ABSTRACT
A growing number of tet(X)-type tigecycline resistance determinants [tet(X1) to tet(X5)] constitutes an expanding family of tetracycline-inactivating enzymes, posing a potential risk to global public health. Here, we report the development of an efficient multiplex PCR method to detect the family of tet(X) variants. This method is successfully applied in the screen and validation of tet(X) genes in the field and clinic bacterial samples. In addition, we found that the formerly proposed tet(X1) is a premature truncated version by the inappropriate annotation, and fixed this error. Overall, it might be the first genetic tool for the detection of different Tet(X) members.
Introduction
Antimicrobial resistance is one of the biggest challenges to one health covering environmental, animal and human sectors [Citation1]. Tigecycline, the third generation of tetracycline [Citation2], is regarded as a final line of defense (somewhat next to polymyxin) against multidrug-resistant pathogens [Citation3]. Because that tigecycline primarily targets protein synthesis, the sporadic cases of tigecycline resistance are generally associated with ribosome protection as well as active efflux pumps [Citation3,Citation4]. In fact, the discovery of Tet(X)-type enzyme represents a distinct mechanism of tigecycline resistance in that they can destruct/inactivate all the known derivatives of tetracycline [Citation5–Citation7]. Among them, the paradigm version refers to Tet(X) [1167 bp, 388aa] that is an NADP-dependent oxidoreductase carried by a transposon of the obligate anaerobe Bacteroides fragilis [Citation8,Citation9]. Subsequently, Whittle et al. [Citation10] reported two additional tetX variants [designated as tet(X1) and tet(X2)] are neighbored in an operon encoded by the conjugative transposon Tn4351 of Bacteriodes. Of note, the multiple sequence alignment analyses suggested that the formerly annotated tet(X1) [1080 bp] is a truncated version lacking its 60 bp at 5ʹ-end (Fig. S1) [Citation10], therefore we fixed this error here by extending it from 1080 bp to 1140 bp (a polypeptide of 379aa, Fig. S2). Unlike that tet(X1) (1140 bp) has only 68.2% similarity to tet(X) at the level of nucleotide acids, tet(X2) (1167 bp) is almost identical to tet(X) [99.8%, with only two substitutions A280G and G1077C in Fig. S3, and E94K and I359M in Fig. S4]. As for Tet(X3) [378aa], it was proposed by Nakamura et al. to be a polypeptide product of tet(X3) [1137 bp] that is only detected in the clinical isolate of Pseudomonas aeruginosa (Acc. no.: AB097942).
Very recently, two different research groups in China reported the emergence of plasmid-borne tet(X4) [and tet(X5)] conferring the appreciably modest level of phenotypic tigecycline resistance [Citation5,Citation6]. Whole-genome sequencing indicated that i) tet(X4) is carried by two distinct plasmids in Escherichia coli, namely the small IncQ1 plasmid pLHM10-1-p6 [12,783 bp] [Citation6] and the mega IncHI2-type plasmid pEC47 [170,312 bp] [Citation5]; ii) tet(X5) [renamed from its formerly duplicated designation of tet(X3)] is harbored by the un-typable megaplasmid p34AB (277,384 bp) in Acinetobacter baumannii strain 34AB [Citation5]. In relative to tet(X) [1167 bp], tetX4 [1158 bp] and tet(X5) [1167 bp] separately displays 94.3% and 87.2% similarity. Despite the limited distribution with a low percentage of detection ratio right now [Citation11], the transferability of plasmid-mediated Tet(X4) [and Tet(X5)] tigecycline resistance still constitutes a risky challenge to clinical anti-infection therapies. Also, we are concerned with the appearance of new tet(X) variants and its expanded dissemination in the near future. Retrospectively, a similar scenario is seen with the MCR family of polymyxin resistance [Citation12–Citation14]. Not only have the new mcr variants already been extended from mcr-1 [Citation1,Citation14,Citation15] to mcr-9 [Citation16,Citation17], but also some of them, like mcr-1 [Citation1,Citation12] and mcr-3 [Citation1,Citation18], have disseminated globally. Thus, it is in great demand to gain insights into the molecular epidemiology of tet(X)-mediated tigecycline resistance.
Here, we report a set of multiplex PCR approaches designed to detect the ongoing Tet(X) family. This method has been successfully applied in the screen of field/clinic samples, validating its efficacy/efficiency. Overall, our work (to some extent) pioneers the development of genetic toolbox used in detection of the expanding family of tet(X) determinants.
Results and discussion
Design and specificity of tet(X) primers
Except that tet(X1) gives the modest level of similarity (68.2%) to tet(X), all the other four variants [tet(X2) to tet(X5)] exhibit relatively high level of identity (85.1% to 99.8%) when compared with tet(X). Though that we recently reported a pair of unique primers specific for the detection of tet(X4) [and tet(X5)] [Citation11], the high level of similarity amongst different tet(X) variants does raise a challenge to design specific primers for each tet(X) variant. In particular, the fact that tet(X2) is almost identical to that of tet(X) with the only two nucleotides’ substitution (A280G and G1077C, in Fig. S3) might render the routine PCR method ineffective to distinguish them. Following rounds of practical trials, we developed an array of combinations of tet(X) primers ()) and optimized the system of PCR reactions ()). As for the five tet(X) [X1 to X5] target genes, the length of resultant DNA fragments ranges from 204 bp to 685 bp ( and )). The trials of optimization revealed that the annealing temperature of 59°C is suitable for the efficient amplification of all the tet(X) variants in single PCR tests, in which bacterial cultures with the tet(X)-bearing plasmid act as templates ()). After separation with an electrophoresis of 2% agarose gel, the various tet(X)-specific PCR fragments with expected sizes are clearly visualized ()). This verified the specificity of all the tet(X) primers we designed ( and ).
Table 1. Primers used in this study
Figure 1. Design and validation of primers specific for five different variants of tet(X) [X1 to X5] (a). Cartoon scheme for physical locations of five pairs of tet(X) primers. The rectangle represents a given tet(X) gene, and an arrow denotes the primer (b). Use of routine PCR to verify the feasibility of tet(X) primers. The PCR products were separated by an electrophoresis with 2.0% agarose gel. Designations: bp, base pair; M, DNA marker (Trans 2K Plus II); the symbol of “–”, negative control
![Figure 1. Design and validation of primers specific for five different variants of tet(X) [X1 to X5] (a). Cartoon scheme for physical locations of five pairs of tet(X) primers. The rectangle represents a given tet(X) gene, and an arrow denotes the primer (b). Use of routine PCR to verify the feasibility of tet(X) primers. The PCR products were separated by an electrophoresis with 2.0% agarose gel. Designations: bp, base pair; M, DNA marker (Trans 2K Plus II); the symbol of “–”, negative control](/cms/asset/9eb7442c-14af-4e84-b091-23557262ab24/kvir_a_1706913_f0001_oc.jpg)
Development of multiplex PCR
The key issue to develop an efficient multiplex PCR is the eliminated possibility that the mixture of multiple sets of primer pairs cross-react. Prior to multiplex PCR, we utilized single PCR to rule out the potential cross-reaction within the pool of tet(X) primers (). In these PCR assays, all the five different bacterial cultures, one of which only harbors a single tet(X) variant on the engineered plasmid (), are tested for each pair of tet(X) primers (). As predicted, the target fragment (486 bp) of the only tet(X1) gene is amplified in the PCR screen containing the pair of primers tet(X1)-F/tet(X1)-R ( and )). In general agreement with that of tet(X1)-F/tet(X1)-R ()), a unique band is exclusively present in our single PCR detections with tet(X2)-F/tet(X2)-R [343 bp] for tet(X2) in ), and tet(X3)-F/tet(X3)-R [685 bp] for tet(X3) in ). In addition, similar scenarios were also seen in the trials with tet(X4)-F/tet(X4)-R [204 bp] for tet(X4) in ), and tet(X5)-F/tet(X5)-R [265 bp] for tet(X5) in ). Evidently, these data supported that our tet(X) primers are qualified to be utilized in the subsequent multiplex PCR assays, ruling out the possibility of cross-reaction.
Table 2. Field and clinical isolates examined in this study
Figure 2. Evaluation of specificity of the five sets of tet(X) [X1 to X5] primers (a). PCR assay suggests that the pair of primer [tet(X1)-F plus tet(X1)-R] is exclusively specific for tet(X1)-containing sample (b). The specificity of the primer [tet(X2)-F plus tet(X2)-R] is validated with PCR assays supplemented with the template of a single colony carrying different tet(X) gene (c). Among five kinds of different samples, only tet(X3)-bearing one is positive in PCR detection with the specific primers of tet(X3)-F plus tet(X3)-R (d). The tet(X4)-harboring sample is exclusively recognized by a single PCR with a pair of primers [tet(X4)-F plus tet(X4)-R] (e). The specificity of the primers [tet(X5)-F plus tet(X5)-R] is unique to the tet(X5)-positive sample PCR products were separated with electrophoresis of 2% agarose gel. Designations: bp, base pair; M, DNA marker (Trans 2K Plus II); the symbol of “–”, negative control
![Figure 2. Evaluation of specificity of the five sets of tet(X) [X1 to X5] primers (a). PCR assay suggests that the pair of primer [tet(X1)-F plus tet(X1)-R] is exclusively specific for tet(X1)-containing sample (b). The specificity of the primer [tet(X2)-F plus tet(X2)-R] is validated with PCR assays supplemented with the template of a single colony carrying different tet(X) gene (c). Among five kinds of different samples, only tet(X3)-bearing one is positive in PCR detection with the specific primers of tet(X3)-F plus tet(X3)-R (d). The tet(X4)-harboring sample is exclusively recognized by a single PCR with a pair of primers [tet(X4)-F plus tet(X4)-R] (e). The specificity of the primers [tet(X5)-F plus tet(X5)-R] is unique to the tet(X5)-positive sample PCR products were separated with electrophoresis of 2% agarose gel. Designations: bp, base pair; M, DNA marker (Trans 2K Plus II); the symbol of “–”, negative control](/cms/asset/49ada51e-6d4e-43d8-a387-1dbf130835a6/kvir_a_1706913_f0002_oc.jpg)
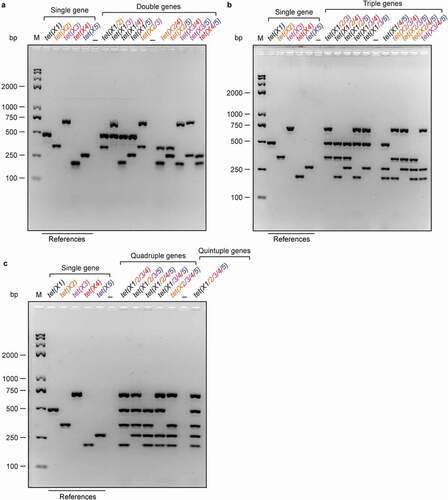
We then attempted to develop a series of multiplex PCR systems. In total, it includes four types of different orientations that separately target double genes, triple genes, quadruple genes, and quintuple genes (). In principle, 10 different sets of primer combinations are set up in the multiplex PCR-based detection of both double genes [tet(X1/2) to tet(X4/5)] in ), and triple genes [tet(X1/2/3) to tet(X3/4/5)] in ). By contrast, five kinds of primer combinations [tet(X1/2/3/4) to tet(X2/3/4/5)] are designed in the multiplex PCR for quadruple genes ()). The only one set of combined primers [tet(X1/2/3/4/5)] is available as for an assay for quintuple genes ()). Of particular being noteworthy, the primers are added in the system of multiplex PCR (50 μl in total) as follows: 10μM for tet(X1)-F/R, 60μM for tet(X2)-F/R, 10μM for tet(X3)-F/R, 60μM for tet(X4)-F/R, and 10μM for tet(X5)-F/R (), respectively. As predicted, the sensitivity of multiplex PCR is determined to be qualified at appreciable level (Figs S5A-B). Consistent with that of single PCR, an annealing temperature (59°C) works well in the aforementioned sets of multiplex PCR experiments with the template of a single colony and/or bacterial cultures (). In general, the mixture of resultant PCR fragments is well separated by the electrophoresis with 2% agarose gel, ranging from 2 bands to 5 bands (–c)). Here, all the PCR products were also demonstrated with direct DNA sequencing. Therefore, we believe that we have success in the development of the multiplex PCR methods suitable for the efficient detection of a family of tet(X) genes.
Figure 3. Establishment of different multiplex PCR assays for the tet(X) family genes (a). Development of multiplex PCR approaches to detect different combinations of double tet(X) genes in a single colony (b). Harnessing multiplex PCR approaches for detection of three tet(X) genes in a single colony (c). Developing efficient multiplex PCR assays to detect quadruple and even quintuple variants of tet(X) from a single colony. The specific PCR products with known sizes in a single PCR act as references. Namely, it denotes 486 bp for tet(X1), 343 bp for tet(X2), 685 bp for tet(X3), 204 bp for tet(X4), and 265 bp for tet(X5) (), respectively. Two percent of agarose gel was applied to separate the mixture of PCR products. Designations: bp, base pair; M, DNA marker (Trans 2K Plus II); the symbol “–”, negative control
Application of multiplex PCR
To further address the feasibility in routine screen, we screened field/clinical samples with this approach of multiplex PCR (). Briefly, bacterial samples examined here consist of 17 field isolates of environmental/animal origins and 8 clinical isolates (). Among the 17 field isolates, the multiplex PCR results suggested that i) seven E. coli isolates are positive for tet(X4); ii) eight Acinetobacter isolates contain tet(X5); and iii) two isolates of Acinetobacter surprisingly coharbor tet(X3) and tet(X5) ( and ,b)). The coexistence of tet(X3) and tet(X5) that initially discovered in multiplex PCR ()) was further consolidated with a single tet(X3) [or tet(X5)]-specific PCR (). By contrast, none of the clinical isolates are positive in the tet(X)-specific multiplex PCR assays ()). It seems likely that tet(X4) is preferentially occupied in E. coli, and tet(X5) is carried in tendency by Acinetobacter (). Indeed, this is generally consistent with recent observations by He et al. [Citation5]. Since we assigned the different genotype of tet(X3) [tet(X4) and tet(X5)] to the field isolates ( and ), we furthered the genotype-to-phenotype relationship in the context of tigecycline resistance. The level of tigecycline resistance in different field isolates varied from 32μg/ml to 64μg/ml (Fig. S6). Together with the fact that expression of tet(X4) only renders the E. coli JM109 resistant to 16μg/ml of tigecycline [Citation11], significantly lower than that of E. coli MG1655 (32 μg/ml, Fig. S6), we favored to believe the possibility that Tet(X)-mediated tigecycline resistance can be strains (and/or species)-dependent, which is somewhat similar to the performance of eptA and arnT in the phenotypic colistin resistance [Citation12]. Not surprisingly, our approach of multiplex PCR was successfully applied in the confirmation of the cooccurrence of tet(X3) and tet(X5) in Acinetobacter. To some extent, it highlights its efficacy in routine surveillance. Despite that we verified the sequence identity of tet(X3) [tet(X5)] in the field isolates, subsequent whole-genome sequencing is needed to finely map the distinct location of tet(X3) [tet(X5)] in plasmids or chromosomes in the near future.
Figure 4. The use of multiplex PCR to screen tet(X) variants in field/clinical isolates. The stains examined here include field isolates and clinical isolates. All the field isolates were provided by the collaborator Dr. Jian Sun from South China Agriculture University, Guangzhou, China (). Among them, 17 are detected to be positive for tet(X) variants, including 8 tet(X5)-positive Acinetobacter isolates (Q477-2, Q478-2, Q479-2, Q480-2, CQW11-1, CQW13-2, CQS29-1 & CQS35-1), and 7 tetX4-bearing E. coli isolates (LHM10-1, G3X16-2, YSP8-1, SS13-1-1, SSS2-2, SSS3-1 & WF90-1), and 2 Acinetobacter isolates coharboring tet(X3) and tet(X5) (). The remaining eight strains (KP-1 to KP-8) refer to clinical isolates of Klebsiella pneumoniae with tigecycline resistance, collected from The Second Affiliated Hospital, Zhejiang University School of Medicine. Electrophoresis of 2% agarose gel was applied to separate the mixture of PCR products. Designations: bp, base pair; M, DNA marker (Trans 2K Plus II); the symbol “–”, negative control; the mixture of tet(X) [X1 to X5] PCR products, positive control
![Figure 4. The use of multiplex PCR to screen tet(X) variants in field/clinical isolates. The stains examined here include field isolates and clinical isolates. All the field isolates were provided by the collaborator Dr. Jian Sun from South China Agriculture University, Guangzhou, China (Table 2). Among them, 17 are detected to be positive for tet(X) variants, including 8 tet(X5)-positive Acinetobacter isolates (Q477-2, Q478-2, Q479-2, Q480-2, CQW11-1, CQW13-2, CQS29-1 & CQS35-1), and 7 tetX4-bearing E. coli isolates (LHM10-1, G3X16-2, YSP8-1, SS13-1-1, SSS2-2, SSS3-1 & WF90-1), and 2 Acinetobacter isolates coharboring tet(X3) and tet(X5) (Table 2). The remaining eight strains (KP-1 to KP-8) refer to clinical isolates of Klebsiella pneumoniae with tigecycline resistance, collected from The Second Affiliated Hospital, Zhejiang University School of Medicine. Electrophoresis of 2% agarose gel was applied to separate the mixture of PCR products. Designations: bp, base pair; M, DNA marker (Trans 2K Plus II); the symbol “–”, negative control; the mixture of tet(X) [X1 to X5] PCR products, positive control](/cms/asset/98506d5b-b194-41c1-b662-71c29a4b9112/kvir_a_1706913_f0004_oc.jpg)
Figure 5. Use of single PCR to re-verify the co-carriage of tet(X3) and tet(X5) in two isolates of CMG11-2 and FS35-1 PCR results of single gene verified that the two strains (CMG11-2 and FS35-1) are consistently positive in tet(X3) [and tet(X5)]-specific molecular detection, whereas negative in tet(X1) [or tet(X2)/tet(X4)]-specific PCR assays. The expected size of PCR products is 685 bp for tet(X3), and 265 bp for tet(X5) (). PCR products were separated with electrophoresis of 2% agarose gel. Designations: bp, base pair; M, DNA marker (Trans 2K Plus II); the symbol “–”, negative control; the mixture of tet(X) [X1 to X5] PCR products, positive control
![Figure 5. Use of single PCR to re-verify the co-carriage of tet(X3) and tet(X5) in two isolates of CMG11-2 and FS35-1 PCR results of single gene verified that the two strains (CMG11-2 and FS35-1) are consistently positive in tet(X3) [and tet(X5)]-specific molecular detection, whereas negative in tet(X1) [or tet(X2)/tet(X4)]-specific PCR assays. The expected size of PCR products is 685 bp for tet(X3), and 265 bp for tet(X5) (Table 1). PCR products were separated with electrophoresis of 2% agarose gel. Designations: bp, base pair; M, DNA marker (Trans 2K Plus II); the symbol “–”, negative control; the mixture of tet(X) [X1 to X5] PCR products, positive control](/cms/asset/40e85da6-17f3-4418-89bd-0efc994fe0ed/kvir_a_1706913_f0005_oc.jpg)
Conclusions
The data shown here represents a timely follow-up study in response to the newly-identification of tet(X4) [tet(X5)] [Citation5,Citation6,Citation19]. It provides an efficient and powerful approach to detect the prevalence of tet(X) family of variants. Similarly, we developed an efficient multiplex PCR method to detect the family of mcr-like genes [Citation20]. As of reporting our method, tet(X4) is only detected in non-human E. coli isolates [Citation5,Citation6,Citation19], whereas tet(X5) is exclusively present in Acinetobacter baumannii [Citation5]. The current situation of tet(X4) [tet(X5)] epidemiology in China is generally consistent with our observation with the multiplex PCR method we developed ( and ). The unusual scenario in our case is the cooccurrence of tet(X3) and tet(X5) in Acinetobacter ( and ), which indicates the complexity and dynamics of tigecycline resistance conferred by Tet(X5) [or Tet(X3)]. Because of the unavailability of tet(X4) [or tet(X5)] distribution beyond China, harnessing this multiplex PCR method to detect tet(X) genes facilitates updating global epidemiology of Tet(X) tigecycline resistance in the context of one health (environmental, animal and clinical sectors).
Both colistin and tigecycline are one of the few clinical options used to combat lethal infections with carbapenem-resistant pathogens [Citation12,Citation21,Citation22]. The resistance to colistin arises primarily from the remodeling of lipid A anchored on the bacterial surface [Citation1], in which EptA/MCR is one of the central players having the activity of lipid A phosphoethanolamine transferase [Citation12]. In contrast, the majority of newly-emerging tigecycline resistance is due to the action of Tet(X)-type tigecycline destructases [Citation5,Citation6,Citation23]. As a member of FAD-dependent, NADP-requiring oxidoreductase [Citation23,Citation24], Tet(X) represents a new resistance mechanism [Citation5,Citation6], threatening the renewed interest of tigecycline [and even the two newly-approved alternatives by FDA (eravacycline [Citation25] and omadacycline [Citation26])] in clinical settings. Moreover, we defined that the FAD-binding cavity and tigecycline-loading channel both are essential for Tet(X4) action and its phenotypic resistance [Citation11]. Because that diversified variants of the MCR family have almost spread to the whole world [Citation27], we are seriously concerned with its potential large-scale dissemination of plasmid-mediated Tet(X) family tigecycline resistance determinants, in spite of its current limited distribution of tet(X4) [Citation11]. Worrisomely, the fact that the members of MCR family have been extended from MCR-1 [Citation15] to MCR-9 [Citation16,Citation17] prompted us to believe that new tet(X) variants [e.g. tet(X6)] other than tet(X4) [or tet(X5)] might be discovered soon. Since that MCR can co-exist with New Delhi Metallo-β-lactamase [NDM-1 [Citation28] or NDM-5 [Citation29,Citation30]] forming a potential superbug insusceptible to both colistin and carbapenem, it is in rational to predict that tet(X) coexists with mcr-like variants and/or blaNDM. However, it requires further experimental demonstration, though that antibiotic exposure can be driving force for the ongoing superbugs with multiple-drug resistance [Citation30].
Materials and methods
Bioinformatic analyses and primer design of tet(X) genes
Five tigecycline resistance genes separately included tet(X1) [Acc. no.: AJ311171], tet(X2) [Acc. no.: AJ311171], tet(X3) [Acc. no.: AB097942], tet(X4) [Acc. no.: MK134376], and tet(X5) [Acc. no.: MK134375]. Multiple alignment of nucleotide sequences was carried out with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/), which provides hints to design primers with primer premier 5.0. The resultant primers () were verified to amplify various target tet(X) fragments in distinct sizes.
Establishment of single and multiplex PCR systems
The mixture of a single PCR reaction (50 μl in total) contained 2× Rapid Taq Master Mix (P222-AA) (Vazyme, China) (25 μl), 10 mM of each primer (1 μl), the template of colony solution (1 μl), and nuclease-free ultra-pure water (22 μl). As we described with mcr-like genes with little change [Citation20], different combinations of various primer pairs [tet(X1)-F/R to tet(X5)-F/R] were utilized in the series of multiplex PCR. Of note, multiple sets of primers were adjusted to different ratio [tet(X1)-F/R, 10μM; tet(X2)-F/R, 60μM; tet(X3)-F/R, 30μM; tet(X4)-F/R 60μM; tet(X5)-F/R,10μM] in all the multiplex PCR trials (50 μl in total). The reaction condition of both single PCR and multiplex PCR comprises i) 3 min of pre-denaturation at 95°C followed by 30 cycles of amplification (15 s of denaturation at 95°C, 15 s of annealing of tet(X) primers with target genes at 59°C, and 20 s of target fragment elongation at 72°C), and a final elongation at 72°C for 5 min. The PCR products were separated by the electrophoresis with 2.0% agarose gel (110 voltages, 35 min). The total time cost in these PCR reactions and electrophoresis is estimated to be around 2 h.
Determination of phenotypic tigecycline resistance
To test the phenotypic tigecycline resistance, all the field isolates that were determined to carry tet(X) genes were evaluated with the method of solid LBA supplemented with varied level of tigecycline (0, 0.5, 1.0, 2.0, 4.0, 8.0, 16.0, 32.0, 64.0, 128.0 and 256.0μg/ml). Log-phase cultures were subjected to series of dilution (10−2, 10−3, 10−4, 10−5, 10−6, and 10−7), and then spotted on the aforementioned LBA plates (5 μl each). Following the overnight incubation at 37°C, bacterial viability was visualized.
Author contributions
YF and CJ designed research; KJ, YX, SS, and YF performed research; YF, CJ, MH, KJ, and YX analyzed data; XJ, and JS contributed reagents and analytic tools; YF, and KJ wrote this draft.
Supplemental Material
Download MS Word (1 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Sun J, Zhang H, Liu YH, et al. Towards understanding MCR-like colistin resistance. Trends Microbiol. 2018;26:794–808.
- Noskin GA. Tigecycline: a new glycylcycline for treatment of serious infections. Clin Infect Dis. 2005;41(Suppl 5):S303–14.
- Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65:232–260.
- Thaker M, Spanogiannopoulos P, Wright GD. The tetracycline resistome. Cell Mol Life Sci. 2010;67:419–431.
- He T, Wang R, Liu D, et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol. 2019;4:1450–1456.
- Sun J, Chen C, Cui CY, et al. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat Microbiol. 2019;4:1457–1464.
- Forsberg KJ, Patel S, Wencewicz TA, et al. The tetracycline destructases: a novel family of tetracycline-inactivating enzymes. Chem Biol. 2015;22:888–897.
- Guiney DG Jr., Hasegawa P, Davis CE. Expression in Escherichia coli of cryptic tetracycline resistance genes from Bacteroides R plasmids. Plasmid. 1984;11:248–252.
- Speer BS, Bedzyk L, Salyers AA. Evidence that a novel tetracycline resistance gene found on two Bacteroides transposons encodes an NADP-requiring oxidoreductase. J Bacteriol. 1991;173:176–183.
- Whittle G, Hund BD, Shoemaker NB, et al. Characterization of the 13-kilobase ermF region of the Bacteroides conjugative transposon CTnDOT. Appl Environ Microbiol. 2001;67:3488–3495.
- Xu Y, Liu L, Sun J, et al. Limited distribution and mechanism of the TetX4 tetracycline resistance enzyme. Sci Bull. 2019;64:1478–1481.
- Zhang H, Srinivas S, Xu Y, et al. Genetic and biochemical mechanisms for bacterial lipid A modifiers associated with polymyxin resistance. Trends Biochem Sci. 2019;44(11):973–988.
- Zhang H, Zong Z, Lei S, et al. A genomic, evolutionary, and mechanistic study of MCR-5 action suggests functional unification across the MCR family of colistin resistance. Adv Sci (Weinh). 2019;6:1900034.
- Ye H, Li Y, Li Z, et al. Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. mBio. 2016;7(2):e00177.
- Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168.
- Kieffer N, Royer G, Decousser JW, et al. mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in Escherichia coli, and its origin. Antimicrob Agents Chemother 2019;63(9):e00965–19. doi:https://doi.org/10.1128/AAC.00965-19.
- Carroll LM, Gaballa A, Guldimann C, et al. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio. 2019;10(3):e00853–19.
- Xu Y, Zhong LL, Srinivas S, et al. Spread of MCR-3 colistin resistance in China: an epidemiological, genomic and mechanistic study. EBioMedicine. 2018;34:139–157.
- Bai L, Du P, Du Y, et al. Detection of plasmid-mediated tigecycline-resistant gene tet(X4) in Escherichia coli from pork, Sichuan and Shandong Provinces, China, February 2019. Euro Surveill. 2019;24:25.
- Lei S, Lv J, Gao S, et al. Developing an efficient multiplex PCR method to detect mcr-like genes. Sci China Life Sci. 2019;62:705–707.
- Moore IF, Hughes DW, Wright GD. Tigecycline is modified by the flavin-dependent monooxygenase TetX. Biochemistry. 2005;44:11829–11835.
- Yahav D, Lador A, Paul M, et al. Efficacy and safety of tigecycline: a systematic review and meta-analysis. J Antimicrob Chemother. 2011;66:1963–1971.
- Yang W, Moore IF, Koteva KP, et al. TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J Biol Chem. 2004;279:52346–52352.
- Volkers G, Palm GJ, Weiss MS, et al. Structural basis for a new tetracycline resistance mechanism relying on the TetX monooxygenase. FEBS Lett. 2011;585:1061–1066.
- Thakare R, Dasgupta A, Chopra S. Eravacycline for the treatment of patients with bacterial infections. Drugs Today. 2018;54:245–254.
- Watkins RR, Deresinski S. Omadacycline: a novel tetracycline derivative with oral and intravenous formulations. Clin Infect Dis. 2019;69(5):890–896.
- Wang Q, Sun J, Li J, et al. Expanding landscapes of the diversified mcr-1-bearing plasmid reservoirs. Microbiome. 2017;5:70.
- Chavda B, Lv J, Hou M, et al. Coidentification of mcr-4.3 and blaNDM-1 in a clinical enterobacter cloacae isolate from China. Antimicrob Agents Chemother. 2018;62(10):e00649–18.
- Sun J, Yang RS, Zhang Q, et al. Co-transfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid in Escherichia coli. Nat Microbiol. 2016;1:16176.
- Mao J, Liu W, Wang W, et al. Antibiotic exposure elicits the emergence of colistin- and carbapenem-resistant Escherichia coli coharboring MCR-1 and NDM-5 in a patient. Virulence. 2018;9:1001–1007.
