ABSTRACT
In the last decades, Candida albicans has served as the leading causal agent of life-threatening invasive infections with mortality rates approaching 40% despite treatment. Candida albicans (C. albicans) exists in three biological phases: yeast, pseudohyphae, and hyphae. Hyphae, which represent an important phase in the disease process, can cause tissue damage by invading mucosal epithelial cells then leading to blood infection. In this review, we summarized recent results from different fields of fungal cell biology that are instrumental in understanding hyphal growth. This includes research on the differences among C. albicans phases; the regulatory mechanism of hyphal growth, extension, and maintaining cutting-edge polarity; cross regulations of hyphal development and the virulence factors that cause serious infection. With a better understanding of the mechanism on mycelium formation, this review provides a theoretical basis for the identification of targets in candidiasis treatment. It also gives some reference to the study of antifungal drugs.
With the increasing number of immunocompromised patients [Citation1], the extensive development of organ transplantation, and the wide use of immunosuppressants and antibiotics in cancer chemotherapy [Citation2], the clinical Candida infection rate increases yearly [Citation3–Citation7]. The detection rate of Candida albicans (C. albicans) was 70%~90% among all candidiasis-causing fungi [Citation8–Citation10]. C. albicans also accounts for a high proportion of candidiasis patients, and the mortality rate of these patients is up to 43.6% due to candidemia [Citation11].
One important characteristic of C. albicans is that it can exist in three phases, budding yeast, pseudohyphae, and hyphae [Citation6]. The plasticity of the mycelial form is a determinant factor of drug resistance and is also an important form during the infection stage [Citation7]. In addition, the transformation of C. albicans from yeast to hypha can help fungi escape the phagocytosis of macrophages, resulting in an increased likelihood of invading host tissues and causing greater damage [Citation12]. Therefore, we summarized recent studies from different fields of fungal cell biology which are instrumental for understanding hyphal growth. This includes research on the differences among C. albicans phases; the regulatory mechanism of hyphal growth, extension, and maintaining cutting-edge polarity and the virulence factors that cause serious infection. With a better understanding of the mechanism on mycelium formation, this review provides a theoretical basis for the identification of targets in candidiasis treatment. It also gives some reference to the study of antifungal drugs.
The differences among yeast, hyphae, and pseudohyphae
C. albicans grows and forms mycelia in changing environments in the host, adapting to a variety of micro-ecological environments. Yeast, hyphae, and pseudohyphae differ in their cell morphology, function, and growth conditions [Citation13–Citation17].
Yeast cells, the default cell morphology under most in vitro conditions, are round or oval, have a unicellular morphology, can be involved in biofilm formation, can be toxic or remain symbiotic in blood, and maintain symbiosis in the oral cavity, skin, and vagina [Citation18–Citation21].
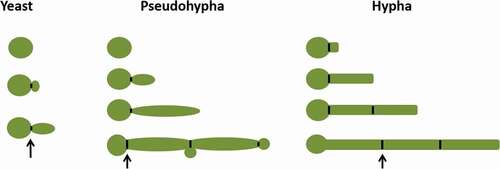
Pseudohyphal cells have long elliptic, multicellular forms, which can be induced at pH 6.0, 35°C, on solid limited nitrogen medium, and via involvement in biofilm formation [Citation22]. In Pseudohyphal cells, there is a constriction at the neck of the bud and mother cell, even at every subsequent septal junction [Citation23]. Pseudohyphae cells can vary widely in width and length so that at one extreme they resemble hyphae, and at the other, they resemble elongated buds of yeast cells. One of the characteristics of pseudohyphae is that the width of each segment that form the mycelia is not constant, being wider at the center than the two ends (). In pseudohyphae, the connection between a mother cell and daughter cell is easily interrupted by mechanical agitation, and this connection is not difficult to interrupt.
Hyphae cells have tubular, multicellular forms, which can be induced by a temperature of 37°C, N-acetyl glucosamine [Citation24–Citation27], embedding matrix [Citation28–Citation30], hypoxia and hypercapnia [Citation31,Citation32], alkaline pH in vitro [Citation33,Citation34], involvement in biofilm formation, and the ability to grow thigmotropically [Citation35]. Hyphae developed from an ungerminated yeast cell, without constriction in the neck of the mother cell and having parallel sides throughout its length. During the hyphal cell cycle, a septin ring will appear in the daughter cell. A simple way to distinguish between hyphae and pseudohyphae is to measure the width of the mycelium, the width of pseudohyphae cells is always larger than the hyphal cells. The width of hyphal cells is about ~2.0 µm on most media. Pseudohyphal cells have a minimum width of 2.8 µm.
Regulation of hyphal morphology
Signaling pathways of hyphal morphogenesis
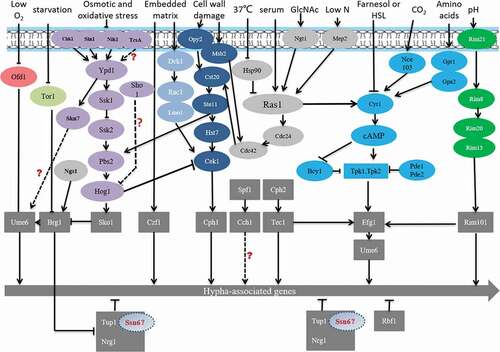
Cek MAPK (mitogen-activated protein kinase) pathway
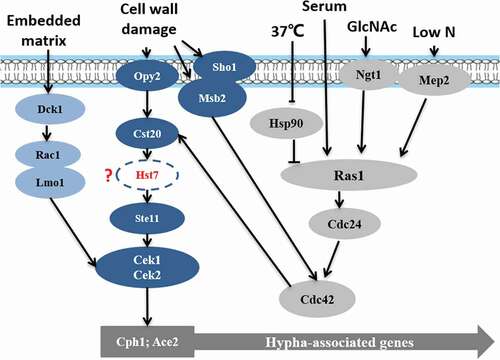
Chen et al. first studied the presence of MAPK in C. albicans [Citation36,Citation37]. The MAPK pathway is induced by factors such as the embedding matrix environment [Citation37], cell wall damage [Citation38], low nitrogen [Citation39]. is a schematic diagram of the Cek MAPK pathway in C. albicans. From the previous study, Opy2, Sho1, Msb2 may form a protein complex that interacts with Cdc42 and Cst20, thus triggering Cek1 phosphorylation [Citation40]. In addition, under osmotic stress, the complex may need to recruit all elements required for polarization/depolarization of cytoskeleton and other related structures such as septin ring [Citation40]. Cells defective in Opy2, Sho1, Msb2 showed lethal phenotypes under osmotic stress, which was presented multinucleated, rounded and abnormally large cells; disorder of actin and myosin cytoskeleton; the septin ring not correctly positioned; and chitin deposition defects. In the previous study, mutant strains formed by deleting CST20, HST7, CEKl, or CPH1 and grown on Spider solid culture medium could not form hyphae [Citation32,Citation41,Citation42]. However, the high expression of CPH1 can mitigate the loss of CEK1, CST20 [Citation43–Citation45]. Recent studies suggested that Hst7 may lie between Cst20 and Cek1/Cek2 [Citation37]. Moreover, CPH1 overexpression can induce the expression of CEK1, CEK2, and genes that are similar to the pheromones of Saccharomyces Cerevisiae, indicating that they may be regulated by the MAPK pathway [Citation46–Citation49]. Ace2, a transcription factor, has multiple functions in cell separation, hyphal morphogenesis, and biofilm formation and works with Efg1, Brg1, and Bcr1 to regulate transcription [Citation50].
Figure 2. Cek MAPK pathway. The Cek1 mitogen-activated protein kinase pathway (MAPK pathway, dark blue) is induced by the embedded matrix environment (light blue), cell wall damage (dark blue), and low nitrogen (gray) and eventually leads to mycelium formation via the phosphorylation of transcription factor Cph1, Ace2
cAMP-PKA (cyclic adenosine monophosphate—protein kinase A) pathway
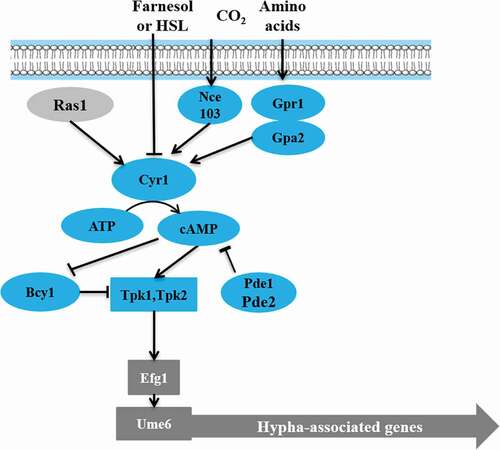
The cAMP-PKA pathway plays a key role in C. albicans growth, morphogenesis, glycogen synthesis, mitochondrial activity, and energy metabolism [Citation51–Citation53]. cAMP is necessary for the activation of PKA pathway in response to various environmental stimuli, including serum, N-Acetylglucosamine (GlcNAc), amino acids, and carbon dioxide. Intracellular cAMP levels are regulated by phosphodiesterase (Pde2) and adenylyl cyclase (Cyr1) [Citation54–Citation56] (). In the mouse model of systemic infection, loss of PDE2 resulted in reduced both hyphal growth and virulence [Citation57].
In C. albicans, Tpk1 and Tpk2 have distinct roles in hyphal development under different filamentous induction conditions [Citation58–Citation61]. On solid medium, Tpk1 is involved in filamentation, while in the liquid medium, Tpk2 is involved in hyphal growth [Citation51,Citation59,Citation61,Citation62]. Interestingly, the C. albicans tpk1/tpk1, tpk2/tpk2 double deletion mutants showed no hyphal growth under various test conditions [Citation63]. However, bcy1Δ/Δ was acquired in a tpk2Δ/Δ background, which reduced PKA activity, with two distinct phenotypes: smooth and filamentous [Citation64,Citation65]. Smooth bcy1Δ/Δ cells showed defects in hyphal growth, while filamentous bcy1Δ/Δ exhibited hyphal growth indistinguishable from the wild type [Citation65]. Furthermore, the cAMP-PKA pathway may be a promising target for antifungal drug development since it is conserved in eukaryotic cells and regulates virulence in C. albicans.
pH-response pathway
Phenotypic plasticity is one of the most prominent characteristics of C. albicans. Different cell forms play different roles in infection and adaptation to different host niches. Acidic pH values inhibit the transformation of yeast into filamentous cells, while neutral and alkaline pH values promote the formation of filamentous cells [Citation66,Citation67]. C. albicans can also actively regulate the environmental pH through the metabolism of effective nutrients.
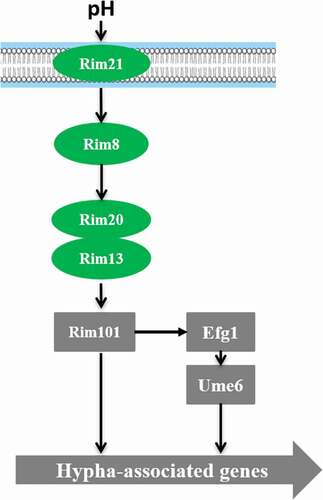
shows a schematic representation of the Rim101‑pH sensing pathway in C. albicans. Rim101 regulates pH-controlled morphologic transition and virulence. Deletion of RIM101 inhibits alkaline pH-induced filamentation [Citation68]. PHR1 and PHR2 are two genes encoding beta-1,3- and beta-1, 6-glucan crosslinking glycosidases in C. albicans, which are regulated differently by Rim101 under different pH conditions [Citation68]. PHR1 is expressed at pH >5.5, while PHR2 is expressed at pH<5.5. The phr1Δ/Δ mutant exhibits growth defects in alkaline pH conditions, while the phr2Δ/Δ mutant grows poorly in acidic pH conditions. Rim101 has been shown to be processed into different versions of the protein under acidic and alkaline conditions [Citation69]. Rim101 is processed to a 74-kD active form at alkaline pH, while to a 65-kD protein with unknown function at acidic condition [Citation69]. The Rim101 activation process also requires three other proteins, Rim8, Rim20, and Rim13 [Citation70,Citation71] (). Studies show that the absence of any one of these four types of proteins can lead to loss of the ability to form hyphae and affect the virulence and adhesion to epithelial cells [Citation71–Citation74].
Hog MAPK pathway
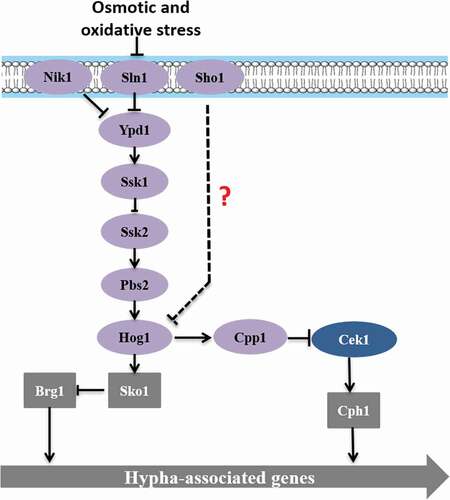
The Hog MAPK pathway (), which is a histidine-kinase pathway that was first found in bacteria, primarily regulates the bacterial resistance and the degradation of aromatic compounds. In C. albicans, the Hog MAPK pathway can regulate mycelium formation, which affects the virulence of the strain, and may even participate in regulating the oxidative stress response [Citation38,Citation75]. C. albicans accumulates glycerol by partially Hog1-dependent pathway under osmotic stress [Citation76]. Hog1 is phosphorylated by hyperosmotic shock and then transferred to the nucleus [Citation77]. Hog1 inhibits the yeast to hyphae transition by downstream elements. This repression is independent of Efg1 and Cph1 since a triple mutant of hog1, efg1 and cph1 shows the hyperfilamentous phenotype characteristic of hog1 single mutant [Citation78]. Deng and Lin found that either HOG1 or a phosphatase gene CPP1 null mutant from both white and opaque cells represented a hyperfilamentous morphology, and HOG1 and CPP1 genes are involved in regulating Cek1 phosphorylation, indicating that the Cpp1 phosphatase may act as a key intermediary between the Hog1 and Cek1 in C. albicans [Citation79]. The Hog pathway is also involved in cell wall biogenesis [Citation78]. In addition, the two-component phosphorelay system Sln1-Ypd1-Ssk1 has also been reported to activate Hog1 under different stress. Sln1, a membrane-bound sensing protein, which is autophosphorylated on a histidine residue. The Sho1 branch is formed by Sho1 transmembrane protein. However, the role of Sln1and Sho1 as upstream components of the Hog pathway is still uncertain, since no clear phenotype has been observed either alone or in combination with other histidine kinase analyses [Citation80]. Studies have shown that the SSK1 deletion significantly increases the risk of Candida cell killing by multicore neutrophils [Citation32,Citation45]. The absence of any protein in the pathway does not affect the cellular response to osmotic pressure, but it can weaken the mycelium formation in Spider culture medium and the toxicity in a mouse system infection model [Citation45,Citation81].
Tup1-mediated negative regulatory pathway
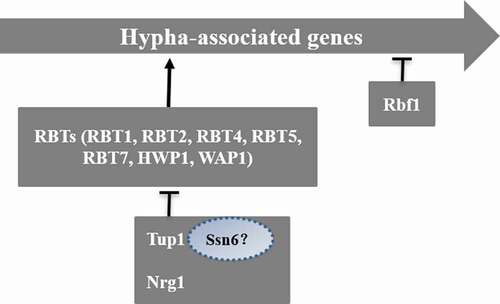
The negative regulation of mycelium formation is primarily mediated by the universal transcription factor Tup1 in synergism with Nrg1 or mycelial growth regulation factor Rfg1 [Citation82] (). Tup1, a sequence-specific DNA-binding protein, can repress transcription, and Tupl-deficient strains can form hyphae effectively without any specific induction conditions [Citation83,Citation84]. RBTs repressed by Tup1 include RBT1, RBT2, RBT4, RBT5, RBT7, HWP1, and WAP1. Rfg1 is a DNA-binding protein that acts downstream of the Tup1 pathway [Citation85]. The absence of Rfg1 can promote the formation of mycelia and the expression of some mycelium-specific RBT genes [Citation83,Citation86]. Nrgl is a zinc finger DNA-binding protein, and its absence can also promote mycelium formation and the expression of some RBTs. Recent studies have shown that the Nrg1 mRNA level in C. albicans cells decreases at 37°C, thus weakening the inhibition of hyphae formation [Citation84]. A DNA microarray analysis found that there are 61 genes whose expression rises significantly during the process of hypha formation when cells are exposed to 37°C and serum factor [Citation86]. Approximately half of these genes are inhibited by Tup1, Nrg1, and Rfg1, indicating that this pathway plays a very important negative regulatory role in mycelium formation [Citation82,Citation84,Citation85]. Cells that lack these repressors will grow to become pseudohyphae, and the expression of specific mycelium genes can be suppressed [Citation83,Citation86]. The prevailing view is that Tup1 and Ssn6 were required for Nrg1-mediated inhibition in C. albicans [Citation87]. However, whether Ssn6 can independently regulate hypha-specific genes under certain conditions remains to be proved.
Regulation of hypha elongation
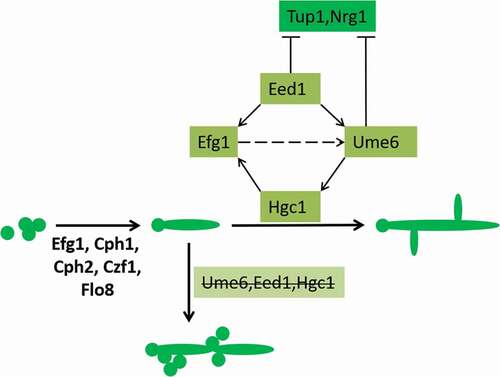
Ume6, Eed1, and Hgc1 play important roles in hypha elongation, as shown in [Citation88–Citation90]. The transcription factors Efg1, Cph1, Cph2, Czf1, and Flo8 are also involved in this elongation process. Eed1 is required for UME6 expression and plays an important role in mycelial maintenance [Citation90]. In an efg1-/- mutant strain, EED1 overexpression will partially compensate for mycelium formation. Ume6 can compensate for the phenotype of an eed1-/- mutant strain. Therefore, Eed1 is located between Efg1 and Ume6 in the pathway [Citation90]. Ume6 and Eed1 can also negatively regulate Tup1 and Nrg1 [Citation91] and indirectly promote the formation of mycelia. Studies have shown that tube formation in eed1-/- and ume6-/- strains in liquid culture medium at the first stage simply remain in the induction stage, and the cells cannot continue to grow [Citation91]. On a solid Spider culture medium, these mutant strains can only form smooth colonies without mycelia [Citation91]. Hgc1 is the periodic protein partner of Cdc28 (also known as Cdk1) in the cell cycle. Hgc1 has multiple roles in cell polarity growth, which inhibits mycelial cell secretion. hgc1-/- mutant strain cells can only form very short bud tubes [Citation92]. In a ume6-/- mutant strain, HGC1 expression can be induced but cannot be maintained. Therefore, the maintenance of HGC1 expression depends on Ume6 [Citation88,Citation93].
Mechanism of mycelial polar growth
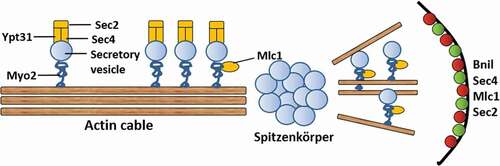
When mycelium elongation, the tip needs to maintain polar growth, which requires the continuous transport of vesicle secretion factors of the membrane envelope to the polarity growth site. These vesicles provide materials to elongate the membrane and enzymes to synthesize new cell walls. The specific process is as follows (): the Golgi body secretes vesicles, Ypt31 on the vesicle surface activates Sec2, and Sec2 activates Sec4 to form a complex. This complex can combine with myosin Myo2 and actin filaments and then be transport to the tip. Myosin Mlc1 can also provide power for transport. Studies have found that blocking the action of actin can cause the hyphal tip to swell because polarity growth is transformed in all directions [Citation94,Citation95]. Deleting the genes for actin Sla1, Sla2, Pan1, Wal1, Vrp1, Myo5 can cause defects in mycelial growth and endocytosis [Citation96–Citation99]. Formin Bni1 and vesicle-related proteins, such as Sec4, Mlc1, and Sec2, aggregate at one point on the tip of the mycelium during three-dimensional rendering. This localization pattern suggests the presence of Spitzenkörper, a structure that was known to drive the tip growth of filaments [Citation100]. The Spitzenkörper is a subterminal region with abundant vesicles that are both exocytic and endocytic in origin. It is thought to be the physical representation of an imaginary vesicle supply center in a model where secretory vesicles radiate from a point source, keeping a certain distance from the end of hyphal. In addition, the mycelial surface has a crescent distribution of exocyst components and polar bodies. The exocyst components help vesicle exit from Spitzenkörper and reach the top surface of the mycelium [Citation100].
Cross regulations of hyphal development
Except for the classic mycelium formation regulatory pathway mentioned above, there are other cross-regulatory pathways to maintain hyphal development (). The previous study found that Spf1, which involved in cell calcium survival pathway, regulates hyphal development and biofilm formation and virulence [Citation101]. Ume6 is a master regulator of filamentation in vitro conditions which null mutants lead to a well-documented filamentation defects. However, Witchley et al. [Citation102] reported ume6 null mutants have the ability to carry out normal morphogenesis in gut by activating the expression of a pro-inflammatory secreted protease, Sap6, and a hyphal cell surface adhesin, Hyr1. The results suggest that Ume6 is not essential for morphogenesis in the gut. At least one fungal filamentous activator must be able to compensate for the loss of Ume6 to regulate gut filamentation. In addition, Su et al. [Citation103,Citation104] found that hyphal induction without inoculation is triggered by Brg1-mediated removal of Nrg1 inhibition. GlcNAc, as well as serum or neutral pH, stimulates the filamentation of log phase cells by down-regulating NRG1 via transcription [Citation105]. Instead of cAMP-PKA pathway, GlcNAc sensor Ngs1 binds to GlcNAc and activates its N-acetyltransferase activity, leading to BRG1 expression. The increased BRG1 level could down-regulate NRG1 transcription, leading to hyphal growth. Skn7 is a conservative fungal heat shock factor-type transcriptional regulator that is part of a two-component signal transduction system. Previous studies have also revealed a close functional interaction between Skn7 and major morphogenetic regulators (including Efg1, Cph1, and Ume6), suggesting that Skn7 is part of the transcriptional pathway that controls filamentous growth of C. albicans [Citation106].
Virulence factors after mycelium formation
The signal transduction pathway described above is a transcription program of a specific mycelial gene. Increases in gene expression during the process of hypha formation can be identified using gene chip technology [Citation107,Citation108]. Highly expressed genes encode virulence factors such as cell wall-dissolving enzymes and adhesion factors such as secreted aspartic proteases (Saps) and agglutinin-like sequence (Als) that allow C. albicans to adhere to, invade, and damage epithelial cells [Citation109]. These factors can enhance the virulence of Candida albicans. Although these proteins are associated with hyphal properties, none is necessary for the formation and maintenance of mycelium.
Invasive enzymes
After mycelium formation, C. albicans destroys the host cell membranes by producing Saps. Saps have an extensive substrate specificity of human proteins, such as albumin, hemoglobin, keratin, collagen, laminin, firestone, mucin, and almost all immunoglobulin, including immunoglobulin A, which is resistant to most of the bacterial proteases. There are 10 genes in SAP family (SAP1–SAP10) which have a vital role in virulence of C. albicans by degrading host tissue proteins as well as adhere to epithelial host tissue. The researchers detected the presence of SAP1 to SAP7 genes in all susceptible or drug-resistant C. albicans isolates. Previous studies have shown that Sap1~3 has a direct effect on tissue damage during superficial infections, while Sap4~6 seems to be more important for penetration into deeper tissues and interactions with the cellular defense [Citation110]. Jessica N. Witchley [Citation102] has reported that Ume6, a main regulator of filamentation, inhibits gut colonization by activating the expression of SAP6 and HYR1, rather than by effects on cell shape. SAP6 deletion strains exhibit increased colonization fitness, whereas SAP6-overexpression strains exhibit attenuated in the gut. Recently Sap8 has been found to be involved in the cleavage of a signaling glycoprotein, which leads to the activation of the Cek1-MAPK pathway [Citation111]. Previous data provided the evidence of SAP7-10 expression in C. albicans strains under human serum influence and hypothesized that SAP7 is essential for C. albicans survival and helps the cells to escape from the bloodstream. Thus, SAP7 may help the fungus to cause systemic infections. The expression of SAP9 in serum was associated with hyphal invasive growth at the site of infection [Citation112]. Previous studies revealed a correlation between prevalence of SAP9 and SAP10 genes and the strong biofilm producers by C. albicans isolates, as 66.7% of these isolates have both Sap9 and Sap10. Sap 9 and Sap10 enzymes maintain cell surface integrity of the Candida cell wall and promote biofilm formation [Citation113]. Recent studies have found that patients infected with fluconazole-resistant C. albicans can enhance the Sap production when treated with this drug [Citation110].
Adhesion
Other major virulence factors expressed after hypha formation are the adhesions, which include agglutinin 1, Als1, mycelium 1, and integrin 1 [Citation114–Citation117]. During the process of infection, they mediated the adhesion of C. albicans to the cells, providing conditions for additional invasion [Citation28,Citation118]. The most common type of adhesion is Als. ALS1 mediates adherence to epithelial cells by encoding a cell surface protein. In C. albicans, disruption of both copies of ALS1 reduces adherence to endothelial cells by 35%, and overexpression of this gene increases adherence by 125%. Recently, some researchers demonstrated that Als3 acts as a type of fungal invasion by binding to the E-, N-calcium adhesive proteins on host cell surface, mediating endocytosis effects, and then invading the host cell [Citation119–Citation121]. Only Als1 and Als5 in Als family have a function similar to that of Als3. C. albicans strains lacking ALS5, ALS6, or ALS7 have a common phenotype of increased adhesion and slightly slower growth. Transcription levels of these genes were consistently lower than other ALS genes. Phenotypic results for ALS5, ALS6, and ALS7 suggested that their proteins have redundant functions or even that they have a role in multiprotein complex [Citation122]. Interestingly, previous studies presented here highlight a number of characteristics of these genes and proteins that remain to be explored in C. albicans [Citation122]. Als9, Als2, and Als4 have not been tested experimentally [Citation109].
Summary
This review described the differences among different phases; the growth, extension and cutting-edge polarity of the mycelial regulatory mechanism, and the hyphal virulence factors. The precise regulatory mechanism of mycelium formation was detailed, and a theoretical basis was provided to identify targets for candidiasis treatment.
Although recent advances in fungal cell biology have begun to illuminate the mechanism of hyphal growth, much remains to be understood. One of the remaining issues is the role of Spitzenkörper in the hyphal tip. How Spitzenkörper exactly focusses on the hyphal tip is unclear. Moreover, the interaction between the carrier and the function of motility proteins is not fully understood. Clearly, motor proteins depend on each other to form functional networks, but how their activity is fine-tuned is unclear. Thus, the secretory mechanism of cytoskeletal polarization, which will be meaningful to elucidate the mechanism of apical protein synthesis in further studies. Although gene chip technology has identified many genes, few genes are involved in the polarity of hyphal growth, which requires additional research.
Disclosure statement
The authors have no conflicts of interest to declare.
Additional information
Funding
References
- McCullough MJ, Clemons KV, Stevens DA. Molecular epidemiology of the global and temporal diversity of Candida albicans. Clin Infect Dis. 1999;29(5):1220–1225.
- Lalla RV, Latortue MC, Hong CH, et al. A systematic review of oral fungal infections in patients receiving cancer therapy. Support Care Cancer. 2010;18(8):985–992.
- Tati S, Davidow P, McCall A, et al. Candida glabrata binding to Candida albicans hyphae enables its development in oropharyngeal candidiasis. PLoS Pathog. 2016;12(3):e1005522.
- Warenda AJ, Konopka JB, Pringle J. Septin function in Candida albicans morphogenesis. Mol Biol Cell. 2002;13(8):2732–2746.
- Lewis MAO, Williams DW. Diagnosis and management of oral candidosis. Br Dent J. 2017;223(9):675–681.
- Saghrouni F, Ben Abdeljelil J, Boukadida J, et al. Molecular methods for strain typing of Candida albicans: a review. J Appl Microbiol. 2013;114(6):1559–1574.
- Thomson DD, Wehmeier S, Byfield FJ, et al. Contact-induced apical asymmetry drives the thigmotropic responses of Candida albicans hyphae. Cell Microbiol. 2015;17(3):342–354.
- Zhou PR, Hua H, Liu XS. Quantity of Candida colonies in saliva: A diagnostic evaluation for oral candidiasis. Chin J Dent Res. 2017;20(1):27–32.
- Kauffman CA. Fungal infections. Proc Am Thorac Soc. 2006;3(1):35–40.
- Sobel JD. Vulvovaginal candidosis. Lancet. 2007;369(9577):1961–1971.
- Moran C, Grussemeyer CA, Spalding JR, et al. Comparison of costs, length of stay, and mortality associated with Candida glabrata and Candida albicans bloodstream infections. Am J Infect Control. 2010;38(1):78–80.
- Uwamahoro N, Verma-Gaur J, Shen -H-H, et al. The pathogen Candida albicans hijacks pyroptosis for escape from macrophages. MBio. 2014;5(2):e00003–14.
- Grinceviciene S, Donders GG. Comment on treatment for recurrent vulvovaginal candidiasis. Am J Obstet Gynecol. 2017;216(4):426–427.
- Green L, Dolen WK. Chronic candidiasis in children. Curr Allergy Asthma Rep. 2017;17(5):31.
- Furuichi Y, Kasai Y, Takeuchi H, et al. Narrow-band imaging can increase the visibility of fibrin caps after bleeding of esophageal varices: a case with extensive esophageal candidiasis. Clin J Gastroenterol. 2017;10(4):331–335.
- Dimopoulos G, Matthaiou DK, Righi E, et al. Elderly versus non-elderly patients with intra-abdominal candidiasis in the ICU. Minerva Anestesiol. 2017;83(11):1126–1136.
- Calderone RA, Fonzi WA. Virulence factors of Candida albicans. Trends Microbiol. 2001;9(7):327–335.
- Andes DR, Safdar N, Baddley JW, et al. Impact of treatment strategy on outcomes in patients with candidemia and other forms of invasive candidiasis: a patient-level quantitative review of randomized trials. Clin Infect Dis. 2012;54(8):1110–1122.
- Antinori S, Milazzo L, Sollima S, et al. Candidemia and invasive candidiasis in adults: A narrative review. Eur J Intern Med. 2016;34:21–28.
- Samaranayake LP, MacFarlane TW. The adhesion of the yeast Candida albicans to epithelial cells of human origin in vitro. Arch Oral Biol. 1981;26(10):815–820.
- Esteves J, Cabrita J, Nobre G, [Yeast-type fungi. Significance of their presence in normal and pathological human biological products]. Cah Med, 1971. 12( 12): p. 1069–1075.
- Soll DR, Stasi M, Bedell G. The regulation of nuclear migration and division during pseudo-mycelium outgrowth in the dimorphic yeast Candida albicans. Exp Cell Res. 1978;116(1):207–215.
- Sudbery PE. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol Microbiol. 2001;41(1):19–31.
- Cassone A, Sullivan PA, Shepherd MG. N-acetyl-D-glucosamine-induced morphogenesis in Candida albicans. Microbiologica. 1985;8(1):85–99.
- Castilla R, Passeron S, Cantore ML. N-acetyl-D-glucosamine induces germination in Candida albicans through a mechanism sensitive to inhibitors of cAMP-dependent protein kinase. Cell Signal. 1998;10(10):713–719.
- Simonetti N, Strippoli V, Cassone A. Yeast-mycelial conversion induced by N-acetyl-D-glucosamine in Candida albicans. Nature. 1974;250(5464):344–346.
- Torosantucci A, Angiolella L, Filesi C, et al. Protein synthesis and amino acid pool during yeast-mycelial transition induced by N-acetyl-D-glucosamine in Candida albicans. J Gen Microbiol. 1984;130(12):3285–3293.
- Kong EF, Tsui C, Kucharíková S, et al. Commensal protection of Staphylococcus aureus against antimicrobials by Candida albicans biofilm matrix. MBio. 2016;7(5). DOI:https://doi.org/10.1128/mBio.01365-16.
- Li F, Majd H, Weir MD, et al. Inhibition of matrix metalloproteinase activity in human dentin via novel antibacterial monomer. Dent Mater. 2015;31(3):284–292.
- Namba N, Yoshida Y, Nagaoka N, et al. Antibacterial effect of bactericide immobilized in resin matrix. Dent Mater. 2009;25(4):424–430.
- Chang Q, Ornatsky OI, Koch CJ, et al. Single-cell measurement of the uptake, intratumoral distribution and cell cycle effects of cisplatin using mass cytometry. Int J Cancer. 2015;136(5):1202–1209.
- de Dios CH, Roman E, Alonso Monge R, et al. The role of MAPK signal transduction pathways in the response to oxidative stress in the fungal pathogen Candida albicans: implications in virulence. Curr Protein Pept Sci. 2010;11(8):693–703.
- Muhlschlegel FA, Fonzi WA. PHR2 of Candida albicans encodes a functional homolog of the pH-regulated gene PHR1 with an inverted pattern of pH-dependent expression. Mol Cell Biol. 1997;17(10):5960–5967.
- Saporito-Irwin SM, Birse CE, Sypherd PS, et al. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol Cell Biol. 1995;15(2):601–613.
- Gow NA, Perera TH, Sherwood-Higham J, et al., Investigation of touch-sensitive responses by hyphae of the human pathogenic fungus Candida albicans. Scanning Microsc, 1994. 8( 3): p. 705–710.
- Iralu V. Formation of aerial hyphae in Candida albicans. Appl Microbiol. 1971;22(3):482–488.
- Chen J, Wang Q, Chen JY, CEK2, a novel MAPK from Candida albicans complement the mating defect of fus3/kss1 mutant. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai), 2000. 32( 3): p. 299–304.
- Cheetham J, Smith DA, da Silva Dantas A, et al. A single MAPKKK regulates the Hog1 MAPK pathway in the pathogenic fungus Candida albicans. Mol Biol Cell. 2007;18(11):4603–4614.
- Roman E, Alonso-Monge R, Gong Q, et al. The Cek1 MAPK is a short-lived protein regulated by quorum sensing in the fungal pathogen Candida albicans. FEMS Yeast Res. 2009;9(6):942–955.
- Herrero-de-Dios C, Alonso-Monge R, Pla J. The lack of upstream elements of the Cek1 and Hog1 mediated pathways leads to a synthetic lethal phenotype upon osmotic stress in Candida albicans. Fungal Genet Biol. 2014;69:31–42.
- Correia I, Alonso-Monge R, Pla J. MAPK cell-cycle regulation in Saccharomyces cerevisiae and Candida albicans. Future Microbiol. 2010;5(7):1125–1141.
- Moyes DL, Runglall M, Murciano C, et al. A biphasic innate immune MAPK response discriminates between the yeast and hyphal forms of Candida albicans in epithelial cells. Cell Host Microbe. 2010;8(3):225–235.
- Moyes DL, Murciano C, Runglall M, et al. Candida albicans yeast and hyphae are discriminated by MAPK signaling in vaginal epithelial cells. PLoS One. 2011;6(11):e26580.
- Moyes DL, Murciano C, Runglall M, et al. Activation of MAPK/c-Fos induced responses in oral epithelial cells is specific to Candida albicans and Candida dubliniensis hyphae. Med Microbiol Immunol. 2012;201(1):93–101.
- Puri S, Kumar R, Chadha S, et al. Secreted aspartic protease cleavage of Candida albicans Msb2 activates Cek1 MAPK signaling affecting biofilm formation and oropharyngeal candidiasis. PLoS One. 2012;7(11):e46020.
- Cullen PJ, Edgerton M. Unmasking fungal pathogens by studying MAPK-dependent cell wall regulation in Candida albicans. Virulence. 2016;7(5):502–505.
- Puri S, Lai WKM, Rizzo JM, et al. Iron-responsive chromatin remodelling and MAPK signalling enhance adhesion in C andida albicans. Mol Microbiol. 2014;93(2):291–305.
- Parrino SM, Si H, Naseem S, et al. cAMP-independent signal pathways stimulate hyphal morphogenesis in Candida albicans. Mol Microbiol. 2017;103(5):764–779.
- Cao C, Wu M, Bing J, et al. Global regulatory roles of the cAMP/PKA pathway revealed by phenotypic, transcriptomic and phosphoproteomic analyses in a null mutant of the PKA catalytic subunit in Candida albicans. Mol Microbiol. 2017;105(1):46–64.
- van Wijlick L, Swidergall M, Brandt P, et al. Candida albicans responds to glycostructure damage by Ace2-mediated feedback regulation of Cek1 signaling. Mol Microbiol. 2016;102(5):827–849.
- Giacometti R, Kronberg F, Biondi RM, et al. Catalytic isoforms Tpk1 and Tpk2 of Candida albicans PKA have non-redundant roles in stress response and glycogen storage. Yeast. 2009;26(5):273–285.
- Sun W, Zhang L, Lu X, et al. The synergistic antifungal effects of sodium phenylbutyrate combined with azoles against Candida albicans via the regulation of the Ras–cAMP–PKA signalling pathway and virulence. Can J Microbiol. 2019;65(2):105–115.
- Lin CJ, Chen YL. Conserved and divergent functions of the cAMP/PKA signaling pathway in Candida albicans and Candida tropicalis. J Fungi (Basel). 2018;4(2):68.
- Grahl N, Demers EG, Lindsay AK, et al. Mitochondrial activity and Cyr1 are key regulators of Ras1 activation of C. albicans virulence pathways. PLoS Pathog. 2015;11(8):e1005133.
- Klengel T, Liang W-J, Chaloupka J, et al. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr Biol. 2005;15(22):2021–2026.
- Xu XL, Lee RTH, Fang H-M, et al. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe. 2008;4(1):28–39.
- Wilson D, Tutulan-Cunita A, Jung W, et al. Deletion of the high-affinity cAMP phosphodiesterase encoded by PDE2 affects stress responses and virulence in Candida albicans. Mol Microbiol. 2007;65(4):841–856.
- Pan X, Heitman J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19(7):4874–4887.
- Sonneborn A, Bockmuhl DP, Gerads M, et al. Protein kinase A encoded by TPK2 regulates dimorphism of Candida albicans. Mol Microbiol. 2000;35(2):386–396.
- Cloutier M, Castilla R, Bolduc N, et al. The two isoforms of the cAMP-dependent protein kinase catalytic subunit are involved in the control of dimorphism in the human fungal pathogen Candida albicans. Fungal Genet Biol. 2003;38(1):133–141.
- Giacometti R, Kronberg F, Biondi RM, et al. Candida albicans Tpk1p and Tpk2p isoforms differentially regulate pseudohyphal development, biofilm structure, cell aggregation and adhesins expression. Yeast. 2011;28(4):293–308.
- Bockmuhl DP, Krishnamurthy S, Gerads M, et al. Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Mol Microbiol. 2001;42(5):1243–1257.
- Almeida L, Oshiro Júnior J, Silva M, et al. Tablet of Ximenia Americana L. developed from mucoadhesive polymers for future use in oral treatment of fungal infections. Polymers (Basel). 2019;11(2):379.
- Cassola A, Parrot M, Silberstein S, et al. Candida albicans lacking the gene encoding the regulatory subunit of protein kinase a displays a defect in hyphal formation and an altered localization of the catalytic subunit. Eukaryot Cell. 2004;3(1):190–199.
- Ding X, Cao C, Zheng Q, et al. The regulatory subunit of Protein Kinase A (Bcy1) in Candida albicans plays critical roles in filamentation and white-opaque switching but is not essential for cell growth. Front Microbiol. 2016;7:2127.
- Biswas S, Van Dijck P, Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev. 2007;71(2):348–376.
- Davis D. Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr Genet. 2003;44(1):1–7.
- Davis D, Wilson RB, Mitchell AP. RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol Cell Biol. 2000;20(3):971–978.
- Li M, Martin SJ, Bruno VM, et al. Candida albicans Rim13p, a protease required for Rim101p processing at acidic and alkaline pHs. Eukaryot Cell. 2004;3(3):741–751.
- Eriksson TJ, Ramadas SN, Dixon SM. Experimental and simulation characterisation of flexural vibration modes in unimorph ultrasound transducers. Ultrasonics. 2016;65:242–248.
- Boikov DA, Locke JB, James KD, et al. In vitro activity of the novel echinocandin CD101 at pH 7 and 4 against Candida spp. isolates from patients with vulvovaginal candidiasis. J Antimicrob Chemother. 2017;72(5):1355–1358.
- Vylkova S, Lorenz MC, Krysan DJ. Modulation of phagosomal pH by Candida albicans promotes hyphal morphogenesis and requires Stp2p, a regulator of amino acid transport. PLoS Pathog. 2014;10(3):e1003995.
- Paranjape V, Datta A. Role of nutritional status of the cell in pH regulated dimorphism of Candida albicans. FEMS Microbiol Lett. 1991;80(2–3):333–336.
- Huang X, Schulte RM, Burne RA, et al. Characterization of the arginolytic microflora provides insights into pH homeostasis in human oral biofilms. Caries Res. 2015;49(2):165–176.
- Yin Z, Stead D, Walker J, et al. A proteomic analysis of the salt, cadmium and peroxide stress responses in Candida albicans and the role of the Hog1 stress-activated MAPK in regulating the stress-induced proteome. Proteomics. 2009;9(20):4686–4703.
- Kayingo G, Wong B. The MAP kinase Hog1p differentially regulates stress-induced production and accumulation of glycerol and D-arabitol in Candida albicans. Microbiology. 2005;151(9):2987–2999.
- Smith DA, Nicholls S, Morgan BA, et al. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol Biol Cell. 2004;15(9):4179–4190.
- Eisman B, Alonso-Monge R, Román E, et al. The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot Cell. 2006;5(2):347–358.
- Deng FS, Lin CH. Cpp1 phosphatase mediated signaling crosstalk between Hog1 and Cek1 mitogen-activated protein kinases is involved in the phenotypic transition in Candida albicans. Med Mycol. 2018;56(2):242–252.
- Roman E, Nombela C, Pla J. The Sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen Candida albicans. Mol Cell Biol. 2005;25(23):10611–10627.
- Bohringer M, Pohlers, S., Schulze, S,et al. Candida albicans infection leads to barrier breakdown and a MAPK/NF-kappaB mediated stress response in the intestinal epithelial cell line C2BBe1. Cell Microbiol. 2016;18(7):889–904.
- Braun BR, Johnson AD. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277(5322):105–109.
- Zeidler U, Lettner T, Lassnig C, et al. UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans. FEMS Yeast Res. 2009;9(1):126–142.
- Sharma M, Manoharlal R, Puri N, et al. Antifungal curcumin induces reactive oxygen species and triggers an early apoptosis but prevents hyphae development by targeting the global repressor TUP1 in Candida albicans. Biosci Rep. 2010;30(6):391–404.
- Kebaara BW, Langford ML, Navarathna DHMLP, et al. Candida albicans Tup1 is involved in farnesol-mediated inhibition of filamentous-growth induction. Eukaryot Cell. 2008;7(6):980–987.
- Banerjee M, Thompson DS, Lazzell A, et al. UME6 a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol Biol Cell. 2008;19(4):1354–1365.
- Garcia-Sanchez S, Mavor AL, Russell CL, et al. Global roles of Ssn6 in Tup1- and Nrg1-dependent Gene regulation in the fungal pathogen, Candida albicans. Mol Biol Cell. 2005;16(6):2913–2925.
- Mendelsohn S, Pinsky M, Weissman Z, et al. Regulation of the Candida albicans hypha-inducing transcription factor Ume6 by the CDK1 cyclins Cln3 and Hgc1. mSphere. 2017;2(2). DOI:https://doi.org/10.1128/mSphere.00248-16.
- Banerjee M, Uppuluri P, Zhao XR, et al. Expression of UME6, a key regulator of Candida albicans hyphal development, enhances biofilm formation via Hgc1- and Sun41-dependent mechanisms. Eukaryot Cell. 2013;12(2):224–232.
- Martin R, Moran GP, Jacobsen ID, et al. The Candida albicans-specific gene EED1 encodes a key regulator of hyphal extension. PLoS One. 2011;6(4):e18394.
- Childers DS, Kadosh D, Sturtevant J. Filament condition-specific response elements control the expression of NRG1 and UME6, key transcriptional regulators of morphology and virulence in Candida albicans. PLoS One. 2015;10(3):e0122775.
- Wang Y. Hgc1-Cdc28-how much does a single protein kinase do in the regulation of hyphal development in Candida albicans? J Microbiol. 2016;54(3):170–177.
- Carlisle PL, Kadosh D. Candida albicans Ume6, a filament-specific transcriptional regulator, directs hyphal growth via a pathway involving Hgc1 cyclin-related protein. Eukaryot Cell. 2010;9(9):1320–1328.
- Crampin H, Finley, K., Gerami-Nejad, M,et al. Candida albicans hyphae have a Spitzenkorper that is distinct from the polarisome found in yeast and pseudohyphae. J Cell Sci. 2005;118(13):2935–2947.
- Hazan I, Liu H. Hyphal tip-associated localization of Cdc42 is F-actin dependent in Candida albicans. Eukaryot Cell. 2002;1(6):856–864.
- Reijnst P, Walther A, Wendland J. Functional analysis of Candida albicans genes encoding SH3-domain-containing proteins. FEMS Yeast Res. 2010;10(4):452–461.
- Asleson CM, Bensen ES, Gale CA, et al. Candida albicans INT1-induced filamentation in Saccharomyces cerevisiae depends on Sla2p. Mol Cell Biol. 2001;21(4):1272–1284.
- Martin R, Hellwig D, Schaub Y, et al. Functional analysis of Candida albicans genes whose Saccharomyces cerevisiae homologues are involved in endocytosis. Yeast. 2007;24(6):511–522.
- Walther A, Wendland J. Polarized hyphal growth in Candida albicans requires the Wiskott-Aldrich Syndrome protein homolog Wal1p. Eukaryot Cell. 2004;3(2):471–482.
- Jones LA, Sudbery PE. Spitzenkörper, exocyst, and polarisome components in Candida albicans hyphae show different patterns of localization and have distinct dynamic properties. Eukaryot Cell. 2010;9(10):1455–1465.
- Yu Q, Wang H, Xu N, et al. Spf1 strongly influences calcium homeostasis, hyphal development, biofilm formation and virulence in Candida albicans. Microbiology. 2012;158(9):2272–2282.
- Witchley JN, Penumetcha P, Abon NV, et al. Candida albicans morphogenesis programs control the balance between gut commensalism and invasive infection. Cell Host Microbe. 2019;25(3):432–443 e6.
- Su C, Lu Y, Liu H. N-acetylglucosamine sensing by a GCN5-related N-acetyltransferase induces transcription via chromatin histone acetylation in fungi. Nat Commun. 2016;7(1):12916.
- Su C, Yu J, Sun Q, et al. Hyphal induction under the condition without inoculation in Candida albicans is triggered by Brg1-mediated removal of NRG1 inhibition. Mol Microbiol. 2018;108(4):410–423.
- Su C, Yu J, Lu Y. Hyphal development in Candida albicans from different cell states. Curr Genet. 2018;64(6):1239–1243.
- Basso V, Znaidi S, Lagage V, et al. The two-component response regulator Skn7 belongs to a network of transcription factors regulating morphogenesis in Candida albicans and independently limits morphogenesis-induced ROS accumulation. Mol Microbiol. 2017;106(1):157–182.
- Ramage G, Coco B, Sherry L, et al. In vitro Candida albicans biofilm induced proteinase activity and SAP8 expression correlates with in vivo denture stomatitis severity. Mycopathologia. 2012;174(1):11–19.
- Chen YC, Wu -C-C, Chung W-L, et al. Differential secretion of Sap4-6 proteins in Candida albicans during hyphae formation. Microbiology. 2002;148(11):3743–3754.
- Fan Y, He H, Dong Y, et al. Hyphae-specific genes HGC1, ALS3, HWP1, and ECE1 and relevant signaling pathways in Candida albicans. Mycopathologia. 2013;176(5–6):329–335.
- Costa CR, Jesuíno RSA, de Aquino Lemos J, et al. Effects of antifungal agents in sap activity of Candida albicans isolates. Mycopathologia. 2010;169(2):91–98.
- Carvalho-Pereira J, Vaz C, Carneiro C, et al. Genetic variability of Candida albicans Sap8 propeptide in isolates from different types of infection. Biomed Res Int. 2015;2015:148343.
- Staniszewska M, BONDARYK M, MALEWSKI T, et al. Quantitative expression of Candida albicans aspartyl proteinase genes SAP7, SAP8, SAP9, SAP10 in human serum in vitro. Pol J Microbiol. 2014;63(1):15–20.
- Kadry AA, El-Ganiny AM, El-Baz AM. Relationship between Sap prevalence and biofilm formation among resistant clinical isolates of Candida albicans. Afr Health Sci. 2018;18(4):1166–1174.
- Arai T, UEDA T, SUGIYAMA T, et al. Inhibiting microbial adhesion to denture base acrylic resin by titanium dioxide coating. J Oral Rehabil. 2009;36(12):902–908.
- Tsutsumi C, Takakuda K, Wakabayashi N. Reduction of Candida biofilm adhesion by incorporation of prereacted glass ionomer filler in denture base resin. J Dent. 2016;44:37–43.
- Vasilas A, Molina L, Hoffman M, et al. The influence of morphological variation on Candida albicans adhesion to denture acrylic in vitro. Arch Oral Biol. 1992;37(8):613–622.
- Varghese N, Yang S, Sejwal P, et al. Surface control of blastospore attachment and ligand-mediated hyphae adhesion of Candida albicans. Chem Commun (Camb). 2013;49(88):10418–10420.
- Ai R, Wei J, Ma D, et al. A meta-analysis of randomized trials assessing the effects of probiotic preparations on oral candidiasis in the elderly. Arch Oral Biol. 2017;83:187–192.
- Mushi MF, Bader O, Taverne-Ghadwal L, et al. Oral candidiasis among African human immunodeficiency virus-infected individuals: 10 years of systematic review and meta-analysis from sub-Saharan Africa. J Oral Microbiol. 2017;9(1):1317579.
- Mackenzie A, Marshall NW, Hadjipanteli A, et al. Characterisation of noise and sharpness of images from four digital breast tomosynthesis systems for simulation of images for virtual clinical trials. Phys Med Biol. 2017;62(6):2376–2397.
- Kullberg BJ, Vasquez J, Mootsikapun P, et al. Efficacy of anidulafungin in 539 patients with invasive candidiasis: a patient-level pooled analysis of six clinical trials. J Antimicrob Chemother. 2017;72(8):2368–2377.
- Zhao X, Oh SH, Hoyer LL. Deletion of ALS5, ALS6 or ALS7 increases adhesion of Candida albicans to human vascular endothelial and buccal epithelial cells. Med Mycol. 2007;45(5):429–434.