following infection with AB. Mortality reached 73.3% following infection with the WT strain, and reached 53.3% within 24 h. By comparison, mortality following infection with the wza knockout strain was 26.7%, and was only 20% within 24 h. Kaplan-Meier survival curve analysis showed that the mortality of G. mellonella was significantly lower when infected with the wza knockout strain (p = 0.010). Furthermore, infection with the AB ATCC17978 control strain was significantly lower than with the WT strain (p = 0.031). Similar results were obtained for the mouse infection experiment (). The mortality of mice infected with the WT strain reached 70% and the mortality rate was 40% within 24 h. By contrast, the mortality of mice infected with the wza knockout strain was only 20%. Kaplan-Meier survival curve analysis showed that the mortality of mice infected with the wza knockout strain was significantly lower than that of mice infected with the WT strain (p = 0.033).
Figure 5. Bacterial infection of G. mellonella and mice
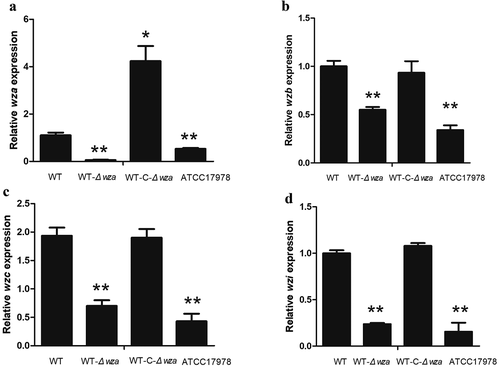
Expression of wzb, wzc, and wzi in wza knockout and complementation strains
Analysis of the expression levels of genes up- and downstream of wza showed that wzb (p = 0.003), wzc (p = 0.002), and wzi (p = 0.002) were significantly decreased in the wza knockout strain, and expression of all three genes was restored to WT levels in the wza complementation strain (). These results indicate that the Wza protein affects the expression of wzb, wzc, and wzi in the Wzy transport pathway.
Figure 6. Expression of wza, wzb, wzc, and wzi in wza gene knockout and complementation strains
Discussion
Acinetobacter baumannii is an important nosocomial pathogen worldwide that causes a variety of serious infections. Due to the extensive use of antibiotics, the detection rate of carbapenem-resistant, multi-drug-resistant, and even totally drug-resistant strains is increasing, and control of infections faces enormous challenges. Unlike other Gram-negative opportunistic pathogens such as E. coli and K. pneumoniae, AB has long been considered an opportunistic pathogen with low virulence, but reports of highly virulent strains are increasing, consistent with an increase in their prevalence. Thus, it is important to study the virulence factors and virulence mechanisms of AB to prevent and control possible epidemics. An in-depth study of the virulence of AB could result in new treatments for multi-drug-resistant AB infections.
In the present study, the MLST of the SKLX024256 strain was ST195, belonging to the CC92 clone complex. At present, ST191, ST195, and ST208 are the most prevalent strains of carbapenem-resistant AB in China [Citation33]. Studies have shown that ST195 clinical isolates are positive for biofilm formation genes (such as those encoding Csu and Bap), and also carry virulence genes such as pga and ompA, which are rare in less virulent isolates. Additionally, biofilm formation, phospholipase C production, hemolytic activity, and toxin production are higher than in non-virulent strains. This indicates that clinically isolated ST195 strains possess high virulence [Citation34].
Highly virulent AB strains have stronger biofilm formation, cell adhesion, and cell invasion abilities than non-virulent strains [Citation35], and these abilities are closely related to the bacterial capsule. Capsular polysaccharide has many important functions in bacteria [Citation1]: it plays an important role in the fight against phagocytosis by phagocytes in the host immune system [Citation36, Citation2] it promotes adhesion between bacteria, and between bacteria and cells, thereby promoting biofilm formation and colonization in different living environments [Citation37]; and [Citation3] it protects bacteria against harmful substances such as host lysozyme and complement factors.
Studies have shown that Wza and OmpA are expressed strongly in a variety of highly toxic AB strains [Citation38]. Both Wza and protein tyrosine kinase (PTK, Wzc) genes are essential for the polymerization and assembly of capsular polysaccharides, and when mutated using transposons, capsule integrity is compromised [Citation18]. In the present study, we constructed a wza knockout strain, and found the strain to be capsule deficient with the thickness of its cell wall reduced by 50% compared with the WT strain, and biofilm formation was reduced. These results suggest that Wza affects the formation of bacterial capsules. Russo et al constructed the wza mutant and found that the wza mutants were capsule deficient [Citation18], Skerniškytė J et al also constructed a wza mutant and found it was a CPS-negative phenotype [Citation26]. These studies were consistent with the results of the cell wall we assessed by TEM. Previous bacterial studies also showed that expression of Wza is positively correlated with the expression of capsular polysaccharides, and deletion of this gene in Lactobacillus johnsonii completely prevents capsular polysaccharide formation [Citation39]. Furthermore, a recombinant plasmid carrying the gene encoding wza completely restored extracellular polysaccharide biosynthesis [Citation39]. Additionally, when the wza gene is mutated in Bacillus amylolytica, biofilm formation is reduced, and biofilm structure is incomplete [Citation40].
In a previous study, normal mice died within 48 h of infection with the highly virulent AB5075 strain at a dose of 1.0 × 105 CFU/mL, but survival was decreased to 33% when infected with a wza mutant strain [Citation3]. In our study,we found the wza knockout strain had significantly lower cell adhesion (3.69 × 108 vs. 2.47 × 109; p = 0.005) and significantly lower survival compare to that of WT (2.5% VS 42.3%, p = 0.001), but the growth rate displayed no difference. These results were consistent with Russo et al 2010 results [Citation18]. These results suggest that wza knockout may disrupt the capsule on the bacterial surface, which may weaken bacterial resistance to harsh external environments and cell adhesion ability, thereby weakening the ability of bacteria to survive and infect the host. Furthermore, our in vivo virulence tests showed that the mortality of G. mellonella and mice infected with the wza knockout strain was significantly lower than that of animals infected with the WT strain.
Wzy capsular polysaccharide synthesis-dependent pathway is crucial for capsular polysaccharide assembly [Citation41]. Ribonucleotide precursors in the cytoplasm assemble four monosaccharide repeating units of the polymer through a series of enzymes including integral membrane proteins and peripheral membrane protein lipids (undecanoic acid diphosphate; und-PP receptor) [Citation42]. The und-PP-linked repeating units are flipped through the inner membrane by the Wzx protein. Under the action of Wzy, polymerization on the periplasmic surface forms long-chain polymers, and high-level Wzy-dependent polymerization requires activation of the tetrameric Wzc protein [Citation43]. The Wza protein undergoes autophosphorylation to facilitate assembly of the capsular polysaccharide [Citation44]. The Wzb phosphatase anchors the catalytic domain on the surface of Wzc, dephosphorylates Wzc, and controls Wzc phosphorylation and dephosphorylation, thereby regulating the degree of polymerization and yield of capsular polysaccharide [Citation45]. Polymers exported to the surface require the outer membrane Wza eight polymer complex, and Wza and Wzc proteins interact to form a complex that spans the periplasm [Citation46–Citation48]. Thus, the activities of Wzc and Wzb play an important role in the synthesis of capsules. Reports indicate that co-expression of these two proteins is essential for capsule assembly in E. coli K30 [Citation49,Citation50].
Additionally, Wzc regulates the export of capsular polysaccharide by controlling the opening and closing of the Wza protein [Citation51,Citation52]. Studies have shown that the Wzi protein is a key factor in the initial extracellular anchoring of capsular polysaccharide [Citation53]. This creates a template for carbohydrate transfer to hydrophobic adventitia to form capsules. Wzi acts as an agglutinin on the cell surface and combines with capsular polysaccharides immediately after transport through Wza. Capsular polysaccharides can be synthesized and transported to extracellular sites, but cannot form capsules with the functional Wzi outer membrane protein [Citation54,Citation55].
Based on the bacterial toxicity test results of our study, Russo et al. 2010 [Citation18] and Skerniskyte et al. 2019 [Citation26], we found that after wza knockout, bacteria couldn’t form capsule layer, which was easily killed by the immune system of the host, and what effect does wza have on the wzy pathway? whether the absence of Wza affects the expression of other molecules involved in the synthesis of Wzy capsular polysaccharides has not been reported. We found wza knockout led to deceases in the expression of wzb, wzc, and wzi, suggesting that knockout of wza gene, the bacterial capsular polysaccharide can not be transported to the outer membrane, in order to avoid the accumulation of excessive capsular polysaccharide in the cells, the expression of the capsular export genes are down-regulated. This indicates that Wza not only controls the output channel of capsular polysaccharides, but also indirectly affects the expression of these three proteins that are crucial for the Wzy capsule formation pathway, thereby affecting the synthesis, transport, and extracellular fixation of capsular polysaccharides. Complementation of the wza gene restored expression of these three genes to WT strain levels
Conclusions
Herein, we constructed wza knockout and complementation strains using clinical isolates of AB SKLX024256. Our results showed that after wza knockout, the capsule is deficient and the thickness of its cell wall is reduced, biofilm formation ability is weakened, adhesion to A549 cells is diminished, and resistance to serum complement killing and oxidants is compromised. The mortality of G. mellonella and mice was also significantly decreased following infection with the wza knockout strain, compared with the WT strain. Therefore, Wza is a conserved capsular polysaccharide transport protein affecting the formation of the bacterial capsules. It is important for the virulence of AB, and provides a potential target for new antibacterial therapies.
Author contributions
YX and TN developed the concept and designed the experiments. TN, LG, QL, and KZ performed experiments. TN and LG performed statistical analysis. YX, WY, YC and CH gave conceptual advice. TN and LG wrote the paper. All authors discussed the results and implications and commented on the manuscript at all stages.
Ethics/Copyright
This study was approved by Ethics Committee of the First Affiliated Hospital,College of Medicine, Zhejiang University, Reference Number:2017-699.
Supplemental Material
Download MS Word (14.8 KB)Acknowledgments
This work was supported by grants from the National Key Research and Development Program of China (No. 2017YFC1200203) and the key research and development program of Zhejiang province (No. 2015C03032) and Medical Science and Technology Project of Zhejiang Province(No.2020KY735). We also thank Beibei Wang in the Center of Cryo-Electron Microscopy (CCEM), Zhejiang University for her technical assistance on Transmission Electron Microscopy.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Data
The supplementary data for this article can be accessed here.
Additional information
Funding
References
- Higgins PG, Dammhayn C, Hackel M, et al. Global spread of carbapenem-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2010;65:233–238.
- Boucher HW, Talbot GH, Bradley JS, et al. Bad bugs, no drugs: no ESKAPE! An update from the infectious diseases Society of America. Clin Infect Dis. 2009;48:1–12.
- Jacobs AC, Thompson MG, Black CC, et al. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. MBio. 2014 May 27;5(3):e01076–14.
- Harris G, Kuo Lee R, Lam CK, et al. A mouse model of Acinetobacter baumannii-associated pneumonia using a clinically isolated hypervirulent strain. Antimicrob Agents Chemother. 2013;57:3601–3613.
- Jones CL, Clancy M, Honnold C, et al. Fatal outbreak of an emerging clone of extensively drug-resistant Acinetobacter baumannii with enhanced virulence. Clin Infect Dis. 2015;61:145–154.
- Liu D, Liu ZS, Hu P, et al. Characterization of surface antigen protein 1 (SurA1) from Acinetobacter baumannii and its role in virulence and fitness. Vet Microbiol. 2016;186:126–138.
- Choi CH, Lee JS, Lee YC, et al. Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. BMC Microbiol. 2008;8:216.
- Lee JS, Choi CH, Kim JW, et al. Acinetobacter baumannii outer membrane protein A induces dendritic cell death through mitochondrial targeting. J Microbiol. 2010 Jun;48(3):387–392.
- Jacobs AC, Hood I, Boyd KL, et al. Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect Immun. 2010 May;78(5):1952–1962.
- Geisinger E, Isberg RR. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog. 2015;11:e1004691.
- Brossard KA, Campagnari AA. The Acinetobacter baumannii biofilm-associated protein plays a role in adherence to human epithelial cells. Infect Immun. 2012;80:228–233.
- Iwashkiw JA, Seper A, Weber BS, et al. Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog. 2012;8:e1002758.
- Bentancor LV, Camacho-Peiro A, Cagla Bozkurt-Guzel GB, et al. Identification of Ata, a multifunctional trimeric autotransporter of Acinetobacter baumannii. J Bacteriol. 2012;194:3950–3960.
- Tomaras AP, Dorsey CW, Edelmann RE, et al. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology. 2003;149:3473–3484.
- Gaddy JA, Arivett BA, McConnell MJ, et al. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect Immun. 2012;80(3):1015–1024.
- King LB, Pangburn MK, McDaniel LS. Serine protease PKF of Acinetobacter baumannii results in serum resistance and suppression of biofilm formation. J Infect Dis. 2013 Apr;207(7):1128–1134.
- Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2018;16:91–102.
- Russo TA, Luke NR, Beanan JM, et al. The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect Immun. 2010;78:3993–4000.
- Kenyon JJ, Hall RM. Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PLoS One. 2013 Apr 16;8(4):e62160.
- Hu D, Liu B, Dijkshoorn L, et al. Diversity in the major polysaccharide antigen of Acinetobacter baumannii assessed by DNA sequencing, and development of a molecular serotyping scheme. PLoS One. 2013;8(7):e70329.
- Dong C, Beis K, Jutta Nesper AL, et al. The structure of Wza, the translocon for group 1 capsular polysaccharides in Escherichia coli, identifies a new class of outer membrane protein. Nature. 2006;444:226–229.
- Drummelsmith J, Whitfield C. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane. Embo J. 2000;19:57–66.
- Lin CL, Chen FH, Huang LY, et al. Effect in virulence of switching conserved homologous capsular polysaccharide genes from Klebsiella pneumoniae serotype K1 into K20. Virulence. 2017;8:487–493.
- Yi H, Yuan B, Liu J, et al. Identification of a wza-like gene involved in capsule biosynthesis, pathogenicity and biofilm formation in Riemerella anatipestifer. Microb Pathog. 2017;107:442–450.
- Hernández-Robles MF, Álvarez-Contreras AK, Juárez-García P, et al. Virulence factors and antimicrobial resistance in environmental strains of Vibrio alginolyticus. Int Microbiol. 2016;19:191–198.
- Skerniškytė J, Krasauskas R, Péchoux C, et al. Surface-related features and virulence among Acinetobacter baumannii clinical isolates belonging to international clones I and II. Front Microbiol. 2019;9:3116.
- Zhou K, Tang X, Wang LX, et al. An emerging clone (ST457) of Acinetobacter baumannii Clonal Complex 92 with enhanced virulence and increasing endemicity in South China. Clinl Infect Dis. 2018;67:S179–S188.
- Liu X, Ji L, Wang X, et al. Role of RpoS in stress resistance, quorum sensing and spoilage potential of Pseudomonas fluorescens. Int J Food Microbiol. 2018 Apr 2;270:31–38.
- Jie MIN, Kaiyu WANG, Tao LIU, et al. Construction and identification of invF gene deleted Yersinia ruckeri and its biological characteristics. J Fish China. 2017;41(12):1858–1866.
- Chen Z, Wang L, Zhang Y, et al. Establishment of a method for gene complementation in Vibrio parahaemolyticus. Nan Fang Yi Ke Da Xue Xue Bao. 2014 Jan;34(1):70–74.
- Hua Y, Sun Q, Wang X, et al. Construction of enterohemorrhagic Escherichia coli O157: h7strains with espF gene deletion and complementation. Nan Fang Yi Ke Da Xue Xue Bao. 2015 Nov;35(11):1546–1551.
- Yu DJ, Yu YS, Fang X, et al. Establishment of mice pneumonla model with imipenem-resistant Acinetobacter baumannii. China J Lab Med. 2010;33:771–775.
- Chang Y, Luan G, Ying X, et al. Characterization of carbapenem-resistant Acinetobacter baumannii isolates in a Chinese teaching hospital. Front Microbiol. 2015;6:910.
- Ali HM, Salem MZM, El-Shikh MS, et al. Investigation of the virulence factors and molecular characterization of the clonal relations of multidrug-resistant acinetobacter baumannii isolates. J AOAC Int. 2017;100:152–158.
- Giannouli M, Antunes LC, Marchetti V, et al. Virulence-related traits of epidemic Acinetobacter baumannii strains belonging to the international clonal lineages I-III and to the emerging genotypes ST25 and ST78. BMC Infect Dis. 2013;13:282.
- Brazeau C, Gottschalk M, Vincelette S, et al. In vitro phagocytosis and survival of Streptococcus suis capsular type 2 inside murine macrophages[J]. Microbiology. 1996;142:1231–1237.
- Costerton JW, Cheng KJ, Geesey GG, et al. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464.
- Tayabali AF, Nguyen KC, Shwed PS, et al. Comparison of the virulence potential of Acinetobacter strains from clinical and environmental sources. PLoS One. 2012;7:e37024.
- Dertli E, Mayer MJ, Colquhoun IJ, et al. EpsA is an essential gene in exopolysaccharide production in Lactobacillus johnsonii FI9785. Microb Biotechnol. 2016;9:496–501.
- Liu J, He D, Li XZ, et al. Gamma-polyglutamic acid (gamma-PGA) produced by Bacillus amyloliquefaciens C06 promoting its colonization on fruit surface. Int J Food Microbiol. 2010;142:190–197.
- Islam ST, Lam JS. Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can J Microbiol. 2014;60(11):697–716.
- Chakraborty AK, Friebolin H, Stirm S. Primary structure of the Escherichia coli serotype K30 capsular polysaccharide. J Bacteriol. 1980;141:971–972.
- Collins RF, Konstantinos Beis BR, Clarke RC, et al. Periplasmic protein-protein contacts in the inner membrane protein Wzc form a tetrameric complex required for the assembly of Escherichia coli group 1 capsules. J Biol Chem. 2006;281:2144–2150.
- Wugeditsch T, Paiment A, Hocking J, et al. Phosphorylation of Wzc, a tyrosine autokinase, is essential for assembly of group 1 capsular polysaccharides in Escherichia coli. J Biol Chem. 2001;276:2361–2371.
- Paiment A, Hocking J, Whitfield C. Impact of phosphorylation of specific residues in the tyrosine autokinase, Wzc, on its activity in assembly of group 1 capsules in Escherichia coli. J Bacteriol. 2002;184:6437–6447.
- Drummelsmith J, Whitfield C. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane. EMBO J. 2000;19:57–66.
- Nesper J, Hill CM, Paiment A, et al. Translocation of group 1 capsular polysaccharide in Escherichia coli serotype K30. Structural and functional analysis of the outer membrane lipoprotein Wza. J Biol Chem. 2003;278:49763–49772.
- Beis K, Collins RF, Ford RC, et al. Three-dimensional structure of Wza, the protein required for translocation of group 1 capsular polysaccharide across the outer membrane of Escherichia coli. J Biol Chem. 2004;279:28227–28232.
- Hagelueken G, Huang H, Mainprize IL, et al. Crystal structures of Wzb of Escherichia coli and CpsB of Streptococcus pneumoniae, representatives of two families of tyrosine phosphatases that regulate capsule assembly. J Mol Biol. 2009;392:678–688.
- Wugeditsch T, Paiment A, Hocking J, et al. Phosphorylation of Wzc, a tyrosine autokinase, is essential for assembly of group 1 capsular polysaccharides in Escherichia coli. J Biol Chem. 2001;276:2361–2371.
- Collins RF, Beis K, Dong C, et al. The 3D structure of a periplasm-spanning platform required for assembly of group 1 capsular polysaccharides in Escherichia coli. Proc Natl Acad Sci U S A. 2007;104:2390–2395.
- Reid AN, Whitfield C. Functional analysis of conserved gene products involved in assembly of Escherichia coli capsules and exopolysaccharides: evidence for molecular recognition between Wza and Wzc for colanic acid biosynthesis. J Bacteriol. 2005;187:5470–5481.
- Bushell SR, Mainprize IL, Wear MA, et al. Wzi is an outer membrane lectin that underpins group 1 capsule assembly in Escherichia coli. Structure. 2013;21:844–853.
- Dou H, Jiang M, Peng H, et al. pH-dependent self-assembly: micellization and micelle-hollow-sphere transition of cellulose-based copolymers. Angew Chem Int Ed Engl. 2003;42:1516–1519.
- Long LX, Yuan XB, Chang J, et al. Self-assembly of polylactic acid and cholesterol-modified dextran into hollow nanocapsules. Carbohydr Polym. 2012;87:2630–2637.
- Demarre G, Guérout AM, Matsumoto-Mashimo C, et al. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPα) conjugative machineries and their cognate Escherichia coli host strains. Res Microbiol. 2005;156:245–255.
- Luo P, He X, Liu Q, et al. Developing universal genetic tools for rapid and efficient deletion mutation in vibrio species based on suicide T-vectors carrying a novel counterselectable marker, vmi480. PLoS One. 2015;10:e0144465.
- Hua Y, Sun Q, Wang X, et al. Construction of enterohemorrhagic Escherichia coli O157: h7strains with espF gene deletion and complementation. Nan Fang Yi Ke Da Xue Xue Bao. 2015;35:1546–1551.