ABSTRACT
The two leading yeast pathogens of humans, Candida albicans and Cryptococcus neoformans, cause systemic infections in >1.4 million patients worldwide with mortality rates approaching 75%. It is thus imperative to study fungal virulence mechanisms, efficacy of antifungal drugs, and host response pathways. While this is commonly done in mammalian models, which are afflicted by ethical and practical concerns, invertebrate models, such as wax moth larvae and nematodes have been introduced over the last two decades. To complement existing invertebrate host models, we developed fifth instar caterpillars of the Tobacco Hornworm moth Manduca sexta as a novel host model. These caterpillars can be maintained at 37°C, are suitable for injections with defined amounts of yeast cells, and are susceptible to the most threatening yeast pathogens, including C. albicans, C. neoformans, C. auris, and C. glabrata. Importantly, fungal burden can be assessed daily throughout the course of infection in a single caterpillar’s feces and hemolymph. Infected caterpillars can be rescued by treatment with antifungal drugs. Notably, these animals are large enough for weight to provide a reliable and reproducible measure of fungal disease and to facilitate host tissue-specific expression analyses. M. sexta caterpillars combine a suite of parameters that make them suitable for the study of fungal virulence.
Introduction
Fungal infections pose a serious threat to human health and well-being worldwide. Each year, as many, if not more patients, die of fungal infections than of malaria or tuberculosis [Citation1]. The leading yeast pathogens, Candida albicans and Cryptococcus neoformans, account for ~1,400,000 life-threatening infections worldwide with mortality rates of up to 70% [Citation1,Citation2]. Candidemia, most frequently caused by C. albicans, is the fourth most common cause of nosocomial blood stream infections, only surpassed by infections with Staphylococci and Enterococcus spp. Disturbingly, candidemia incidence rates are on the rise. Within less than ten years, they increased by 36% [Citation3]. Although cryptococcosis incidence rates are on the decline in North America, cryptococcosis as an AIDS-defining illness is responsible for 15% of all AIDS-related deaths worldwide [Citation3,Citation4]. This dire situation is further confounded by the emergence of drug-resistant yeast species, such as C. glabrata and C. auris. Patients at risk of developing invasive candidemia are often prophylactically treated with fluconazole and echinocandins as a first line defense strategy [Citation5]. Yet, C. glabrata, the most common non-albicans Candida species associated with nosocomial blood stream infections [Citation6], is intrinsically less susceptible to azole drugs and acquires resistance to echinocandins rapidly [Citation7]. The rapid global spread of multidrug-resistant C. auris has further exacerbated the threat posed by fungal infections. C. auris was first reported in 2009 in Japan [Citation8]. In 2015, C. auris arrived in Europe causing an outbreak involving 72 patients in a cardio-thoracic hospital in London [Citation9]. C. auris outbreaks have been reported from South Korea, India, Spain, Columbia, Switzerland, Germany, Israel, Kuwait, and Oman [Citation10]. Most concerningly, up to 25% of C. auris isolates are multidrug resistant, with some strains being resistant to three of the four drug classes available for the treatment of systemic candidemia. In addition to the unacceptably high burden on human health, fungal infections substantially increase health care costs. Treatment of candidemia requires extended hospitalization, resulting in additional costs of up to 45,000 USD in adult patients or up to 119,000 USD in pediatric patients [Citation11].
It is thus imperative to investigate fungal virulence and host response mechanisms. This is traditionally done in mammalian models. Mice are by far the most frequently deployed host model. The tail vein infection model of candidemia, the gastrointestinal infection model of candidemia, the Candida vaginitis model [Citation12] and the mouse inhalation model of cryptococcosis [Citation13] are used extensively. While mice outnumber rabbits, specific aspects of fungal disease, such as chronic cryptococcal meningitis [Citation14], and Candida keratitis [Citation15] are often studied in rabbits. Both mammalian models combine a number of features that make them particularly amenable for the study of fungal diseases, such as susceptibility, availability of knockout mutants, and histology comparable to human disease. Yet, using mammals is ethically controversial, costly, and requires extensive board certifications and documentation.
In an effort to reduce the usage of mammalian hosts, alternative invertebrate models have been developed and successfully used over the past two decades. The most commonly employed invertebrate species include the nematode Caenorhabditis elegans, the fly Drosophila melanogaster, and larvae of the Greater Wax moth Galleria mellonella. All three can be easily maintained in the laboratory, are much less expensive than mice or rabbits, and have been used for the study of diverse yeast pathogens, such as C. neoformans [Citation16,Citation17], C. albicans [Citation17–Citation19], C. parapsilosis [Citation19–Citation21], C. glabrata [Citation20,Citation22]. Of note, invertebrate models differ in their applicability and the best suitable model should be carefully selected [Citation23]. Unlike mammalian models, these invertebrates do not have adaptive immunity but all share components of the innate immune system [Citation24,Citation25], some of which are conserved with mammals. This includes the Toll-like receptors found in the fly [Citation26] and the homolog of the MKK3/6 kinase in the nematode [Citation27]. Ironically, it is the Toll-like receptors that protect flies from infections with C. neoformans [Citation28], C. albicans [Citation29], and C. glabrata [Citation30] and the MKK3/6 homolog SEK-1 protects the nematode from bacterial invaders [Citation27]. Thus, to increase susceptibility of flies and nematodes to fungal pathogens, Toll and sek-1 [Citation31] mutants are required. A key limitation for the study of human pathogens, is the inability of nematodes and flies to survive human body temperature. Only Galleria can withstand 37°C [Citation17]. Due to their long-standing history as eukaryotic models, well-curated genomes and genome databases exist for the nematode and the fly. Although the Galleria genome has been announced recently [Citation32], detailed analyses and annotations are still outstanding. Delivery of an exact inoculum of fungal cells is crucial when comparing mutant and wild-type yeast strains, neither fly nor nematode permit routine direct injection. Galleria can be directly injected with a defined cell number. Extensive stock collections provide fly and nematode strains, while until recently, Galleria larvae had to be purchased from fishing shops. Now, UK-based TruLarv is selling research grade larvae.
Tobacco Hornworms are most common in the southern United States, where they feed on solanaceous plants and are thus considered a plant pest. As an insect model with a long history in research, Manduca sexta has yielded important insights into flight mechanisms, nicotine resistance, hormonal regulation of development, metamorphosis, antimicrobial defenses, and bacterial pathogenesis. M. sexta laboratory stocks have been derived from animals collected in North Carolina, USA [Citation33] and have been maintained in laboratories on both sides of the Atlantic for several decades. M. sexta’s research portfolio includes a draft genome sequence that has been complemented with tissue-specific transcriptomic analyses [Citation34], numerous successful applications of RNAi [Citation35–Citation39]. Down-stream analyses [Citation40] of the animal’s innate immunity [Citation41], which also comprises b-(1,3)-glucan recognition proteins [Citation41], are facilitated by protocols for the efficient extraction of hemocytes. Despite its versatility, M. sexta has yet to be explored for its suitability as a host model for fungal infections.
Here, we aimed to establish M. sexta as a novel model host for the study of fungal virulence. Inbred animals from the University of Bath’s research colony were tested for their ability to live at 37°C, their susceptibility to different yeast species, and the reproducibility of C. albicans mutant phenotypes obtained in mice virulence studies. Indeed, the caterpillars grow at 37°C while maintaining susceptibility to the leading yeast pathogens C. albicans, C. neoformans, and the emergent C. auris. Specific C. albicans mutants are just as attenuated in their virulence in M. sexta as they are in mice. To expand M. sexta’s applicability as a host model, we developed an infection protocol that permits screening of fungal burden throughout the course of infection in a single animal and uses weight as a proxy measure for virulence in addition to survival. M. sexta can furthermore be used to test efficacy of common antifungal drugs and to query the host transcriptional response to systemic yeast infections. Our results define M. sexta characteristics that recommend these caterpillars as a nonmammalian host model for the study of fungal virulence with susceptibility to different yeast species.
Materials and methods
Origin of the Bath colony of Manduca sexta
The University of Bath colony has been in continuous culture since 1978 without the addition of animals from elsewhere. Bath’s genetic stock was derived from animals from the Truman-Riddiford laboratories at the University of Washington in Seattle, USA. Their animals date back to the ones originally collected in North Carolina in 1976 [Citation33]. Researchers in Europe wishing to obtain M. sexta caterpillars, please contact [email protected]. Researchers in North America can purchase M. sexta caterpillars from the Carolina Biological Supply Company (item #143,882).
Caterpillar maintenance and yeast culture conditions
M. sexta caterpillars from Bath’s facility were reared to fifth instar under standardized conditions. They were maintained in 125 ml disposable cups (Sarstedt Ltd., Cat. No. 75.1335) on a wheat germ-based diet (Table S1) at a constant temperature of 25°C with 50% humidity and 12 hours light/dark cycles. Three days prior to fungal inoculations, animals were shifted to a formaldehyde-free diet as formaldehyde is toxic to nonmethylotrophic yeast.
For infection assays, yeasts were grown overnight in 50 ml YPD (1% yeast extract, 2% peptone, 2% dextrose) at 30°C and cells were harvested by centrifugation for 3 minutes at 3,000 rpm. The cell pellet was washed twice with 1x phosphate buffered saline (PBS) and suspended in 5 ml 1x PBS. Cells were counted and numbers adjusted as indicated. C. albicans YSD85 () cells were heat-inactivated by incubation at 65°C for 20 minutes. For long-term storage, yeasts were cryo-archived at −80°C in 25% glycerol.
Table 1. Yeast strains used in this study.
Yeast infections and measurements of fungal burden and drug efficacy
100 µl of washed and number-adjusted yeast suspension were injected into each caterpillar’s distal right proleg with a 30G1/2” needle (BD Microlance) and a 1 ml NORM-JECT syringe. Following injection, each animal’s weight was recorded. Animals were scored for survival and weight once daily for three or four days post infection. During the course of the experiment, animals were kept on their regular diet (Table S1), on a 12-hour light/dark cycle at the temperature indicated.
To measure fungal burden in caterpillar feces and hemolymph, six animals were injected with either 100 µl of 1x PBS or 106 cells of the wild type SN95 (YSD89) or the hog1∆/∆ mutant strain YSD883 () and kept at 37°C. On infection day (Day 0), feces and hemolymph were dissolved in 1x PBS and spread without further dilution onto YPD. On Day 1, two animals were selected from each group and their hemolymph and feces were collected daily throughout the course of infection (Days 1–3). To collect hemolymph, animals were first kept on ice for 15 minutes. The “horn” was then surface sterilized with 70% ethanol and its top 1–2 mm clipped with a pair of micro scissors. Hemolymph was collected in a prechilled 1.5 ml Eppendorf tube and cooled immediately to reduce polymerization and melanization. One fecal pellet was collected daily with sterile forceps, weighted and suspended in 500 µl 1x PBS. Prior to diluting, the suspension was thoroughly vortexed for 10 seconds, and centrifuged for 5 seconds using a table-top centrifuge to separate fecal matter. To quantify fungal burden, hemolymph and fecal samples were plated either directly onto YPD-agar with 50 µg/ml Kanamycin or in ten-fold serial dilutions. Agar plates were incubated at 30°C for 48 hours and colonies counted.
To assess the efficacy of commonly used antifungal drugs, animals were infected with 107 cells of YSD85 () or 100 µl PBS and treated with increasing doses of fluconazole and caspofungin (Sigma Aldrich, Inc.) as indicated. Drugs were injected 30 minutes postinfection with an ethanol-sterilized Hamilton syringe in a total volume of 10 µl per animal. Caterpillars were weighed and scored for survival on the day of injection and the following three days.
Statistical analyses
Kaplan-Meier estimators were calculated from the survival data using the Surv function in the “survival” R package [Citation50], implementing right censoring for surviving animals. Survival curves were plotted using the “survminer” R package [Citation51]. Differences between treatments were evaluated with a log rank test comparing either different yeast inoculum sizes to each other or different drug concentrations to the vehicle only controls. Weight and fungal burden were plotted using ggplot2 [Citation52] and weight differences were evaluated using linear models with day postinoculation and the interaction between treatment and days post infection as fixed effects and individual as a random effect using the lme function from the “nlme” R package [Citation53]. The survival and weight analyses code can be found here: https://github.com/hobrien/DiezmannLabManduca/blob/master/R/Results.Rmd. All analyses were conducted in R version 3.6.0.
RNA sequencing and gene expression analysis
Five animals were injected with 100 µl 1x PBS or 106 yeast cells of the wild-type strain SN95 (YSD89) () and maintained on a 12-hour light/dark cycle at 37°C for 24 hours. Three animals were then randomly selected for transcriptomic analyses of their midguts. In preparation for midgut extraction, animals were surface sterilized and midgut tissue extracted and washed as described before [Citation34]. Midgut RNA was extracted using TRIzol reagent (Invitrogen, Inc.), quantified using the Agilent 2100 Bioanalyzer system and used to generate RNA-seq libraries (strand-specific, paired end) using the TruSeq RNA sample prep kit (Illumina).
150 nucleotides of the sequence were determined from both ends of each cDNA fragment using the HiSeq 2500 platform (Illumina). An average of 83.79 million read pairs were obtained for each sample (range: 51.4 M to 109.8 M read pairs). Sequencing reads were aligned to a combination of the Manduca sexta whole genome assembly v1.0 (https://i5k.nal.usda.gov/Manduca_sexta) and the Candida albicans SC5314 genome version 4 (http://fungi.ensembl.org/Candida_albicans_sc5314_gca_000784635) using HISAT2 [Citation54]. HTSeq [Citation55] was used to generate read counts for each gene. RNAseq data are available at NCBI’s Short Read Archive (PRJNA629104).
Statistical analysis of differential gene expression was performed using the DEseq2 package from Bioconductor [Citation56] and the SARTools pipeline [Citation57]. A gene was considered differentially expressed if the FDR-value for differential expression was less than 0.10. A heatmap of all differentially expressed genes was made using the heatmap.2 function from the “gplots” R package [Citation58].
In order to compare the M. sexta transcriptional response to C. albicans infection by to that of a mouse, we generated a blast reference database using the peptide sequences for 67,960 proteins from the mouse genome (GRCm38) available from Ensembl (https://useast.ensembl.org). We downloaded the peptide sequences for 27,403 proteins from the M. sexta database [Citation34]. We computed peptide sequence similarity between the M. sexta proteins and the mouse proteins using the blastp local alignment search tool (ncbi-blast+ v2.8.1) [Citation59]. For each M. sexta protein, we had multiple hits that were ranked based on the e-value computed by the blastp search tool. For the list of differentially expressed M. sexta genes from each comparison, we extracted the mouse orthologs with e-values smaller than 10−20. This list of orthologs was then compared to differential expression lists based on RNA-seq analysis of kidneys [Citation60], tongues [Citation61], and vaginas [Citation62] from C. albicans infected mice. Gene lists were compared using VennDiagram (https://cran.r-project.org/web/packages/VennDiagram/VennDiagram.pdf).
Results
M. sexta caterpillars are susceptible to the leading fungal pathogen C. albicans at 37°C
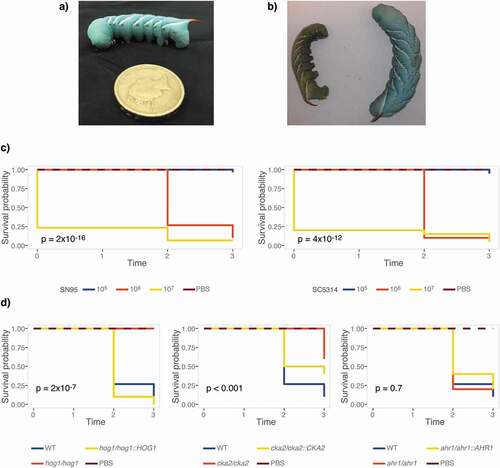
We first aimed to determine if M. sexta fifth instar caterpillars ()), reared and maintained at standard conditions, are susceptible to C. albicans. To do so, groups of ten animals were infected with increasing doses of the widely used C. albicans laboratory strains SC5314 and SN95 [Citation63] (). Animals were kept at 25°C, their standard maintenance temperature, and scored for survival on three consecutive days. Dead animals go limp and turn brown-green in color, which is in stark contrast to the vivid turquoise of live animals ()). Caterpillars infected with C. albicans succumbed in a dose-dependent manner. Both C. albicans strains killed M. sexta efficiently at inocula of 106 or 107 cells per animal ()). To assess if survival measures in caterpillars are comparable to those obtained in the current gold standard, the murine model of systemic candidemia, we tested C. albicans mutants with published phenotypes of either attenuated virulence, such as the hog1∆/∆[Citation64] and ahr1∆/∆[Citation65] mutants, or wild-type levels of virulence, such as cka2∆/∆[Citation66] (). Hog1 is just as essential for virulence in caterpillars as it is in mice. Cka2 is not required to establish systemic infections in mammals but is in caterpillars. Ahr1, while required for virulence in mammals, appears to be dispensable for virulence in caterpillars ()).
Figure 1. M. sexta caterpillars are susceptible to infections with C. albicans at their standard maintenance temperature of 25°C. (a) 13-day old M. sexta fifth instar caterpillar prior to injection, weighing ~2 g. Its distinguishing feature, the reddish horn at the posterior, is clearly visible. (b) 24 hours post injection with the C. albicans wild-type strain SC5314, the dead animal on the left has lost color and turgidity compared to the live PBS control animal on the right. (c) Groups of 10 animals were infected with C. albicans SN95 or SC5314 wild-type strains. Kaplan-Meier curves show dose-dependent killing of caterpillars as indicated by the inset p-values for overall differences, excluding PBS controls. (d) Survival curves of 10 animals per group infected with 107 cells of C. albicans mutants that have attenuated virulence phenotypes in mice or epithelial cell models. The hog1∆/∆ and cka2∆/∆ mutants exhibit attenuated virulence, while virulence of the ahr1∆/∆ mutant is comparable to wild type. The hog1∆/∆ mutant differs significantly from the wild type (p = 0.0001), while the complemented strain hog1/hog1::HOG1 kills M. sexta at a level comparable to that of the wild-type strain (p = 0.21). The cka2∆/∆ mutant is significantly less virulent than the wild type (p = 0.00021), while the complemented strain cka2/cka2::CKA2 is not (p = 0.054). Shown is one of two comparable biological replicates.
Given the importance of temperature for fungal virulence, we investigated if M. sexta retained their susceptibility to C. albicans at human body temperature. We first aimed to establish the effect of raising the temperature from 25°C to 37°C on uninfected caterpillars’ survival and development. All animals survive four-day exposure to 37°C but grow slower than at 25°C (p = 0.0001) (Fig. S1). Next, we quantified survival of animals infected with C. albicans wild type at 37°C ()). 105 C. albicans cells elicit 50% mortality on Day 2, while 106 C. albicans cells lead to 100% mortality on Day 3. In comparison, 107 cells are required for the same outcome at 25°C ()). Increased susceptibility at 37°C may be due to the temperature-dependent expression of C. albicans virulence traits, such as the yeast-to-hyphae transition, which is triggered at 37°C. To exclude the possibility that mortality is due to starvation rather than the outcome of a host-pathogen interaction, we infected caterpillars with live and heat-killed C. albicans wild-type cells and measured survival and weight at 37°C. Only live cells, but not heat-killed yeast cells, kill caterpillars suggesting that killing is not due to nutritional limitations (Fig. S2a). Weight gain, however, was larger in caterpillars injected with heat-killed Candida cells than PBS, indicating that dead yeast cells may be of nutritional value to M. sexta (p < 0.0001) (Fig. S2b).
Figure 2. Elevated temperature increases susceptibility of M. sexta to C. albicans. Groups of 10 animals were injected with increasing doses of the C. albicans wild-type strain SC5314 or with 106 cells of the hog1∆/∆ mutant strain and its complemented control and wild-type progenitor and kept at 37°C for the duration of the experiment. (a) M. sexta caterpillars succumb to infection with 105 cells of the laboratory strain SC5314 at 37°C. (b) Weight gain of caterpillars infected with C. albicans was significantly reduced at all doses relative to PBS controls (**** = P < 0.0001). (c) Attenuated virulence of the hog1∆/∆ mutant is retained at 37°C. (d) Animals infected with the hog1∆/∆ mutant gain significantly more weight than those infected with the wild-type strain RM1000 (**** = P < 0.0001). Infection with the hog1/hog1::HOG1 complemented strain resulted in a lack of weight gain comparable to wild type (p = 0.16). Shown is one of two comparable replicates.
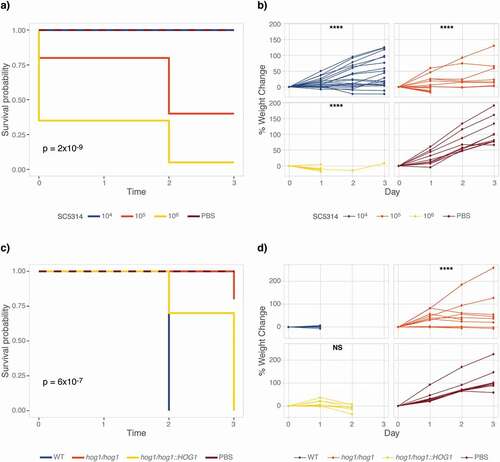
Demonstrating susceptibility of M. sexta caterpillars to C. albicans lent support to their suitability as an alternative host model for the study of fungal virulence and emphasized the need for additional measures of fungal virulence. To add granularity to fungal virulence data collected from M. sexta, we complemented measures of survival with quantifications of weight. To collect weight data, caterpillars were weighed prior to infection and then daily. Weight gain in animals infected with a low dose of 104 cells did not significantly differ from those injected with 1x PBS (p = 0.5847) but caterpillars infected with105 cells per animal exhibited significant weight loss (p = 0.0001). Too few animals survived infection with 106 cells to allow for a meaningful comparison ()). After establishing susceptibility of M. sexta to C. albicans at 37°C, we aimed to validate the attenuated virulence phenotype of the hog1∆/∆ mutant strain. To test this, caterpillars were infected with 106 cells of the wild-type strain, the hog1∆/∆ deletion mutant, and the hog1/hog1::HOG1 complementation strain (). Virulence in the hog1∆/∆ mutant is significantly attenuated at 37°C ()). Weight gain was larger in animals infected with the hog1∆/∆ mutant strain than those infected with the wild-type strain (p < 0.0001), while infection with the complemented strain lead to a comparable lack in weight gain as in wild type infected animals ()).
Measurements of fungal burden in feces and hemolymph obtained from a single animal throughout infection provides a useful measure of virulence
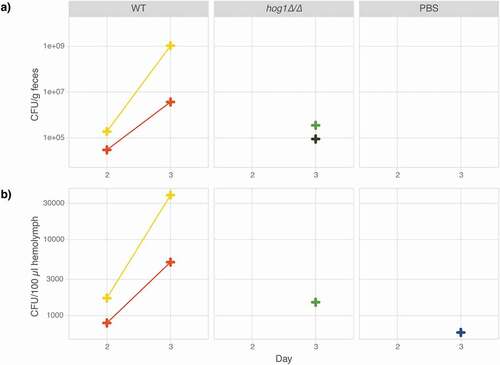
To further expand the applicability of M. sexta caterpillars as a host model for fungal infections, fungal burden in the hemolymph and feces was quantified daily throughout the course of infection. Since the collection of hemolymph or feces does not necessitate killing the animal, data could be collected daily throughout the course of infection for the same caterpillar. Animals infected with the wild type and the hog1∆/∆ mutant () were compared to control animals injected with 1x PBS only and fungal burden measured as colony-forming units (CFUs) in feces and hemolymph in two animals per group. Yeasts were detected in the feces and hemolymph of animals infected with the wild type on Day 2 and CFU counts increased on Day 3 (). In animals infected with the hog1∆/∆ mutant, low CFU counts were detected on Day 3 in feces and hemolymph. Even fewer CFUs were detected in the hemolymph of animals injected with PBS on Day 3. Weight measures showed an inverse correlation growth of the pathogen and the host for the C. albicans wild-type strain (Fig. S3). Thus, measurements of fungal burden, that can be obtained from the same animal throughout the course of infection, provide a valuable parameter for the study of fungal virulence.
Figure 3. Fungal burden in caterpillar feces and hemolymph as a measure of disease progression. Six animals were injected with either 100 µl 1x PBS or 106 cells of the wild-type strain SN95 or its hog1∆/∆ derivative (YSD883) and maintained at 37°C. On Day 1, two surviving animals were selected from each group for CFU analysis. Colony forming units per gram feces (a) and 100 µl hemolymph (b) increase in the wild-type strain between Day 2 and 3 but are not detectable in the hog1∆/∆ mutant nor the control animals until Day 3. Each CFU data point represents the average of at least two (usually three to four) technical replicates.
Caterpillars are suitable for antifungal drug treatment studies
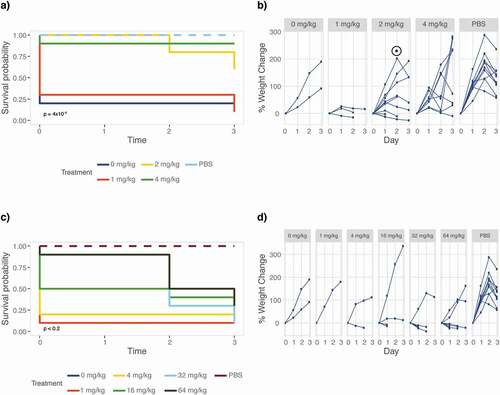
The currently available armamentarium of antifungal drugs is limited and the drugs that are available are often lacking in efficacy. Abilities to study drug efficacy and mode of action in a host model are thus pertinent to the development of novel antifungal drugs. To determine M. sexta’s suitability for drug efficacy testing, we recorded survival and weight of animals infected with the C. albicans wild-type strain SC5314 () that were treated with increasing doses of two common antifungals, fluconazole and caspofungin (). Treatment with the echinocandin caspofungin significantly improves caterpillar survival (p = 4x10−4) and weight gain (p = 0.0047) when compared to untreated animals (). Doses of 2 mg/kg (p = 0.01) and 4 mg/kg (p = 0.002) are most effective in improving survival. Treating animals with fluconazole improves survival rates but caterpillars still grow slower (). Thus treatment with antifungals cures caterpillars from fungal disease.
Figure 4. Antifungal efficacy testing of fluconazole and caspofungin. Groups of ten caterpillars infected with 106 cells of C. albicans strain SC5314 were treated with increasing doses of antifungal drugs fluconazole or caspofungin and maintained at 37°C for the duration of the experiment. Differences between drug treatments were evaluated using a log rank test comparing animals injected with different amounts of drug to those injected with drug solvent (water or DMSO) only, excluding PBS injection controls. Weight gain was compared to the PBS control due to low survival in the solvent control group. (a) Animals were scored daily for survival showing survival improved significantly in animals receiving caspofungin treatment. Inset p value relative to drug solvent control. (b) Rate of weight gain, however, remains low (p < 0.0001) for animals that received 2 or 4 mg/kg caspofungin. The peak and drop in weight, marked with a ⊙, is coinciding with the onset of prepupation. This “pupation drop” is due to the animals refraining from food upon entering the early stages of pupation. (c) Fluconazole treatment improves M. sexta survival but weight gain remains low compared to PBS control infected animals (p < 0.005) for all treatments. (d) Weight data were not analyzed for significance due to the lack of surviving animals in the no treatment group.
M. sexta caterpillars are broadly susceptible to different yeast pathogens
While C. albicans is undoubtedly a major fungal pathogen of humans, clinical manifestations of fungal infections are not limited to C. albicans. To broaden M. sexta’s applicability, we sought to quantify the caterpillars’ susceptibility to other fungal pathogens. To this end, animals were infected with wild-type strains of C. neoformans, C. auris, and C. glabrata (). Type strains of Saccharomyces cerevisiae, the baker’s or brewer’s yeast and Metschnikowia pulcherrima (), a yeast inhabiting fruits and flowers [Citation67], served as reference points for species with attenuated virulence. Based on our initial C. albicans infection assays (), animals were infected with increasing doses of yeast cells starting at 105 cells per animal and up to 109 cells per animal and maintained at 37°C. Groups of ten animals per yeast dose and species were then screened for survival and weight daily ( and ). Notably, only infections with pathogenic yeast species affected survival of caterpillars (). Infections with S. cerevisiae or M. pulcherrima barely affected caterpillar survival. Comparing survival amongst the pathogenic yeast species revealed C. albicans to be the most severe. 107 C. albicans cells result in 100% killing within in one day. 109 C. auris cells were required to kill all animals within four days and the same dose of C. glabrata resulted in 75% killing. 108 C. neoformans cells were required to achieve 50% killing within four days, comparable to C. glabrata. It should be noted that due to the high viscosity of the cell suspension, we could not test higher concentrations than the ones stated here. In addition to assessing survival, each surviving caterpillar’s weight was recorded daily.
Figure 5. Caterpillars are susceptible to common yeast pathogens. Groups of ten animals were infected with increasing numbers of yeast cells as specified and survival was recorded daily. Inset p values represent results from a log rank test of all treatments excluding PBS. Caterpillars are not susceptible to S. cerevisiae or M. pulcherrima but infections with C. neoformans, C. glabrata, C. auris, and C. albicans result in significantly reduced survival rates.
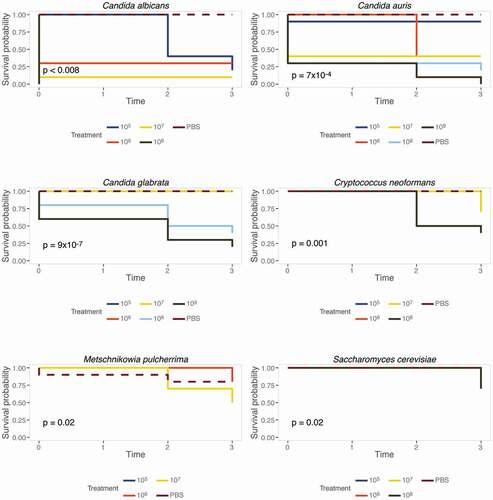
Figure 6. Caterpillar weight as a measure of virulence. Groups of ten animals were infected with increasing amounts of yeast inocula from six different species or PBS and the weight of surviving animals was recorded daily. Weight measures of yeast-infected caterpillars were analyzed for statistical significance revealing that yeast infections affect weight regardless of mortality rates. The slope of each curve at each yeast concentration was compared to the PBS control by fitting a mixed model where caterpillars were treated as a random effect using linear mixed-effects modeling. Statistical significance was assessed as follows: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
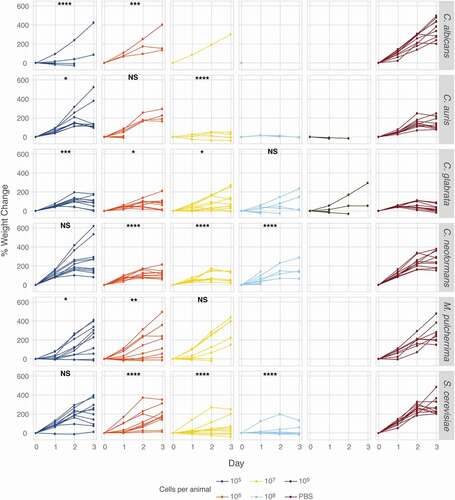
Interestingly, S. cerevisiae and M. pulcherrima did not kill caterpillars as effectively as clinical species, yet, animals infected with S. cerevisiae did not gain weight as well as uninfected control animals (). This is in contrast to animals infected with the C. albicans hog1∆/∆ mutant whose weight gain was significantly different from wild type infected caterpillars (). Infections with C. albicans resulted in severely reduced weight gain, even at the lowest yeast dose tested, while only the highest dose of C. auris elicited a significant reduction in weight gain. C. glabrata infection significantly reduced weight gain at the lowest dose but only slightly at higher doses. Infections with C. neoformans resulted in significantly reduced weight gain at all but the lowest concentration tested. Caterpillars displayed variable degrees of susceptibility to different yeast pathogens and with weight providing an additional measure of host damage. Thus, M. sexta caterpillars are broadly applicable for the study of fungal virulence.
M. sexta transcriptional response profiles differ between uninfected and infected animals
As a proof-of-concept we recorded transcriptional profiles in three animals infected with the wild-type strain SN95 () and compared them to three uninfected control animals. To do so, we collected the mid-gut, rinsed the tissue thoroughly, and extracted RNA, which was then sequenced using Illumina’ HiSeq 2500 platform. With the vast majority of reads mapping to M. sexta rather than C. albicans (Fig. S4), animals infected with Candida displayed a transcriptional profile different from that of uninfected animals (Fig. S5, Table S2). Infection with the wild type elicited quantifiable transcriptional changes in the caterpillars. 157 M. sexta genes were up-regulated and 165 went down when compared to the PBS control animals (Table S2).
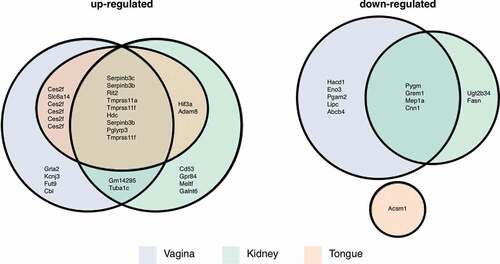
Lastly, we compared our set of differentially expressed M. sexta genes to those identified in three murine candidemia studies. Here, tissue-specific responses in the kidneys [Citation60], tongues [Citation61], and vaginas [Citation62] were recorded for animals infected with Candida wild type (, Table S3). M. sexta genes up-regulated during infection and their homologs in all three mouse tissues include serine peptidase inhibitors (Serpinb3 c, Serpinb3b), a Ras-like protein (Rit2), transmembrane proteases (Tmprss11a, Tmprss11 f), a histidine decarboxylase (Hdc), and a peptidoglycan recognition protein (Pglyrp3). M. sexta genes down-regulated in mouse kidney and vagina include a muscle glycogen phosphorylase (Pygm), a bone morphogenetic antagonist of the DAN family (Grem1), meprin 1 alpha (Mep1a), and calponin 1 (Cnn1). Two of these genes are annotated as functioning in immune system processes (Grem1, Pglyrp3) and three more are relevant for signaling and response to different stimuli (Rit2, Pglyrp3, Pygm). Seven genes, however, are involved in protein metabolic processes (Serpinb3 c, Serpinb3b, Rit2, Tmprss11a, Tmprss11 f, Grem1, Mep1a), suggesting this fundamental process to be of importance in the defense against microbial attacks.
Figure 7. Mouse homologs of M. sexta differentially expressed genes. Numbers of Manduca genes and their murine homologs that were up- or downregulated in mouse vagina, kidney, or tongue and the overlap between the different data sets as depicted in a Euler diagram. Note, in some cases the same mouse gene was the best BLAST match for several M. sexta genes.
Discussion
Invertebrate host models have become valuable alternatives to mammalian hosts for the study of fungal disease. Here, we developed M. sexta caterpillars as a novel invertebrate model showing that they are naturally susceptible to different human pathogenic yeast species, including the emergent multidrug-resistant C. auris. Unlike other host models, these large caterpillars permit daily measures of fungal burden throughout the course of infection in a single animal by either collecting feces or hemolymph. M. sexta can be maintained at 37°C, is large enough to be injected with a specified yeast inoculum and for weight to be a reliable measure of virulence. C. albicans mutant virulence phenotypes found in mice can be replicated and yeast inocula required to elicit a response in caterpillars are comparable to those used in the murine model. Additionally, the caterpillars permit study of antifungal drug efficacy. While these parameters commend M. sexta as a novel host model for the study of fungal virulence, a couple of caveats should be noted. Firstly, as with all animal models, their development follows strict rules. The caterpillars will conclude their fifth instar stadium within 4–5 days, possibly limiting their suitability for some assays. This time span, however, is comparable to the leading insect model, G. mellonella, which has successfully been used for antifungal drug evaluations [Citation68], antibacterial testing [Citation69], and testing of gemini surfactants active against C. albicans [Citation70]. Secondly, about 10% of infected animals exhibited normal growth and survival. It is currently unclear if this is a limitation of the infection protocol or something to do with the underlying biology of the animals. This will need to be factored in when determining sample sizes for future experiments.
This new model system allows for fungal burden to be monitored throughout the course of infection in a single animal via CFU count. This is unlike any other experimental system, where fungal burden is either an endpoint measure in the mouse kidney, in homogenized wax moth larvae [Citation71], nematodes [Citation72], flies [Citation73], or requires genetically modified fluorescent yeast strains for microscopic imaging and analyses [Citation74]. While we did detect very low CFU counts in the PBS control 3 days post infection, we consider this a spurious finding due to cross contamination as preliminary experiments of plating contents of hemolymph and feces of naïve animals did not detect any yeast growth (data not shown). As a consequence, we amended the protocol to include changing gloves when handling animals of different treatment groups.
When reviewing the weight data collected as part of our study, we noticed that while S. cerevisiae has very little effect on caterpillar survival, infections with S. cerevisiae led to reduced caterpillar weight gain. A similar, albeit weaker, pattern was observed for infections with the fruit yeast M. pulcherrima. The dichotomy between survival and weight observed here further emphasizes that fungal virulence comprises more than a measure of survival. It appears that M. sexta would allow discrimination between disease (weight) and death (survival) quantitatively adding further granularity to measuring fungal virulence.
Heat-killed C. albicans cells appear to be nonpathogenic in M. sexta caterpillars. The lack of mortality in response to inoculation with heat-killed yeast cells indicates that yeast viability and proliferation are required for pathogenesis and excludes the possibility of death due to an allergic reaction in response to a large number of fungal cells. This appears to differ from the responses of other host models to fungal pathogens. While susceptibility was reduced, but still measurable, in G. mellonella [Citation75] and the two-spotted cricket [Citation76], heat-killed C. albicans cells elicited 100% mortality in a sepsis-like murine model [Citation77]. In mice, serum levels of β-(1,3)-glucan were elevated in animals injected with heat-killed yeasts when compared to those infected with live cells. Indeed, heat inactivation leads to increased exposure of β-(1,3)-glucan on the C. albicans cell surface [Citation78] and β-(1,3)-glucan activates the innate immune response in invertebrates and mammals [Citation79]. M. sexta’s innate immunity includes a β-(1,3)-glucan recognition protein recognizing S. cerevisiae, which is expressed in the fat body and secreted into the hemolymph [Citation80,Citation81] as well as the antifungal molecule diapausin-1, whose expression is developmentally regulated [Citation82]. The lack of response observed here could be due to insufficient expression of either protein or suboptimal exposure of β-(1,3)-glucan in our heat-killed C. albicans cells.
Measures of the host transcriptional response provide useful insights into the relevance of specific defense mechanisms. Mouse transcriptional profiles in response to C. albicans infections differ between different types of tissues [Citation60–Citation62], which allowed for the identification of tissue-specific response mechanisms such as IL-17 signaling in oral epithelia. Here, we detected numerous differentially expressed genes in the caterpillar midgut when comparing animals infected with C. albicans to the PBS control. Given the size of M. sexta caterpillars, which permits tissue-specific transcriptional analyses [Citation34], this phenomenon could be systematically investigated in caterpillars rather than mammalian hosts, who are burdened by ethic concerns and economic challenges prohibiting large scale or time-course studies. Furthermore, partial conservation of the transcriptional response to C. albicans infections between insects and mammals offers the opportunity to identify and investigate core eukaryotic response mechanisms.
In summary, Manduca sexta caterpillars expand the current repertoire of invertebrate models for the study of fungal disease. They combine a suite of measures that commend it as a new model system. Although, M. sexta genomic and transcriptomic analyses are currently still in their infancy, we would expect that the Tobacco Hornworm’s long history of being an invaluable model for diverse facets of biology will lead to reliable tools in combination with genetic tractability and protocols establishing the fungus’ fate inside the caterpillar.
Supplemental Material
Download Zip (719.9 KB)Acknowledgments
We would like to thank Ewan Basterfield and Chris Apark for expert advice and technical assistance in preparing M. sexta caterpillars. Thanks to all the laboratories who kindly shared their strains with us. This work was supported by an ERC Marie Curie Career Integration Grant to SD as well as undergraduate and postgraduate research funding from the University of Bath.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Brown GD, Denning DW, Gow NAR, et al. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:165rv13.
- Harsha MV, International SV. Emerging fungal pathogens-a major threat to human life. IJPSR. 2017;8:1923–1934.
- Pfaller MA, Diekema DJ. Epidemiology of invasive mycoses in North America. Crit Rev Microbiol. 2010;36:1–53.
- Nyazika TK, Tatuene JK, Kenfak-Foguena A, et al. Epidemiology and aetiologies of cryptococcal meningitis in Africa, 1950–2017: protocol for a systematic review. BMJ Open. 2018;8:e020654.
- Pappas PG, Kauffman CA, Andes D, et al. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Inf Dis. 2009;48:503–535.
- Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–163.
- Jensen RH, Johansen HK, Søes LM, et al. Posttreatment antifungal resistance among colonizing Candida isolates in candidemia patients: results from a systematic multicenter study. Antimicrob Agents Chemother. 2016;60:1500–1508.
- Satoh K, Makimura K, Hasumi Y, et al. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53:41–44.
- Schelenz S, Hagen F, Rhodes JL, et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control. 2016;5:1–7.
- Bidaud AL, Chowdhary A, Dannaoui E. Candida auris: an emerging drug resistant yeast – A mini-review. J Mycol Med. 2018;28:568–573.
- Zaoutis TE, Argon J, Chu J, et al. The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clin Infect Dis. 2005;41:1232–1239.
- Segal E, Frenkel M. Experimental in vivo models of candidiasis. JoF. 2018;4:21.
- Zaragoza O, Alvarez M, Telzak A, et al. The relative susceptibility of mouse strains to pulmonary Cryptococcus neoformans infection is associated with pleiotropic differences in the immune response. Infect Immun. 2007;75:2729–2739.
- Perfect JR, Lang SD, Durack DT. Chronic cryptococcal meningitis: A new experimental model in rabbits. Am J Pathol. 1980;101:177–194.
- Ray WA, O’Day DM, Head WS, et al. Variability in isolate recovery rates from multiple and single breeds of outbred pigmented rabbits in an experimental model of Candida keratitis. Curr Eye Res. 2009;3:949–953.
- Mylonakis E, Ausubel FM, Perfect JR, et al. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. PNAS. 2002;99:15675–15680.
- Fuchs BB, O’Brien E, Khoury EJB, et al. Methods for using Galleria mellonella as a model host to study fungal pathogenesis. Virulence. 2014;1:475–482.
- Pukkila-Worley R, Ausubel FM, Mylonakis E. Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses. PLoS Pathog. 2011;7:e1002074.
- Chamilos G, Lionakis MS, Lewis RE, et al. Drosophila melanogaster as a facile model for large-scale studies of virulence mechanisms and antifungal drug efficacy in Candida species. J Infect Dis. 2006;193:1014–1022.
- Ortega-Riveros M, De-la-Pinta I, Marcos-Arias C, et al. Usefulness of the non-conventional Caenorhabditis elegans model to assess Candida virulence. Mycopathologia. 2017;182:785–795.
- Gago S, García-Rodas R, Cuesta I, et al. Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis virulence in the non-conventional host Galleria mellonella. Virulence. 2014;5:278–285.
- Ames L, Duxbury S, Pawlowska B, et al. Galleria mellonella as a host model to study Candida glabrata virulence and antifungal efficacy. Virulence. 2017;8:1–9.
- Desalermos A, Fuchs BB, Mylonakis E. Selecting an invertebrate model host for the study of fungal pathogenesis. PLoS Pathog. 2012;8:e1002451.
- Mylonakis E, Aballay A. Worms and flies as genetically tractable animal models to study host-pathogen interactions. Infect Immun. 2005;73:3833–3841.
- Vogel H, Altincicek B, Glöckner G, et al. A comprehensive transcriptome and immune- gene repertoire of the lepidopteran model host Galleria mellonella. BMC Genomics. 2011;12:308.
- Lemaitre B, Nicolas E, Michaut L, et al. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983.
- Kim DH, Feinbaum R, Alloing G, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–626.
- Apidianakis Y, Rahme LG, Heitman J, et al. Challenge of Drosophila melanogaster with Cryptococcus neoformans and role of the innate immune response. Eukaryot Cell. 2004;3:413–419.
- Alarco A-M, Marcil A, Chen J, et al. Immune-deficient Drosophila melanogaster: A model for the innate immune response to human fungal pathogens. J Immunol. 2004;172:5622–5628.
- Quintin J, Asmar J, Matskevich AA, et al. The Drosophila Toll pathway controls but does not clear Candida glabrata infections. J Immunol. 2013;190:2818–2827.
- Breger J, Fuchs BB, Aperis G, et al. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 2007;3:e18.
- Lange A, Beier S, Huson DH, et al. Genome sequence of Galleria mellonella (Greater Wax Moth). Genome Announc. 2018;6:61–62.
- Flowers RW, Entomologist RYF. Feeding on non-host plants by the Tobacco Hornworm (Manduca sexta (Lepidoptera: Sphingidae). JSTOR. 1982;65:523–530.
- Kanost MR, Arrese EL, Cao X, et al. Multifaceted biological insights from a draft genome sequence of the Tobacco Hornworm moth, Manduca sexta. Insect Biochem Mol Biol. 2016;76:118–147.
- Flores-Escobar B, Rodríguez-Magadan H, Bravo A, et al. Differential role of Manduca sexta aminopeptidase-N and alkaline phosphatase in the mode of action of Cry1Aa, Cry1Ab, and Cry1Ac toxins from Bacillus thuringiensis. Appl Environ Microbiol. 2013;79:4543–4550.
- Burke WG, Kaplanoglu E, Kolotilin I, et al. RNA interference in the Tobacco Hornworm, Manduca sexta, using plastid-encoded long double-stranded RNA. Front Plant Sci. 2019;10:313.
- Eleftherianos I, Millichap PJ. ffrench-Constant RH, Reynolds SE. RNAi suppression of recognition protein mediated immune responses in the Tobacco Hornworm Manduca sexta causes increased susceptibility to the insect pathogen Photorhabdus. Dev Comp Immunol. 2006;30:1099–1107.
- Eleftherianos I, Gökçen F, Felföldi G, et al.; ffrench-Constant RH, Reynolds SE. The immunoglobulin family protein Hemolin mediates cellular immune responses to bacteria in the insect Manduca sexta. Cell Microbiol. 2007;9:1137–1147.
- Kumar P, Pandit SS, Baldwin IT. Tobacco Rattle Virus vector: A rapid and transient means of silencing Manduca sexta genes by plant mediated RNA interference. PLoS ONE. 2012;7:e31347–10.
- Stoepler TM, Castillo JC, Lill JT, et al. A simple protocol for extracting hemocytes from wild caterpillars. JoVE. 2012;69:1–6.
- Kanost MR, Jiang H, Yu X-Q. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol Rev. 2004;198:97–105.
- Blankenship JR, Heitman J. Calcineurin is required for Candida albicans to survive calcium stress in serum. Infect Immun. 2005;73:5767–5774.
- Bruno VM. Regulation of azole drug susceptibility by Candida albicans protein kinase CK2. Mol Microbiol. 2005;56:559–573.
- Smith DA, Nicholls S, Morgan BA, et al. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol Biol Cell. 2004;15:4179–4190.
- Sniegowski P, Dombrowski P. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation. FEMS Yeast Res. 2002;1:299–306.
- Pitt JI, Miller MW. Sporulation in Candida pulcherrima, Candida reukaufii and Chlamydozyma species: their relationships with Metschnikowia. Mycologia. 1968;60:663–685.
- Dujon B, Sherman D, Fischer G, et al. Genome evolution in yeasts. Nature. 2004;430:35–44.
- Ben-Ami R, Berman J, Novikov A, et al. Multidrug-resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg Infect Dis. 2017;23:1–9.
- Wong B, Perfect JR, Beggs S, et al. Production of the hexitol D-mannitol by Cryptococcus neoformans in vitro and in rabbits with experimental meningitis. Infect Immun. 1990;58:1664–1670.
- Therneau TM, Grambsch PM. Modeling survival data: Extending the Cox model. Springer Science & Business Media New York; 2000.
- Kassambara A, Kosinski M, Biecek P, et al. survminer: drawing survival curves using ‘ggplot2ʹ. [accessed: 22 April 2020]. Available from: http://www.sthda.com/english/rpkgs/survminer/
- Wickham H. ggplot2. New York, NY: Springer-Verlag New York; 2009.
- Pinheiro J, Bates D, DebRoy S, et al. {nlme}: linear and nonlinear mixed effects models. [accessed 22 April 2020]. Available from: https://CRAN.R-project.org/package=nlme
- Kim D, Paggi JM, Park C, et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–915.
- Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169.
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:21–31.
- Varet H, Brillet-Guéguen L, Coppée J-Y, et al. SARTools: a DESeq2- and edger-based R pipeline for comprehensive differential analysis of RNA-seq data. PLoS ONE. 2016;11:e0157022–8.
- Warnes GR, Bolker B, Bonebakker L, et al. gplots: various R programming tools for plotting data. [accessed 22 April 2020]. Available from: https://github.com/talgalili/gplots
- Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421–429.
- Liu Y, Shetty AC, Schwartz JA, et al. New signaling pathways govern the host response to C. albicans infection in various niches. Genome Res. 2015;25:679–689.
- Conti HR, Bruno VM, Childs EE, et al. IL-17 receptor signaling in oral epithelial cells is critical for protection against oropharyngeal candidiasis. Cell Host Microbe. 2016;20:606–617.
- Bruno VM, Shetty AC, Yano J, et al. Transcriptomic analysis of vulvovaginal candidiasis identifies a role for the NLRP3 inflammasome. mBio. 2015;6:3896.
- Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005;4:298–309.
- Alonso-Monge R, Navarro-García F, Molero G, et al. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J Bacteriol. 1999;181:3058–3068.
- Askew C, Sellam A, Epp E, et al. The zinc cluster transcription factor Ahr1p directs Mcm1p regulation of Candida albicans adhesion. Mol Microbiol. 2010;79:940–953.
- Chiang LY, Sheppard DC, Bruno VM, et al. Candida albicans protein kinase CK2 governs virulence during oropharyngeal candidiasis. Cell Microbiol. 2007;9:233–245.
- Vadkertiová R, Molnárová J, Vránová D, et al. Yeasts and yeast-like organisms associated with fruits and blossoms of different fruit trees. Can J Microbiol. 2012;58:1344–1352.
- Li Y, Jiao P, Li Y, et al. The synergistic antifungal effect and potential mechanism of d-penicillamine combined with fluconazole against Candida albicans. Front Microbiol. 2019;10:2853.
- Liu Y, Leung SSY, Guo Y, et al. The capsule depolymerase Dpo48 rescues Galleria mellonella and Mice from Acinetobacter baumannii systemic infections. Front Microbiol. 2019;10:545.
- Wani FA, Amaduddin AB, Sheehan G, et al. Synthesis of novel benzimidazolium gemini surfactants and evaluation of their anti-Candida activity. ACS Omega. 2019;4:11871–11879.
- Lu M, Yu C, Cui X, et al. Gentamicin synergises with azoles against drug-resistant Candida albicans. Int J Antimicrob Agents. 2018;51:107–114.
- Sun L, Liao K, Wang D. Effects of magnolol and honokiol on adhesion, yeast-hyphal transition, and formation of biofilm by Candida albicans. PLoS ONE. 2015;10:e0117695–20.
- Wurster S, Bandi A, Beyda ND, et al. Drosophila melanogaster as a model to study virulence and azole treatment of the emerging pathogen Candida auris. J Antimicrob Chemother. 2019;74:1904–1910.
- Sun L, Zhi L, Shakoor S, et al. microRNAs involved in the control of innate immunity in Candida infected Caenorhabditis elegans. Nature. 2016;6:1–13.
- Cotter G, Doyle S, Kavanagh K. Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunol Med Microbiol. 2000;27:163–169.
- Kochi Y, Matsumoto Y, Sekimizu K, et al. Two-spotted cricket as an animal infection model of human pathogenic fungi. DD&T. 2017;11:259–266.
- Panpetch W, Somboonna N, Bulan DE, et al. Oral administration of live- or heat-killed Candida albicans worsened cecal ligation and puncture sepsis in a murine model possibly due to an increased serum (1→3)-β-D-glucan. PLoS ONE. 2017;12:e0181439–15.
- Wellington M, Dolan K, Krysan DJ. Live Candida albicans suppresses production of reactive oxygen species in phagocytes. Infect Immun. 2008;77:405–413.
- Means TK, Mylonakis E, Tampakakis E, et al. Evolutionarily conserved recognition and innate immunity to fungal pathogens by the scavenger receptors SCARF1 and CD36. J Exp Med. 2009;206:637–653.
- Ma C, Kanost MR. A 1,3-glucan recognition protein from an insect, Manduca sexta, agglutinates microorganisms and activates the phenoloxidase cascade. J Biol Chem. 2000;275:7505–7514.
- Rao X-J, Zhong X, Lin X-Y, et al. Characterization of a novel Manduca sexta beta-1, 3-glucan recognition protein (betaGRP3) with multiple functions. Insect Biochem Mol Biol. 2014;52:13–22.
- Al Souhail Q, Hiromasa Y, Rahnamaeian M, et al. Characterization and regulation of expression of an antifungal peptide from hemolymph of an insect, Manduca sexta. Dev Comp Immunol. 2016;61:258–268.
