ABSTRACT
Streptococcus suis
serotype 2 (SS2) is a serious zoonotic pathogen which causes symptoms of streptococcal toxic shock syndrome (STSS) and septicemia; these symptoms suggest that SS2 may have evade innate immunity. Phagocytosis is an important innate immunity process where phagocytosed pathogens are killed by lysosome enzymes, reactive oxygen, and nitrogen species, and acidic environments in macrophages following engulfment. A previously constructed mutant SS2 library was screened, revealing 13 mutant strains with decreased phagocytic resistance. Through inverse PCR, the transposon insertion sites were determined. Through bioinformatic analysis, the 13 disrupted genes were identified as Cps2F, 3 genes belonging to ABC transporters, WalR, TehB, rpiA, S-transferase encoding gene, prs, HsdM, GNAT family N-acetyltransferase encoding gene, proB, and upstream region of DnaK. Except for the capsular polysaccharide biosynthesis associated Cps2F, the other genes had not been linked to a role in anti-phagocytosis. The survival ability in macrophages and whole blood of randomly picked mutant strains were significantly impaired compared with wild-type ZY05719. The virulence of the mutant strains was also attenuated in a mouse infection model. In the WalR mutant, the transcription of HP1065 decreased significantly compared with wild-type strain, indicating WalR might regulated HP1065 expression and contribute to the anti-phagocytosis of SS2. In conclusion, we identified 13 genes that influenced the phagocytosis resistant ability of SS2, and many of these genes have not been reported to be associated with resistance to phagocytosis. Our work provides novel insight into resistance to phagocytosis, and furthers our understanding of the pathogenesis mechanism of SS2.
Introduction
Streptococcus suis serotype 2 (SS2) is a zoonotic pathogen that has caused severe diseases in humans and pigs, including streptococcal toxic shock syndrome (STSS), sudden death, septicemia, and meningitis; SS2 has resulted in great economic losses in the porcine industry [Citation1,Citation2]. Since SS2 infection can cause STSS, it is plausible that SS2 could successfully evade innate immunity. Phagocytosis is a crucial step of host immunity, and hence, the phagocytosis resistance ability of SS2 has attracted our attention. A transposon mutant library of ZY05719 was previously constructed in our laboratory [Citation3]. This mutant library contained 2400 strains of SS2 mutants in total. The mariner Himar1 transposon was randomly inserted into the genome, and the transposon distribution within the SS2 genome was shown to be random without any hot spots [Citation3]. Transposon mutants have been used to screen for virulence-associated genes by generating altered phenotypes compared to wild-type strains [Citation4–Citation7].
Macrophages have evolved to defend against pathogens by phagocytosis and eliminate intracellular pathogens by lysosomal proteases, phagosome acidification, reactive oxygen and nitrogen species (ROS and RNS), and antimicrobial peptides. However, pathogens have also developed ways to counteract elimination by host cells. For example, pathogens can defend against opsonophagocytosis and other forms of phagocytosis of macrophages. Additionally, they are able to resist lysosome enzymes, reactive oxygen, and nitrogen species (ROS and RNS), and overcome host nutritional limitations by the host [Citation8]. In SS2, capsular polysaccharide plays an important role in phagocytosis resistance [Citation9]. Suilysin is another well-known phagocytic resistance factor that contributes to opsonophagocytosis resistance, phagocytosis resistance, and intracellular survival within phagocytes of SS2 [Citation10]. Sialic acid is a component of the SS2 capsule. Biosynthesis of sialic acid is encoded by neuC gene. Deletion of neuC gene results in decreased phagocytosis resistance of SS2 [Citation11]. A novel transglutaminase of SS2 was shown to be associated with phagocytic resistance [Citation12]. The factor H binding protein (Fhb) of SS2 could bind to human factor H, thus blocking the deposition of complement on the bacteria, which further contributes to phagocytosis resistance ability of SS2 [Citation13].
In this study, we screened 1100 mutant strains from a previously generated mutant library for mutants that led to an increased rate of phagocytosis compared to the wild-type strain ZY05719. We identified 13 phagocytosis susceptibility mutants which could possibly contribute to attenuated virulence. Only one of the 13 genes (capsule biosynthesis gene) has previously been reported for involvement in phagocytosis resistance [Citation14], and the rest remains to be elucidated through further study. WalR was identified in our screen and was reported to be involved in cell wall biosynthesis [Citation15,Citation16]. Cell wall synthesis-related genes were examined, and LysM transcription was shown to increase, along with a corresponding decrease in HP1065 transcription relative to wild-type ZY05719; this might be implicated in the impaired phagocytosis resistance ability of the WalR mutant strain.
Materials and methods
Bacterial strains, cell lines and culture conditions
All the strains and plasmids used in this study are listed in . The wild-type SS2 strain ZY05719 was isolated from a diseased pig from an SS2 outbreak in Ziyang, China. A mutant strain library based on strain ZY05719 was previously constructed in this lab. Briefly, the pMar4 s plasmid containing the TnYLB-1 transposon was transformed into ZY05719 by electroporation. The mutant pool was stored in 50% glycerol at −80°C. Both wild-type SS2 strains and mutant strains were cultured in Todd-Hewitt Broth (THB; Becton Dicksinson) liquid or agar media at 37°C. For growth assay, THB liquid media supplemented with 2% Yeast Extract (THY) was used. Escherichia coli DH5α was maintained in Luria-Bertani (LB) medium. When the pSET2 or pSET4s plasmids were transformed into DH5α and the SS2 mutant strain, the THB medium was supplemented with 50 μg/mL spectinomycin (Spc) or 100 μg/mL Spc, respectively.
Table 1. Bacterial strains and plasmids used in this study.
Raw264.7 macrophage cells derived from mouse Abelson murine leukemia virus-induced tumors were purchased from the American Type Culture Collection (ATCC). Cells were cultured in high-glucose Dulbecco’s modified Eagle medium (DMEM) (Gibco, Invitrogen) supplemented with 10% fetal bovine serum (Gibco, Invitrogen) at 37°C under a 5% CO2 humidified atmosphere.
Phagocytosis assay, intracellular survival assay and adhesion assay in macrophages
A phagocytosis assay was performed according to previous research with a few modifications [Citation17]. In brief, wild-type and each mutant strain were cultured separately to an OD600 of 0.6 ~ 0.8, washed with phosphate-buffered saline (PBS) 3 times and resuspended in DMEM. Raw264.7 cells were cultured to approximately 3.0 × 105 cells/well in a 24-well plate for the phagocytosis assay. Each well of Raw264.7 cells was infected with different single mutant or wild-type strain at a multiplicity of infection (MOI) of 10:1. After incubation for 1 h at 37°C under 5% CO2, the cells were washed with PBS 3 times and incubated with DMEM supplemented with 10 µg/mL penicillin G and 100 µg/mL gentamicin for 1 h to kill extracellular bacteria. The cells were washed with PBS three times, and 1 mL of ddH2O was added to each well to lyse the cells. For the initial semiquantitative screen, 900 µL of ddH2O was removed from the wells and 900 µL of Semisolid Todd-Hewitt agar (THA) were added. The phagocytosed mutant bacteria were compared with the phagocytosed wild-type strain in the 24-well plates. The mutant strains with obviously increased CFUs in the assay were verified with a quantitative phagocytosis assay, which was similar to the initially performed phagocytosis assay except that 1 mL of ddH2O containing the intracellular bacteria, was serially diluted and plated on THA. The phagocytosed bacteria were counted, and significant increases in CFU (P < 0.05) of mutant strains relative to the wild-type control strain indicated attenuated phagocytosis resistance. Assays were performed independently in triplicate.
An intracellular survival assay was conducted as previously described [Citation18]. In brief, the initial steps were the same as the phagocytosis assay. After incubation with antibiotics, cells were lysed at different timepoints of cells (1 h and 3 h), as described in the phagocytosis assay and counted by serial dilution. The intracellular survival rate was calculated as CFU3 h/CFU1 h × 100%. The assay was performed independently three times.
The macrophage adhesion assay was conducted as follows. Raw264.7 cells were pretreated with 1 μg/mL cytochalasin D (Gene Operation) for 30 min at 37°C under 5% CO2 to inhibit actin polymerization. Cells were infected with bacteria at an MOI of 10:1 for 2 h. The wells were then washed with PBS for 5 times. The cells were lysed with 1 mL ddH2O, and the adherent bacteria were plated on THA at a suitable dilution. The colonies derived from bacteria attached to Raw264.7 cells were counted. The assay was performed three independent times.
Defining the transposon insertion sites
Chromosomal DNA was extracted from mutant bacteria with a TIANamp Bacteria DNA Kit (TIANGEN) following the manufacturer’s instructions. The genomes of mutant strains were digested with TaqI (Takara) at 65°C for 5 h as per the manufacturer’s protocols. The fragmented genome was then extracted with a MiniBEST DNA Fragment Purification Kit (Takara). Fragments were circularized with T4 DNA Ligase (Takara) at 16°C overnight. The ligation products were purified and acted as the inverse PCR templates. Inverse PCR was conducted to detect the flanking region of the transposon insertional site. Primers oIPCR1 and oIPCR2 were used in this assay and were listed in . The PCR products were purified and sequenced. BLAST was used to identify the insertional genes from the sequence data. Some BLAST results showed continuous genes that required further confirmation by PCR. The primers for each gene are listed in .
Table 2. Primers used in this study.
Bactericidal assay
The assay was modified based on a previous report [Citation19]. ZY05719 and the mutants were grown to mid-exponential phase and washed 3 times with PBS. The bacteria were then resuspended in PBS to an OD600 of 0.1. A total of 100 μL of bacteria were added to 900 μL of whole pig blood, and incubated together at 37°C for 3 h. The survival rate was calculated by CFU3 h/CFU0 h × 100%. Each assay was performed independently in triplicate.
Survival curve of mice infected with transposon mutant strains
To determine the virulence of the transposon mutants that showed decreased phagocytosis resistance ability, the virulence of M319 (MetQ), M431 (WalR), and M137 (ABC transporter) was determined in mice. Each group contained eight specific pathogen-free (SPF) Balb/c mice (female, 4 weeks old). The aforementioned bacteria were cultured along with wild-type ZY05719 to log phase; the mice were challenged via intraperitoneal injection at a dose of 3 × 108 CFU/mouse. The survival rate was monitored every day for 7 d.
To validate the virulence of the mutant strains, the mice were challenged with two knockout strains ΔMetQ and ΔWalR, as well as their complemented strains CΔMetQ and CΔWalR. The method was the same as previously described.
Growth assay
Mutants with decreased phagocytic resistance and ZY05719 were cultured to an OD600 of 0.6 in THY and inoculated into fresh THY (1:100 dilution). Bacteria were cultured at 37°C with vigorous shaking at 180 rpm. The OD600 was measured hourly for 12 h, and then the 24-h OD600 was measured. All the growth kinetics were measured with three biological replicates. Data were shown as the means with standard deviations.
Transmission electron microscopy
Capsules of ZY05719, M431, and M604 were observed with transmission electron microscopy (TEM) as described previously [Citation20]. The strains were cultured to an OD600 of 0.6–0.8 and centrifuged at 5000 rpm for 5 min, washed three times with PBS and fixed in 2.5% glutaraldehyde overnight. The samples were then dehydrated with propylene oxide for 10 min and embedded in epoxy resin. After fixation, the sections were observed with the Hitachi H-7650 system. The CPS thickness of each strain was measured in 10 different bacteria by Image J software according to the graphical scale. The capsule thicknesses were shown in the histogram.
RNA manipulation and real-time quantitative PCR (qPCR)
Wild-type ZY05719 and M431were grown to exponential phase at an OD600 of 0.6–0.8, and the total RNA was extracted. The RNA extraction was conducted using an E.Z.N.A. Bacterial RNA Kit (Omega). cDNA was synthesized using HiScript II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme) following the manufacturer’s instructions. mRNA levels were determined with a SYBR Premix Ex TaqTM kit (TaKaRa) in the ABI Step One Plus Real-Time PCR System. Primers are listed in . The parC gene was used as a reference [Citation21]. Relative expression of the target gene was calculated using the 2−ΔΔCt method. The experiment was repeated three times independently.
Construction of knockout strains, complemented strains and overexpression strain
To obtain a knockout strain of MetQ, WalR, and HP1065 (ΔMetQ, ΔWalR, ΔHP1065), homologous recombination method was conducted as previously reported [Citation22]. Briefly, the flanking regions of the corresponding genes were amplified (PrimeSTAR HS, Takara) and fused by PCR with the primers listed in . The fusion fragments were ligated to the pSET4s [Citation22] shuttle vector (ClonExpress® MultiS One Step Cloning Kit, Vazyme). Recombinant plasmids were transformed into ZY05719 and the bacteria were cultured on TH agar supplemented with 100 μg/mL spectinomycin at 37°C. The colonies grown on the plate were inoculated and cultured in THB at 28°C for 10 generations. The knockout strains were spectinomycin susceptible and confirmed by PCR.
Complementation strains of ΔMetQ and ΔWalR were constructed by using the shuttle vector pSET2 [Citation23]. Regions containing whole open reading frames (ORFs) as well as putative promoters and terminators of the corresponding genes were amplified using primers CM319 F/R and CM431 F/R. The products were then ligated to the pSET2 plasmid. The recombinant vectors pSET2-319 and pSET2-431 were, respectively, electroporated into ΔMetQ and ΔWalR competent cells to generate the complementation strains CΔMetQ and CΔWalR.
To obtain the LysM2173 overexpression strain (CLysM), pSET2-LysM2173 was first constructed by amplifying LysM2173 as along with the putative promoter and terminator with the primers CMLysMF/R, and then ligating the PCR product to the pSET2 vector. The recombinant plasmid pSET2-LysM2173 was transformed into the wild-type SS2 ZY05719.
Ethics statement
All animal experiments were conducted according to the guidelines of the Institutional Animal Care and Use Committee of Nanjing Agricultural University (Nanjing, China) and approved by the Committee of the Faculty of Veterinary Science of Nanjing Agricultural University (Permit Number: PZ2019132). SPF Balb/c mice were purchased from the Comparative Medicine Center of Yangzhou University and kept in ventilated cages at room temperature of 25°C. After all experiments, surviving mice were anesthetized with isoflurane and euthanized by CO2.
Statistical analysis
Significant differences were determined using unpaired two-tailed Student’s t-test with GraphPad Prism 5 software. Each experiment was performed as three independent repeats. For all tests, a P value < 0.05 was considered statistically significant. Data were expressed as the means with a standard error of the mean (SEM) or the means with standard deviations (SD).
Results
Identification of mutant strains that have decreased phagocytosis resistance capability from mutant library
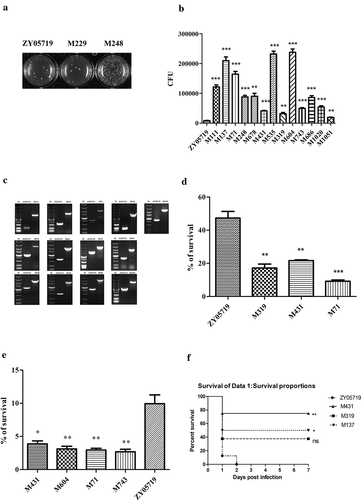
The initial screening was semiquantitative, and the phagocytosis assay was conducted as such to increase throughput. In the last step, 900 μL of the ddH2O which lysed the cells were discarded and 900 μL semi-solid THB were added to the wells directly and plates were cultured at 37°C overnight. Colonies were visualized in the 24-well plates to compare with wild-type ZY05719. The M229 mutant strain had no obvious difference in colony numbers compared with ZY05719, whereas the M248 showed obviously increased CFU relative to ZY05719 in the semi-solid THB ()). After screening approximately 1100 mutant strains from the mutant pool, 13 mutant strains were identified that had a decreased phagocytosis resistance phenotype. These 13 mutants were verified by a quantitative phagocytosis assay to determine the exact phagocytic rate compared with the wild-type strain ()). As shown in ), the mutants of interest exhibited increased phagocytosis rates relative to ZY05719 to different degrees from twofold to 29-fold (P < 0.01). Cps2F mutant (M604) showed the most obvious change in phagocytosed bacteria compared to the wild-type strain. Since capsular polysaccharide (CPS) is an important anti-phagocytic factor, this result was in accordance with the previous report.
Figure 1. Screening of phagocytosis resistance transposon mutants. (a) Semiquantitative phagocytosis assay was performed for the initial screening of mutants with decreased anti-phagocytosis ability. Part of the results were presented. (b) A total of 3.0 × 105 Raw264.7 cells were infected individually with the transposon mutant strains or ZY05719 at an MOI of 10:1 for 1 h. CFUs of phagocytosed mutant strains and ZY05719 recovered from macrophages were determined (c) Confirmation of transposon insertion site. The transposon inserted gene had a 1300 bp size shift compared with the normal gene, which corresponds to the transposon sequence length. Lane M indicated the 5000 bp DNA marker. (d) Three SS2 ZY05719 transposon mutants with decreased anti-phagocytosis ability (M319, M431 and M71) were selected randomly and examined for their survival rate in whole swine blood. The survival ability of the 3 mutants decreased significantly compared to the wild-type strain. (e) Four SS2 ZY05719 transposon mutants with decreased anti-phagocytosis ability (M431, M604, M71 and M743) were selected randomly, and the survival ability in Raw264.7 cells were evaluated. The mutants showed decreased survival in Raw264.7 cells compared to the wild-type strain. Data represent the means with standard error of the mean of three samples. The statistical significance was determined by Student’s t test. * P < 0.05; ** P < 0.01; *** P < 0.001. (f) Virulence to Balb/c mice were determined. Three mutants M431, M319, M137 and wild-type ZY05719 were injected via intraperitoneal at a dose of 3 × 108 per mouse. Each group contained 8 mice. The mutants showed attenuated virulence to different extents compared with ZY05719.
The transposon insertion site in the SS2 genome was examined by amplifying the flanking region of the transposon with primers oIPCR1/2. The PCR products were sequenced, and the sequences were aligned by BLAST. The disrupted genes are listed in . Some sequencing results showed two or more adjacent genes because of the random digest by TaqI and the following circularization, and hence a second PCR was conducted to determine the exact location of the transposon in each sample. The primers used are listed in . All the mutants and ZY05719 were tested with primers specific to the predicted genes, and a 1300 bp size shift was evident, which represented the transposon sequence between the corresponding band of the wild-type strain ()). The results demonstrated that the transposons were inserted within the coding regions of the identified genes.
Table 3. Identification of transposon inserted genes of mutants with decreased anti-phagocytosis ability.
Survival ability in macrophages and whole blood was attenuated in mutant strains
The whole blood contained neutrophils and other polymorphonuclear leukocytes (PMNs) that would also engulf pathogens. Examining the survival ability in whole blood could also indicate phagocytosis resistance to some extent. We discovered that the survival rate of M319, M431, and M71 was significantly reduced compared with the wild-type strain ()), suggesting the survival abilities of the mutants in whole blood were significantly attenuated.
The phagocytosis assay method in our experiment was based on the CFU of surviving bacteria that were engulfed by macrophages. Macrophages could not only internalize the bacteria, but could also eliminate the engulfed bacteria. Therefore, we also compared the survival ability of mutant strains M431, M604, M71, M743, and ZY05719 in the Raw264.7 cells in this assay, and observed that the intracellular survival ability of the mutant strains decreased significantly compared to ZY05719 ().
Transposon mutants with decreased phagocytic resistance impaired the virulence of SS2 in vivo
The process of phagocytosis is an important step in innate immunity. The mutants with attenuated phagocytosis resistance might be more vulnerable in the host. To evaluate the virulence of the strains with reduced phagocytosis resistance ability, we infected Balb/c mice at a dose of 3 × 108 CFU per mouse. In our experiment, we observed that transposon mutant M137-challenged mice showed a 50% survival rate, M431-infected mice showed a 75% survival rate, M319-infected mice showed 37.5% survival rate, while the wild-type ZY05719-challenged mice showed a 0% survival rate ()). The results suggested that the transposon mutants we randomly selected had attenuated virulence compared with the wild-type strain in the mouse infection model to a different extent.
Phagocytic and adhesion to macrophages, and virulence of knockout strains in vivo
From the evaluation of the characteristics of the mutant strains and their previously reported functions, we focused on the MetQ and WalR genes for further research. The anti-phagocytosis phenotype of ΔMetQ, ΔWalR, and their corresponding complementary strains CΔMetQ and CΔWalR were examined. The phagocytosis resistance abilities of ΔMetQ and ΔWalR decreased, and were in accordance with those of M319 and M431, respectively. Phagocytic resistances of CΔMetQ and CΔWalR complementation strains were restored to the level of the wild-type strain (,). Since the phagocytic rates of the wild-type strain and knockout strains were relatively low, M604, which was a capsular polysaccharide (CPS) negative mutant with high phagocytic rate was chosen as a positive control for the phagocytosis assay.
Figure 2. Phagocytosis, adhesion rate and virulence of knockout strains ΔMetQ and ΔWalR. (a) A total of 3.0 × 105 Raw264.7 cells were infected separately with the ΔMetQ, CΔMetQ or ZY05719 at an MOI of 10:1 for 1 h. CFUs of phagocytosed bacteria recovered from macrophages were determined. ΔMetQ showed M319 similar impairment in phagocytosis resistance. CΔMetQ restored the anti-phagocytosis ability to some extent. (b) A total of 3.0 × 105 Raw264.7 cells were infected separately with the ΔWalR, CΔWalR or ZY05719 at an MOI of 10:1 for 1 h. CFUs of phagocytosed bacteria recovered from macrophages were examined. ΔWalR were more susceptible to phagocytosis, whereas CΔWalR recovered the phenotype to the level of ZY05719. (c) Raw264.7 cells were pretreated with 1 μg/mL cytochalasin D for 30 min at 37°C under 5% CO2. Cells were infected with bacteria (ΔMetQ, ΔWalR, CΔMetQ, CΔWalR or wild-type ZY05719) at an MOI of 10:1 for 2 h. Adhesion to macrophages were examined. Except for ΔWalR, adhesion rates were not significantly different from that of ZY05719. Data are shown as the means and standard error of the mean from three independent experiments. * P < 0.05; ** P < 0.01; *** P < 0.001. (d) Balb/c mice were challenged with ΔMetQ, ΔWalR, CΔMetQ, CΔWalR or wild-type ZY05719 via intraperitoneal injection at a dose of 3 × 108 per mouse. Each group contained 8 mice. Virulence of ΔMetQ and ΔWalR were evaluated and the virulence to Balb/c mice were attenuated to different extent compared with ZY05719. The results were similar with the virulence of the corresponding transposon mutant strains M319 and M431. Complemented strains recovered the phenotype to the level of wild-type ZY05719. P < 0.05; ** P < 0.01; *** P < 0.001.
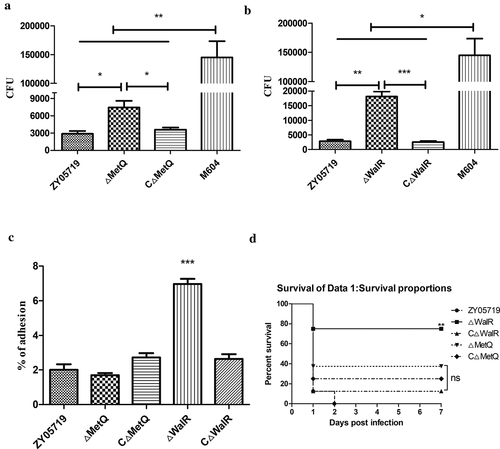
The adhesion to macrophages was examined in ΔMetQ, ΔWalR, CΔMetQ, and CΔWalR, and it showed that only the adhesion rates of ΔWalR varied significantly among the strains ().
To validate the virulence results of transposon mutant strains, two knockout strains ΔMetQ, and ΔWalR, as well as the corresponding complemented strains CΔMetQ and CΔWalR were compared with wild-type strain ZY05719 for their virulence in Balb/c mice. Measures of their virulence were similar to the corresponding transposon mutants M431 and M319, in which the ΔWalR showed 75% survival rate, ΔMetQ-infected mice showed 37.5% survival, wild-type showed 0% survival rate. CΔWalR-challenged mice showed 12.5% survival, and CΔMetQ-challenged mice showed 25% survival ().
Growth characteristics of mutant strains
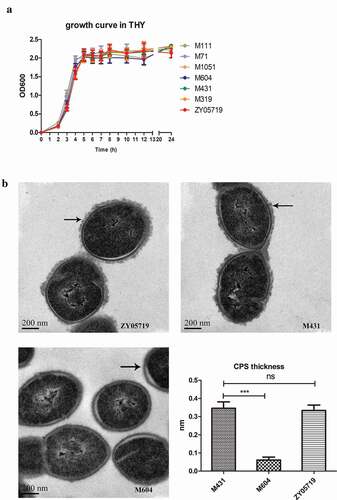
To determine whether the growth kinetics of mutants contribute to the decreased anti-phagocytic ability and virulence, the growth characteristics of transposon mutants were measured in THY. Results showed that the growth of the mutant strains did not have obvious differences compared with wild-type ZY05719 (). Hence, the growth kinetics did not contribute to the changes in phagocytic resistance and pathogenesis of the mutants.
Figure 3. Growth kinetics in THY and CPS biosynthesis. (a) A subset of the 13 mutant strains were examined for their growth characteristics in THY at 37°C with vigorous shaking. There were no obvious differences in growth characteristics among mutants and wild-type ZY05719. Data are shown as the means with standard deviations from three independent experiments. (b) TEM images show the capsule of M431, M604 and ZY05719. Black arrows indicate the CPS of SS2, and the scale bars are 200 nm. CPS thickness of M431 and ZY05719 did not change obviously. CPS of M604 could hardly be visualized. The capsule thicknesses were quantified and shown in histogram. Data are shown as the means and standard error of the mean. * P < 0.05; ** P < 0.01; *** P < 0.001.
Transmission electron microscopy (TEM) observation of capsular polysaccharide (CPS)
The SS2 capsule is a well-known anti-phagocytic factor. The capsule was examined in mutant stains to determine if capsule changes resulted in decreased phagocytic resistance ability. TEM was used to observe the capsule structures of two mutant strains, M431 and M604, and the wild-type ZY05719. M604 was the Cps2F insertional mutant strain. The results showed that the capsule of M604 could hardly be observed. However, the CPS thickness of M431 showed no significant changes compared with that of the wild-type strain (). The CPS thickness was also measured using Image J software and shown in histogram (). Therefore, factors other than CPS contributed to the anti-phagocytic ability of SS2 and these will require further study.
Peptidoglycan biosynthesis-related gene expression in M431
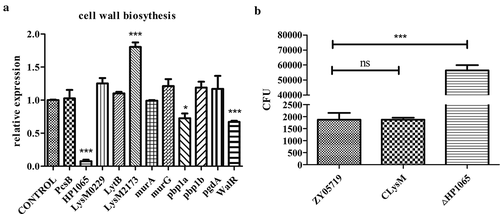
M431 is the WalR insertional mutant strain, and WalR was reported to be associated with cell wall hemeostasis [Citation16]. We examined transcription of the peptidoglycan biosynthesis-related genes PcsB, HP1065, two LysM genes (known to encode the peptidoglycan-binding domain proteins), LytB, murA, murG, pbp1a, pbp1b, and pgdA. Since M431 is the WalR insertional mutant, we also examined the transcription of WalR, and the mRNA level of WalR was decreased 0.67-fold relative to wild-type ZY05719 (P < 0.0001); these results showed that M431 is a WalR knock-down strain. M431 was more sensitive to phagocytosis compared with ZY05719 in the previously described experiment. In our experiment, we discovered that the LysM peptidoglycan-binding domain protein LysM2173 (ZY05719_10260) increased approximately 1.8-fold (P < 0.001), HP1065 (ZY05719_01065, hypothetical protein 1065) decreased significantly (P < 0.001), pbp1a (penicillin-binding protein) decreased 0.72-fold compared to ZY05719 (P < 0.05) (). We speculated that WalR negatively regulates LysM2173 and activates HP1065 and pbp1a in SS2, which would thereby affect the phagocytosis resistance of SS2.
Figure 4. Peptidoglycan biosynthesis-related genes in M431. (a) Ten peptidoglycan biosynthesis-related genes were examined by qPCR for their mRNA levels in M431 and wild-type strain ZY05719. Levels of HP1065 and pbp1a decreased significantly in M431 compared with those in ZY05719, while levels of LysM2173 increased significantly. Each sample was assayed in two replicates and the data represented the means and standard error of the mean from three independent experiments. * P < 0.05; ** P < 0.01; *** P < 0.001. (b) Deletion of HP1065 (ΔHP1065) and overexpression of LysM2173 in wild-type strain ZY05719 (CLysM), were constructed and the phagocytosis assay was conducted. Raw264.7 cells were infected separately with the ΔHP1065, CLysM or ZY05719 at an MOI of 10:1 for 1 h. CFUs of phagocytosed bacteria recovered from macrophages were examined. Phagocytosed ΔHP1065 increased significantly compared with ZY05719. Whereas, engulfed CLysM did not change significantly compared with wild-type ZY05719. Data represent the means and standard error of the mean from three independent experiments. * P < 0.05; ** P < 0.01; *** P < 0.001.
Knockout of HP1065 impaired phagocytosis resistance of SS2.
We discovered that LysM2173 transcription increased while that of HP1065 decreased in the WalR mutant strain, which led us to speculate that overexpression of LysM2173 and deletion of HP1065 might impair the phagocytosis resistance of SS2. The knockout strain of HP1065 (ΔHP1065) significantly attenuated the phagocytosis resistance ability of SS2. However, we constructed a LysM2173 overexpression strain, CLysM, and the phagocytosis rate of CLysM did not change significantly compared with that of the wild-type strain (). Hence, HP1065 might contribute to the phagocytosis resistance ability of SS2, M431 led to the downregulation of HP1065 which could have resulted in the decreased anti-phagocytosis of SS2. Meanwhile, the upregulation of LysM2173 was not involved with the phagocytosis resistance of SS2.
Discussion
Streptococcus suis is an important zoonosis pathogen that causes sudden death, acute sepsis, arthritis, and meningitis in humans and pigs; it has caused great losses in the swine industry, and posed a threat to public health. The symptoms of SS2 infection suggest the possibility that SS2 can evade innate immunity. The phagocytosis process plays an important role in innate immunity, and it is also involved in the acquired immune response by antigen presentation. Actin cytoskeleton rearrangement is the basis of the phagocytosis processes. Actin recruitment is regulated by Rho family GTPases, including RhoA, Rac, and Cdc42 [Citation24]. Phagocytic pathways are initiated by various receptors, Arp2/3, and WASP/WAVE family proteins are triggered to further activate the actin remodeling [Citation25]. Effective phagocytosis by macrophages also includes elimination phagocytosed bacteria by lysosome enzymes, acidic environments, ROS and RNS [Citation26]. In response, pathogens have evolved to counteract host cell-mediated phagocytosis. Some pathogen bacteria produce proteins capable of hijacking the host proteins to impact the actin cytoskeleton. Type III secretion system effectors of enteropathogenic Escherichia coli (EPEC) and enterohaemorrhagic Escherichia coli (EHEC), such as EspH, could interfere with the cytoskeleton dynamics by binding to RhoGEF and resulted in inhibition of Rho activation and actin dynamics [Citation27]. YopT, a type III secretion system effector of Yersinia, could cleave Rho GTPases and result in disruption of actin cytoskeleton and contribute to the anti-phagocytic ability of Yersinia [Citation28]. There are various activating and inhibitory immunoreceptors in neutrophils that modulate the hemeostasis of neutrophil function. Furthermore, some pathogens have evolved to manipulate the associated pathways of neutrophils to evade the immune processes. For example, sialylated capsular polysaccharide of Group B Streptococci (GBS) can interact with inhibitory Siglec-9 and inhibit the phagocytosis process. β-protein of GBS can bind Siglec-5 to dampen macrophage phagocytosis [Citation29,Citation30]. When Escherichia coli bind to inhibitory FcγRIII, phagocytosis of Escherichia coli mediated by class A scavenger receptor MARCO is inhibited [Citation31]. Some bacteria can counteract the host cells when engulfed by macrophages or even survive within professional phagocytes. For Mycobacterium tuberculosis, a membrane-associated protein Rv3671 c was involved with resistance to acidic environments [Citation32]. The KatB gene of Mycobacterium tuberculosis has the function of catalase, peroxidase, and peroxynitritase, and was shown to be involved with the resistance of phagocyte oxidative burst [Citation33]. There were a number of factors that contributed to the resistance to opsonophagocytosis and phagocytosis in Streptococcus. In group A Streptococci, hyaluronic acid capsule and surface M protein were famous for their anti-phagocytosis ability [Citation34,Citation35]. In SS2, it has been reported that a variety of factors could help escape the complement deposition and activation. CPS is the well-established anti-phagocytosis factor [Citation9]. IdeSsuis could degrade porcine IgM to facilitate evasion of complement opsonization and killing [Citation36]. However, mechanism of SS2 resistance to non-opsonophagocytosis are unknown and remain to be elucidated. Therefore, we screened for more pathogenesis-associated genes via a SS2 mutant library constructed previously in this lab [Citation3].
In this study, the SS2 mutant pool was screened for genes related to phagocytosis resistance and 13 related genes were identified. Among the 13 disrupted genes, Cps2F (M604) is a member of the capsular saccharide biosynthesis locus of SS2. The capsule of SS2 is a well-known virulence factor and is famous for its anti-phagocytic ability in Streptococcus [Citation37,Citation38]. Nonencapsulated SS2 were shown to be easily phagocytosed by macrophages and to be avirulent in murine and pig models of infection [Citation9,Citation39]. GNAT family N-acetyltransferase (M743) and HsdM (M678) might be involved in posttranslational modifications (PTMs) of bacterial proteins. In Salmonella typhimurium, acetylation of the pathogenicity island 1 regulator HilD could stabilize the HilD protein and reduce its affinity for DNA-binding [Citation40]. An amino acid mutation at 94 residue was identified in glutamate 5-kinase (proB) (M1020) in a small colony variant (SCV) of S. pneumoniae [Citation41]. The bacterial genome contains a number of ABC transporters that are involved in nutrient uptake, antibiotic resistance, protein excretion, and other functions [Citation42]. In our screening, we identified genes related to the phagocytosis resistance of SS2; three ABC transporter components were identified, including methionine ABC transporter substrate-binding protein MetQ (M319), a branched-chain amino acid ABC transporter substrate-binding protein (M137) and an unknown ABC transporter permease (M535). Many ABC transporters are associated with the full virulence of pathogenic bacteria [Citation43,Citation44]. In our experiments, mice infected with M319 and M137 had an increased survival rate relative to wild-type. MetQ is the substrate-binding protein of the methionine ABC transporter, which is responsible for methionine uptake from the environment. Neisseria gonorrhoeae MetQ is associated with survival in macrophages and monocytes as well as the ability to adhere to epithelial cells [Citation45]. Through in vivo-induced antigen technology (IVIAT), MetQ expression was shown to increase during Streptococcus suis infection, indicating that MetQ might be a potential virulence factor, and could facilitate an important pathogenesis mechanism of SS2 [Citation46]. In Streptococcus pneumoniae, the branched-chain amino acid ABC transporter livJHMGF was necessary for its full virulence [Citation47]. MetQ and livM (branched-chain amino acid ABC permease) have been reported to be essential for replication in cerebrospinal fluid (CSF) in Streptococcus pneumoniae [Citation48]. In this study, however, except for Cps2F gene, the other 12 genes have not reported for their anti-phagocytosis ability.
Knockout strains ΔMetQ and ΔWalR, as well as corresponding complementary strains CΔMetQ and CΔWalR, were examined for their phagocytosis resistance ability and attachment ability to macrophages. Rates of phagocytosis increased in the knockout strains and were restored to wild-type levels in the complementary strains. However, the adhesion rates of the knockout strains and complementary strains did not correlate with the phagocytic rates, indicating that attachment to macrophages might not contribute to the phagocytosis of SS2; this finding was in agreement with a previous report [Citation49]. Growth kinetics in THY were determined for the transposon mutants, and there were no significant differences in growth characteristics among the mutant strains and wild-type strain, indicating the phagocytosis resistance of SS2 was not metabolic dependent process. The capsule of SS2 is well-known for its anti-phagocytic ability. Therefore, the capsules of two mutant strains M431 and M604, and wild-type ZY05719 were examined by TEM and we discovered no obvious differences in capsule thickness between M431 and ZY05719. M604 is a Cps2F insertional mutant, and CPS could hardly be detected in M604. The result indicated that capsule of SS2 was not the only reason for its anti-phagocytosis ability.
M431 is a WalR insertional mutant, and WalR is part of the two-component signal transduction system WalRK (YycFG), which was reported to be related to peptidoglycan formation and cell wall hemeostasis. In S. aureus, YycFG regulates cell wall metabolism, including autolysins (atlA), transglycosylase (IsaA, SceD), and CHAP amidase domain-containing proteins [Citation15,Citation50]. In Streptococcus pneumoniae, the WalRK system is essential for growth and plays an important role in virulence and competence [Citation51]. WalRK of S. pneumoniae could activate the PcsB, pspA, lytB, and LysM repeat proteins [Citation52]. WalRK could also indirectly regulate fatty acid biosynthesis in S. pneumoniae [Citation53]. In Streptococcus mutans, WalR directly regulates comCDE, nlmC, and copY [Citation54], which are involved in competence and biofilm formation. S. mutans WalKR could also regulate atlA and autolysis [Citation55]. In our experiment, the cell wall homeostasis-associated genes were examined, and we observed that the HP1065 and pbp1a transcripts were significantly reduced in M431. However, one of the peptidoglycan-binding proteins, LysM2173, was negatively regulated by WalR, which was in contrast with a previous report [Citation52,Citation56]. The knockout strain ΔHP1065 was more sensitive to phagocytosis, indicating that the disruption of the HP1065 gene might result in the attenuated phagocytosis resistance in M431. HP1065 showed 29% identity to choline-binding surface protein A (PspA) of S. pneumoniae. PspA is a virulence factor in S. pneumoniae that are involved in the evasion of the complement system [Citation57]. PspA could also contribute to the resistance of killing by neutrophil extracellular traps (NETs) [Citation58]. It was also suggested that LysM contributed to the phagocytic resistance of SS2. LysM deletion strain were more easily ingested by macrophages [Citation59]. In our experiment, the transcription of LysM increased in M431. However, overexpression of LysM in ZY05719 did not impair the phagocytosis resistant ability of SS2 in our experiment.
In summary, we screened approximately 1100 mutants from the SS2 mutant library constructed by our lab for phagocytic resistance-related genes and identified 13 gene disruptions that led to increased susceptibility to phagocytosis by macrophages. Randomly selected mutants from these 13 mutants showed decreased survival ability in whole blood and attenuated virulence in mice, indicating that anti-phagocytosis of SS2 was related to the pathogenesis of SS2. The WalR mutant (M431) downregulated the transcription of HP1065. ΔHP1065 showed attenuated phagocytosis resistance relative to the wild-type strain, indicating WalR might regulate the expression of HP1065 and thereby contribute to the phagocytic resistance. Even though most of the screened genes have been suggested to be associated with the phagocytosis resistance of SS2, the mechanism of their functions is not well understood. Therefore, screening was the first step to detect more phagocytosis-related genes that were not discovered before, and further research is needed for each gene to explore its role in phagocytosis resistance and the pathogenesis mechanism.
Author contributions
HF, XP and HZ conceived this experiment. XP and ML conducted the experiments. XP analyzed the data. XP fulfilled the manuscript. All authors approved the final manuscript.
Acknowledgments
We thank for all the staff for the help and suggestions in the experiment.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Feng Y, Zhang H, Wu Z, et al. Streptococcus suis infection: an emerging/reemerging challenge of bacterial infectious diseases? Virulence. 2014;5(4):477–497.
- Gottschalk M, Xu J, Calzas C, et al. Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol. 2010;5(3):371–391.
- Liu R, Zhang P, Su Y, et al. A novel suicide shuttle plasmid for Streptococcus suis serotype 2 and Streptococcus equi ssp. zooepidemicus gene mutation. Sci Rep. 2016;6:27133.
- Cameron DE, Urbach JM, Mekalanos JJ. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc Natl Acad Sci U S A. 2008;105(25):8736–8741.
- Jung H-J, Littmann ER, Seok R, et al. Genome-wide screening for enteric colonization factors in carbapenem-resistant ST258 Klebsiella pneumoniae. mBio. 2019;10(2):e02663–02618.
- Le Breton Y, Mistry P, Valdes KM, et al. Genome-wide identification of genes required for fitness of group A Streptococcus in human blood. Infect Immun. 2013;81(3):862–875.
- Pang M, Xie X, Dong Y, et al. Identification of novel virulence-related genes in Aeromonas hydrophila by screening transposon mutants in a Tetrahymena infection model. Vet Microbiol. 2017;199:36–46.
- Flannagan RS, Heit B, Heinrichs DE. Antimicrobial mechanisms of macrophages and the immune evasion strategies of Staphylococcus aureus. Pathogens. 2015;4(4):826–868.
- Smith HE, Damman M, van der Velde J, et al. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect Immun. 1999;67(4):1750–1756.
- Benga L, Fulde M, Neis C, et al. Polysaccharide capsule and suilysin contribute to extracellular survival of Streptococcus suis co-cultivated with primary porcine phagocytes. Vet Microbiol. 2008;132(1–2):211–219.
- Lecours MP, Fittipaldi N, Takamatsu D, et al. Sialylation of Streptococcus suis serotype 2 is essential for capsule expression but is not responsible for the main capsular epitope. Microbes Infect. 2012;14:11.
- Yu J, Pian Y, Ge J, et al. Functional and structural characterization of the antiphagocytic properties of a novel transglutaminase from Streptococcus suis. J Biol Chem. 2015;290(31):19081–19092.
- Pian Y, Gan S, Wang S, et al. Fhb, a novel factor H-binding surface protein, contributes to the antiphagocytic ability and virulence of Streptococcus suis. Infect Immun. 2012;80(7):2402–2413.
- Lakkitjaroen N, Takamatsu D, Okura M, et al. Capsule loss or death: the position of mutations among capsule genes sways the destiny of Streptococcus suis. FEMS Microbiol Lett. 2014;354(1):46–54.
- Dubrac S, Msadek T. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J Bacteriol. 2004;186(4):1175–1181.
- Howell A, Dubrac S, Andersen KK, et al. Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol Microbiol. 2003;49(6):1639–1655.
- Segura M, Gottschalk M, Olivier M. Encapsulated Streptococcus suis inhibits activation of signaling pathways involved in phagocytosis. Infect Immun. 2004;72(9):5322–5330.
- Tang Y, Zhang X, Wu W, et al. Inactivation of the sodA gene of Streptococcus suis type 2 encoding superoxide dismutase leads to reduced virulence to mice. Vet Microbiol. 2012;158(3–4):360–366.
- Yamaguchi M, Terao Y, Ogawa T, et al. Role of Streptococcus sanguinis sortase A in bacterial colonization. Microbes Infect. 2006;8(12–13):2791–2796.
- Hyams C, Yuste J, Bax K, et al. Streptococcus pneumoniae resistance to complement-mediated immunity is dependent on the capsular serotype. Infect Immun. 2010b;78(2):716–725.
- Wu Z, Wu C, Shao J, et al. The Streptococcus suis transcriptional landscape reveals adaptation mechanisms in pig blood and cerebrospinal fluid. RNA. 2014;20(6):882–898.
- Takamatsu D, Osaki M, Sekizaki T. Thermosensitive suicide vectors for gene replacement in Streptococcus suis. Plasmid. 2001;46(2):140–148.
- Takamatsu D, Osaki M, Sekizaki T. Construction and characterization of streptococcus suis–Escherichia coli shuttle cloning vectors. Plasmid. 2001;45(2):101–113.
- Caron E, Hall A. Identifification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science. 1998;282(5394):1717–1721.
- May RC, Machesky LM. Phagocytosis and the actin cytoskeleton. J Cell Sci. 2001;114(Pt 6):1061–1077.
- Flannagan RS, Cosío G, Grinstein S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol. 2009;7(5):355–366.
- Caron E, Crepin VF, Simpson N, et al. Subversion of actin dynamics by EPEC and EHEC. Curr Opin Microbiol. 2006;9(1):40–45.
- Shao F, Merritt PM, Bao Z, et al. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell. 2002;109(5):575–588.
- Carlin AF, Chang YC, Areschoug T, et al. Group B Streptococcus suppression of phagocyte functions by protein-mediated engagement of human Siglec-5. J Exp Med. 2009a;206(8):1691–1699.
- Carlin AF, Uchiyama S, Chang YC, et al. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood. 2009b;113(14):3333–3336.
- Pinheiro da Silva F, Aloulou M, Skurnik D, et al. CD16 promotes Escherichia coli sepsis through an FcR gamma inhibitory pathway that prevents phagocytosis and facilitates inflammation. Nat Med. 2007;13(11):1368–1374.
- Vandal OH, Pierini LM, Schnappinger D, et al. A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis. Nat Med. 2008;14(8):849–854.
- Ng VH, Cox JS, Sousa AO, et al. Role of KatG catalase‐peroxidase in mycobacterial pathogenesis: countering the phagocyte oxidative burst. Mol Microbiol. 2004;52(5):1291–1302.
- Fischetti VA. Streptococcal M protein: molecular design and biological behavior. Clin Microbiol Rev. 1989;2(3):285–314.
- Moses A, Wessels E, Zalcman MR, et al. Relative contributions of hyaluronic acid capsule and M protein to virulence in a mucoid strain of the group A Streptococcus. Infect Immun. 1997;65(1):64–71.
- Seele J, Beineke A, Hillermann L, et al. The immunoglobulin M-degrading enzyme of Streptococcus suis, IdeSsuis, is involved in complement evasion. Vet Res. 2015;46(1):45.
- Hyams C, Camberlein E, Cohen JM, et al. The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect Immun. 2010a;78(2):704–715.
- Marques MB, Kasper DL, Pangburn MK, et al. Prevention of C3 deposition by capsular polysaccharide is a virulence mechanism of type III group B streptococci. Infect Immun. 1992;60(10):3986–3993.
- Brazeau C, Gottschalk M, Vincelette S, et al. In vitro phagocytosis and survival of Streptococcus suis capsular type 2 inside murine macrophages. Microbiology. 1996;142(Pt 5):1231–1237.
- Sang Y, Ren J, Qin R, et al. Acetylation regulating protein stability and DNA-binding ability of HilD, thus modulating Salmonella typhimurium virulence. J Infect Dis. 2017;216(8):1018–1026.
- Tikhomirova A, Trappetti C, Standish AJ, et al. Specific growth conditions induce a Streptococcus pneumoniae non-mucoidal, small colony variant and determine the outcome of its co-culture with Haemophilus influenzae. Pathog Dis. 2018;76(7). DOI:https://doi.org/10.1093/femspd/fty074
- Sutcliffe IC, Russell R. Lipoproteins of gram-positive bacteria. J Bacteriol. 1995;177(5):1123.
- Brown JS, Gilliland SM, Holden DW. A Streptococcus pneumoniae pathogenicity island encoding an ABC transporter involved in iron uptake and virulence. Mol Microbiol. 2001;40(3):572–585.
- Miyaji EN, Dias WO, Gamberini M, et al. PsaA (pneumococcal surface adhesin A) and PspA (pneumococcal surface protein A) DNA vaccines induce humoral and cellular immune responses against Streptococcus pneumoniae. Vaccine. 2001;20(5–6):805–812.
- Semchenko EA, Day CJ, Seib KL. MetQ of Neisseria gonorrhoeae is a surface-expressed antigen that elicits bactericidal and functional blocking antibodies. Infect Immun. 2017;85(2):e00898–00816.
- Gu H, Zhu H, Lu C. Use of in vivo-induced antigen technology (IVIAT) for the identification of Streptococcus suis serotype 2 in vivo-induced bacterial protein antigens. BMC Microbiol. 2009;9(1):201.
- Basavanna S, Khandavilli S, Yuste J, et al. Screening of streptococcus pneumoniae abc transporter mutants demonstrates that livjhmgf, a branched-chain amino acid ABC transporter, is necessary for disease pathogenesis. Infect Immun. 2009;77(8):3412–3423.
- Molzen T, Burghout P, Bootsma H, et al. Genome-wide identification of Streptococcus pneumoniae genes essential for bacterial replication during experimental meningitis. Infect Immun. 2011;79(1):288–297.
- Segura M, Gottschalk M. Streptococcus suis interactions with the murine macrophage cell line j774: adhesion and cytotoxicity. Infect Immun. 2002;70(8):4312–4322.
- Utsumi R. Bacterial signal transduction: networks and drug targets. Vol. 631. Springer, New York; 2008.
- Wagner C, Saizieu A, Schonfeld A, et al. Genetic analysis and functional characterization of the Streptococcus pneumoniae vic operon. Infect Immun. 2002;70(11):6121–6128.
- Ng WL, Robertson GT, Kazmierczak KM, et al. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol Microbiol. 2003;50(5):1647–1663.
- Mohedano ML, Overweg K, De La Fuente A, et al. Evidence that the essential response regulator YycF in Streptococcus pneumoniae modulates expression of fatty acid biosynthesis genes and alters membrane composition. J Bacteriol. 2005;187(7):2357–2367.
- Senadheera DB, Cordova M, Ayala EA, et al. Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. J Bacteriol. 2012;194(6):1307–1316.
- Ahn S-J, Burne RA. Effects of oxygen on biofilm formation and the AtlA autolysin of Streptococcus mutans. J Bacteriol. 2007;189(17):6293–6302.
- Ng WL, Tsui H-CT, Winkler ME. Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae. J Bacteriol. 2005;187(21):7444–7459.
- Tu AH, Fulgham RL, McCrory MA, et al. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect Immun. 1999;67(9):4720–4724.
- Martinez PJ, Farhan A, Mustafa M, et al. PspA facilitates evasion of pneumococci from bactericidal activity of neutrophil extracellular traps (NETs). Microb Pathog. 2019;136:103653.
- Xu B, Zhang P, Li W, et al. hsdS, Belonging to the type i restriction-modification system, contributes to the Streptococcus suis serotype 2 survival ability in phagocytes. Front Microbiol. 2017;8:1524.
