ABSTRACT
Avian leucosis (AL) is a disease characterized by tumors and is caused by the avian leukosis virus (ALV). Because of the high variability of viruses and complex pathogenic mechanisms, screening and breeding J subgroup of ALV (ALV-J) resistant avian breeds is one of the strategies for prevention and treatment of AL, thus screening of significant immune markers is needed to promote the development of disease-resistant breeds. In this study, data-independent acquisition (DIA) technology was used to detect the DEPs of three breeds of chicken according to different comparison to investigate the potential markers. Results showed special DEPs for spleen development of each breed were detected, such as PCNT, DDB2, and ZNF62. These DEPs were involved in intestinal immune network used in production of IgA signaling pathways and related to immune response which can be used as potential markers for spleen development in different breeds. The DEPs such as RAB44 and TPN involved in viral myocarditis, transcriptional misregulation in cancer, and tuberculosis can be used as potential markers of spleen immune response after ALV-J infection in chickens. Pair-wise analysis was performed for the three breeds after the infection of ALV-J. The proteins such as RFX1, TAF10, and VH1 were differently expressed between three breeds. These DEPs involved in antigen processing and expression, acute myelogenous leukemia, and viral carcinogenesis can be used as potential immune markers after ALV-J infection of different genetic backgrounds. The screening of potential markers at protein level provides a strong theoretical research basis for disease resistance breeding in poultry.
Introduction
Avian leukosis virus (ALV) is a tumor-causing pathogen that causes great harm to the poultry breeding industry in China [Citation1,Citation2]. The occurrence of Avian leucosis (AL) has become increasingly common in China [Citation3–Citation5]. Therefore, research into ALV via various technologies is vital for poultry development. ALV is a double-stranded RNA virus that not only causes tumors in multiple tissues but also causes immune suppression [Citation4,Citation6]. Studies have shown that the pathogenic rate of J subgroup of ALV is different in chickens of different ages and strains. After experimental inoculation, the incidence of disease varies, which is greatly related to the viral strain, chicken breed, inoculation route, dose and chicken’s age [Citation7,Citation8]. With the development of genomics, transcriptome, proteomics, and bioinformatics, a lot of research into the pathogenic mechanisms of ALV-J, the evolution of the virus, and the resistance to ALV-J in chickens have been done at the macro level [Citation9,Citation10].
The occurrence of certain functions and/or the formation and development of diseases in an organism may be reflected in changes in the abundance of protein content. Data-independent acquisition (DIA) can be used to quantify on a large scale while maintaining targeted protein quantitative technical accuracy and sensitivity advantages [Citation11]. The regulatory mechanism of protein level of ALV-J is little [Citation12–Citation14]. Improvements in the accuracy and depth of proteomic technology have allowed more research into the pathogenic mechanisms and molecular regulatory network of ALV-J. However, due to variabilities in virulence, the omics results of various studies are not suitable for integrated analysis. Therefore, studying omics differences in virus-infected organisms from different genetic backgrounds will provide a strong theoretical basis for breeding disease resistance in poultry. The purpose of this study was to examine the sensitivity of different breeds of chickens infected with ALV-J and use DIA technology to detect differentially expressed proteins (DEPs) in the spleens of these chickens.
Materials and methods
Animals for proteomics analysis
All animal care and experimental procedures were reviewed and approved by the Animal Care and Use Committee (#YYS130125) of Animal Care Advisory at Sichuan Agricultural University. This study was carried out in strict accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals of the State Council of the People’s Republic of China. Since the NX0101 strains were from broiler breeds [Citation15], susceptible breeds of Avian broilers were added as controls. Avian broilers (A) were provided by the Zheng Da Company, Chengdu, China. Tibetan chickens (T) were provided by Maoxian farm, Aba, China. Pengxian yellow chickens (P) were provided by Tianhua Company, Chengdu, China. The hens were maintained separately in two pathogen-free negative pressure isolators for poultry (Strong Star Equipment Technology Co, Qingdao, China), and fed according to the chicken feeding management procedures. All chickens used in this study were detected by PCR technology (the specific primer sequences were F: 5ʹ-GCTGCCATCGAGGTTACT-3ʹ; R: 5ʹ-AGTTGTCAGGGAATCGAC-3ʹ) and ALV test kit (IDEXX, Westbrook, USA) to make sure exogenous ALV was free.
Sample preparation and collection for proteomics analysis
After hatching, the female chicks at 1 day age of three breeds were both divided into injected virus groups and control groups. Each chick of injected virus group was injected with 100 μl ALV-J based on the TCID50 of the virus (approximately 104 TCID50/100 μl); each chick of control groups was injected with 100 μl DMEM. Three chickens were euthanized and the spleens, thymus, and bursa of fabricius were harvested on days 7, 15, 30, and 45 post infection of each group. Based on the immune index (the weight of spleen divided by the body weight), the spleens of chickens euthanized on days 15 were used for the DIA study. The DIA samples from were named as follows: the Pengxian chickens, Tibetan chickens, and Avian broiler treated with ALV-J were 15PI, 15TI, and 15AI, respectively. The Pengxian chickens, Tibetan chickens, and Avian broiler control groups were 15PC, 15TI, and 15AC, respectively. Statistical analyses were conducted via one-way ANOVA using SAS 8.0 software for Windows and the figures were made using GraphPad Prism 5.0. All data are expressed as mean ± SEM. P-values <0.05 were considered statistically significant.
DIA procedures
The spleen protein digestion procedure was performed according to the literature, with minor modifications. The mixtures were placed into a Tissue Lyser for 2 min at 50 Hz to release proteins. After centrifugation with 25,000 g at 4°C for 20 min, the supernatant was transferred into a new tube, reduced with 10 mM dithiothreitol (DTT) at 56°C for 1 hand alkylated by 55 mM iodoacetamide (IAM) in the dark at room temperature for 45 min. Following centrifugation (25,000 g, 4°C, 20 min), the supernatant containing proteins was quantified by Bradford and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Trypsin Gold (Promega, Madison, WI, USA) was used to digest the proteins at 37°C. The peptides were desalinated and vacuum dried. The peptides were separated on a Shimadzu LC-20AB HPLC Pump system coupled with a high pH RP column. The eluted peptides were pooled as 10 fractions and vacuum-dried. The peptides separated in liquid phase were ionized by nanoESI source and then entered the Q-Exactive HF (Thermo Fisher Scientific, San Jose, CA, USA) for DIA mode detection. This process was mainly based on a high-resolution mass spectrometer to produce sample data. For large-scale DIA data, the MaxQuant [Citation16] and Spectronaut [Citation17] used constructed spectrum image database information to complete the deconvolution extraction of data, and the mProphet algorithm was used to complete the analysis and quality control of data by Msstats software package [Citation18]. The biological repetition of each group was executed the DIA procedure, respectively. Based on the quantitative results, differential proteins between different comparison groups were searched and functional analysis of differentially enriched proteins was performed by R software packages.
Multiple reaction monitoring (MRM) procedures
Protein extraction, quality control, and enzymatic hydrolysis were carried out according to the above methods. Samples were digested as described and spiked with 50 fmol of β-galactosidase for data normalization. MRM analyses were performed on a QTRAP5500 mass spectrometer (AB SCIEX, Foster City, CA, USA) equipped with an LC-20AD nanoHPLC system (Shimadzu, Kyoto, Japan). A spectral library of MS/MS data was generated on a TripleTOF5600 (AB SCIEX, Foster City, CA, USA) and searched using Mascot v2.3 (Matrix Science, London, UK) against aHomo database (35985 entries). The data file was imported into Skyline software where a library was built. The peptides were selected for MRM method development according to the following criteria: (1) peptides with unique sequences in the database; (2) a maximum peptide m/z of <1250 (limitation of Quadrupole scan), with a peptide length range of 5–40 aa; (3) no Methionine in peptides; (4) Carbamidomethyl present on Cysteine and no variable modifications in peptides; and (5) no missed cleavage of trypsin. The chromatograms of all transitions generated on QTRAQP5500 were input to Skyline. The MRM method of a given protein was successfully developed only if the protein had at least one unique peptide which (1) was identified with the MS/MS spectral library (cutoff score >0.95), (2) had >5 fragment ions with the same elution profile and in the same ratios as the spectral library, and (3) had an accurate retention time (less than ± 2 min deviation from the predicted retention time). Statistical analysis was performed using one-way analysis of variance (ANOVA). P-values <0.05 were considered statistically significant.
Results
Disease statistics
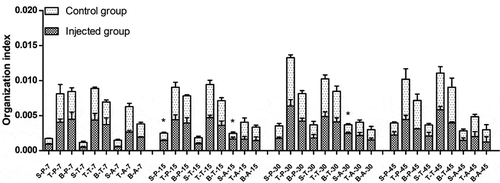
Organization indexes at different times in the injected and control groups of different breeds are shown in . Since the NX0101 strains came from broiler breeds [Citation15], susceptible breeds Avian broilers were added. On day 15, the organotin index of spleen function was significantly higher in both Pengxian yellow chickens and Avian broilers injected with ALV-J than in the control group of each breed (P < 0.05). On day 30, the organotin index of spleen function was significantly higher in Avian broilers injected with ALV-J than in the control group (P < 0.05). There were no significant differences in the thymus or bursa of fabricius among the three breeds on days 7, 15, 30, or 45 of injected with ALV-J groups and control groups. There were no significant differences in the spleen, thymus, or bursa of fabricius on days 7, 15, 30, or 45 in Tibetan chickens injected with ALV-J compared to Tibetan chickens in the control group.
Figure 1. Organization index of infected and control groups of different breeds at different times. S: spleen; T: thymus; B: bursa of fabricius. P: Pengxian yellow chicken; T: Tibetan chicken; A: Avian broilers. All values are represented as the mean ± SEM (n = 3). (*) represents statistical significance (P < 0.05).
Identification of proteins
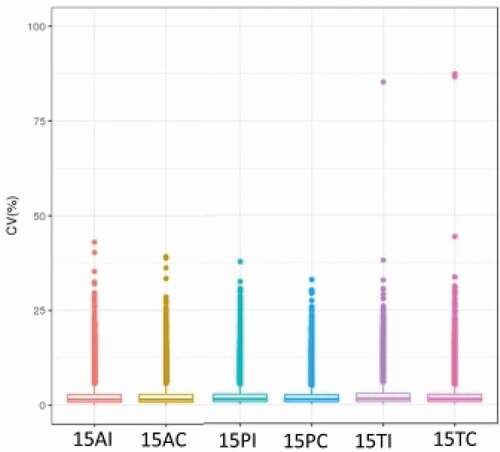
Approximately 30,000 peptides and 6,000 proteins were identified in each sample. shows the coefficient of variation (CV) within groups that was used to evaluate the quality of the data. The data had high reliability and could be further analyzed. briefly summarizes the peptide number and protein number for each sample. Fold change ≥2 and P < 0.05 were used as the screening criteria for significantly differentially expressed proteins (DEPs). The results are shown in . There were 57 down-regulated proteins and 43 up-regulated proteins between the 15AC group and 15PC group, 233 down-regulated proteins, and 119 up-regulated proteins between the 15AC group and 15TC group, and 140 down-regulated proteins and 113 up-regulated proteins between 15PC group and 15TC group.
Table 1. Overview of quantitative results of each sample.
Table 2. Statistical list of differential proteins.
Figure 2. CV profile of each groups. The horizontal axis is the sample groups, and the vertical axis is the corresponding CV.
A total of 65 down-regulated proteins and 38 up-regulated proteins between the 15AI group and 15AC group, 25 down-regulated proteins, and 51 up-regulated proteins between the 15PI group and 15PC group, and 48 down-regulated proteins and 44 up-regulated proteins between the 15TI group and 15TC group. There were 192 down-regulated proteins and 69 up-regulated proteins between the Avian broilers and Pengxian yellow chickens, 387 down-regulated proteins, and 153 up-regulated proteins between the Avian broilers and Tibetan chickens, and 141 down-regulated proteins and 99 up-regulated proteins between the Pengxian yellow chickens and Tibetan chickens.
The analysis of DEPs in spleen development of three breeds
The three breeds were compared for spleen development analysis and the protein interaction network, GO function enrichment, and pathways of DEPs were analyzed. The results are shown in , and . shows the DEPs of |log2FC| >3.
Table 3. Statistical list of pathway enrichment of differentially expressed proteins (DEPs) in the spleens of three breeds (|log2FC|>3).
Figure 3. Protein interaction network analysis of DEPs of spleen development of three breeds. A: 15AC-VS-15PC; B: 15AC-VS-15 TC; C: 15PC-VS-15 TC. “ – ”activation; “ – ”inhibition; “ – ”binding; “ – ”catalysis; “ – ”phenotype; “ – ”posttranslational modification; “ – ”reaction; “ – ”transcriptional regulation; “→”positive; “—”negative; “ – ∙”unspecified.

Figure 4. GO Enrichment analysis of DEPs in the spleens of three breeds. A: 15AC-VS-15PC; B: 15AC-VS-15 TC; C: 15PC-VS-15 TC. Cluster frequency means the ratio of annotation is the same GO term between all of DEPs and all of the proteins.
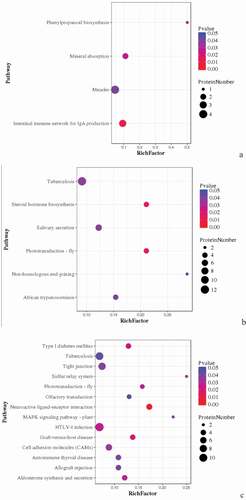
Figure 5. Pathway enrichment analyses of DEPs in spleens of three breeds. A: 15AC-VS-15PC; B: 15AC-VS-15 TC; C: 15PC-VS-15 TC. The enrichment factor is the number of DEPs annotated to the pathway divided by all of the identified proteins annotated to the pathway. The higher the value, the higher the proportion of differentially expressed proteins annotated to this pathway. The dot size in the figure represents the number of DEPs annotated to this pathway.
There were interactions between TACR1 and ANXA1, INTS8 and POLR2B, CNOT11 and CNOT2, and PTGES2 and AARSD1 of Avian broilers and Pengxian yellow chickens and DDB2 was positively regulated by MCRS1 ( (A)). The spleen developmental DEPs of Avian broilers and Pengxian yellow chickens were formed interactions network by UBE2V2 and FBXL20 as the center, PCNT, HAUS2, and CEP290 as the center, MRPS7, ERAL1, and MRPS18A as the center, RBM5, PHF5A, and PLRG1 protein interactions as the center and CDK14, ZNF148, and ORC2 as the center. There was negative regulation or positive regulation between DEPs, such as LMO7 negatively regulated FBXL20 and KDM5A positively regulated NUP153 ( (B)). The spleen developmental DEPs of Pengxian yellow chickens and Tibetan chickens were formed interactions network by MRPS14, OASL, and RNF7 as the center, CTTN, TACR1, and CLTA as the center and CEP290, PCNT and HAUS2 as the center. There was negative regulation or positive regulation between DEPs, such as TFDP1 negatively regulated RB1 and RB1 positively regulated SMC5 ( (C)).
(A) shows the GO enrichments of DEPs in the spleen development of Avian broilers and Pengxian yellow chickens. DEPs were enriched in cell receptor complex, alpha-beta T cell receptor complex, HOPS complex (cellular component); histone acetyltransferase activity, phosphatidylinositol binding and carbohydrate binding (molecular function); regulation of signal transduction, fatty acid transport, and Notch signaling pathway (biochemical process).
(B) shows the GO enrichments of DEPs in the spleen development of Avian broilers and Tibetan chickens. DEPs were enriched in transcriptional repressor complex and nuclear origin of replication recognition complex (cellular component); serine-type endopeptidase inhibitor activity and low-density lipoprotein receptor activity (molecular function); convergent extension involved in axis elongation, histone H2A acetylation, and visual perception (biochemical process).
(C) shows the GO enrichments of DEPs in the spleen development of Pengxian yellow chickens and Tibetan chickens. DEPs were enriched in AP-1 adaptor complex, trans-Golgi network transport vesicle, and HAUS complex (cellular component); histone tyrosine kinase activity, protein tyrosine kinase activity, and stem cell factor receptor activity (molecular function); response to antibiotics, response to lipopolysaccharides, defense response to bacterium and response to inorganic substances (biochemical process).
KEGG pathway enrichment was analyzed for DEPs. DEPs involved in the intestinal immune network for IgA production, mineral absorption, and phenylpropanoid biosynthesis, such as BLB, Mx1, and RAB44 were compared in spleen development between Avian broilers and Pengxian yellow chickens () ( (A)). DEPs involved in tuberculosis, African trypanosomiasis and non-homologous end-joining, such as RAB44, VH1, MATN3, DDB2, and VCAM1 were compared in spleen development between Avian broilers and Tibetan chickens () ( (B)). DEPs involved in tuberculosis, neuroactive ligand–receptor interaction, MAPK signaling pathway, HTLV-I infection, graft-versus-host disease, autoimmune thyroid disease and allograft rejection, such as BF2, DHX36, CNPY3, and HYAL1 were compared in spleen development between Pengxian yellow chickens and Tibetan chickens () ( (C)).
The analysis of DEPs of infected with ALV-J groups and control groups of three breeds
The proteomics of infected with ALV-J groups and control groups of three breeds were compared. Protein interaction network, GO function enrichment, and pathway of DEPs were analyzed. The results are shown in , and . shows the DEPs of |log2FC| >3.
Table 4. Statistical list of pathway enrichment of DEPs in spleens of three breeds after infection with ALV-J (|log2FC|>3).
Figure 6. Protein interaction network analysis of DEPs of spleen of injected groups and control groups of three breeds. A: 15AI-VS-15AC; B: 15PI-VS-15PC; C: 15 TI-VS-15 TC. “ – ”activation; “ – ”inhibition; “ – ”binding; “ – ”catalysis; “ – ”phenotype; “ – ”posttranslational modification; “ – ”reaction; “ – ”transcriptional regulation; “→”positive; “—”negative; “ – ∙”unspecified.
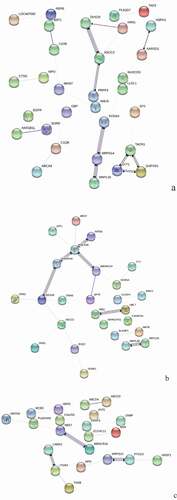
Figure 7. GO Enrichment analysis of DEPs in spleens of injected and control groups of three breeds. A: 15AI-VS-15AC; B: 15PI-VS-15PC; C: 15 TI-VS-15 TC. Cluster frequency means the ratio of annotation is the same GO term between all DEPs and all proteins.
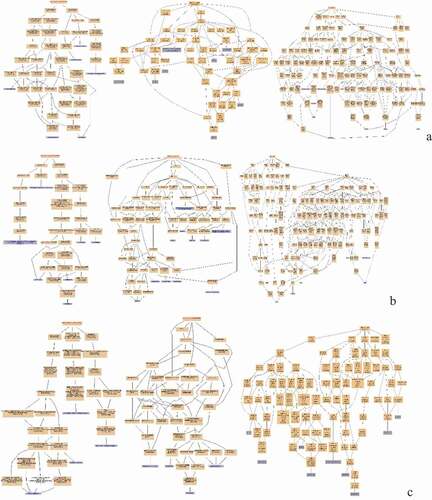
Figure 8. Pathway Enrichment Analysis of DEPs of spleens of injected and control groups of three breeds. A: 15AI-VS-15AC; B: 15PI-VS-15PC; C: 15 TI-VS-15 TC. The enrichment factor is the number of DEPs annotated to the pathway divided by all of the identified proteins annotated to the pathway. The higher the value, the higher the proportion of differentially expressed proteins annotated to this pathway. The dot size in the figure represents the number of DEPs annotated to this pathway.
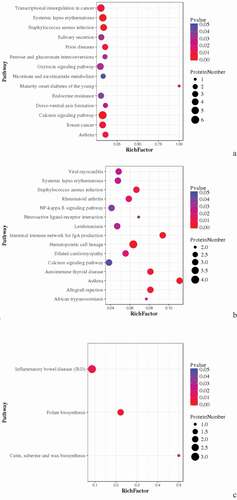
For the DEPs of Avian broilers infected with ALV-J groups and control groups, TACR1, SYT1, and GAPVD1 were interacted with each other, KCNA3, MRPS14, and MRPL50 were interacted with each other, and PRPF3, ASCC3, and DHX29 were interacted with each other ( (A)). For the DEPs of Pengxian yellow chickens infected with ALV-J groups and control groups, there were interactions between DDX49, SNRNP40, POLR2B, and INTS9 proteins, UBL7 and MIB1 proteins, and MRPL50 and MRPL30 proteins ( (B)). There were interactions between NEK7 and WBSCR16, MRPS14 and PTCD3, and LAMA2 and ITGB3 of the DEPs of Tibetan chicken infected with ALV-J groups and control groups ( (C)).
(A) shows the GO enrichments of DEPs in infected with ALV-J groups and control groups of Avian broilers. DEPs were enriched in integral to plasma membrane protein, MHC class II protein complex and transport vesicle (cellular component); guanylate cyclase activity, ligand-gated sodium channel activity, and voltage-gated potassium channel activity (molecular function); regulation of protein transport, negative regulation of neurotransmitter secretion, and negative regulation of protein import into nucleus (biochemical process).
(B) shows the GO enrichments of DEPs in infected with ALV-J groups and control groups of Pengxian yellow chickens. DEPs were enriched in transcription factor complex, MHC class II protein complex, and integral to plasma membrane (cellular component); chromatin binding, ATP-dependent helicase activity, and DNA-dependent ATPase activity (molecular function); positive regulation of phagocytosis, negative regulation of growth of symbiont in host and positive regulation of viral transcription (biochemical process).
(C) shows GO the enrichments of DEPs in infected with ALV-J groups and control groups of Tibetan chickens. DEPs were enriched in secretory granule membrane, specific granule, and alveolar lamellar body membrane (cellular component); histone methyltransferase activity and Rho guanyl-nucleotide exchange factor activity (molecular function); regulation of exocytosis, regulation of gene expression by genetic imprinting and histone methylation (biochemical process).
KEGG pathway enrichment was analyzed for DEPs. DEPs in infected with ALV-J groups and control groups of Avian broilers were involved in transcriptional misregulation in cancer, salivary secretion, prion diseases, endocrine resistance, breast cancer, and asthma such as CDC42SE2, XRN1, MKI67, and RAB44 () ( (A)). DEPs in infected with ALV-J groups and control groups of Pengxian yellow chickens were involved in viral myocarditis, staphylococcus aureus infection, NFκB signaling pathway, intestinal immune network for IgA production, hematopoietic cell lineage, and autoimmune thyroid disease, such as KDM5A and RAB44 () ( (B)). DEPs in infected with ALV-J groups and control groups of Tibetan chicken were involved in inflammatory bowel disease and folate biosynthesis, such as PLEKHA8, HBBR, and ABCD3 () ( (C)).
The analysis of DEPs of each pairwise comparison of three breeds after ALV-J infection
Pairwise analysis was performed for the three breeds under the stimulation of ALV-J infection. Protein interaction network, GO functional enrichment, and pathway enrichment analysis were carried out for the obtained DEPs. The results are shown in , and . shows the DEPs of |log2FC |> 3.
Table 5. Statistical list of pathway enrichment of DEPs in spleens of ALV-J infected comparable groups (|log2FC|>3).
Figure 9. Protein interaction network analysis of DEPs of each comparison group. A: 15A-VS-15P; B: 15A-VS-15 T; C: 15P-VS-15 T. “ – ”activation; “ – ”inhibition; “ – ”binding; “ – ”catalysis; “ – ”phenotype; “ – ”posttranslational modification; “ – ”reaction; “ – ”transcriptional regulation; “→”positive; “—”negative; “ – ∙”unspecified.

Figure 10. GO enrichment analysis of DEPs of each comparison group. A: 15A-VS-15P; B: 15A-VS-15 T; C: 15P-VS-15 T. Cluster frequency means the ratio of Annotation is the same GO term between all DEPs and all proteins.
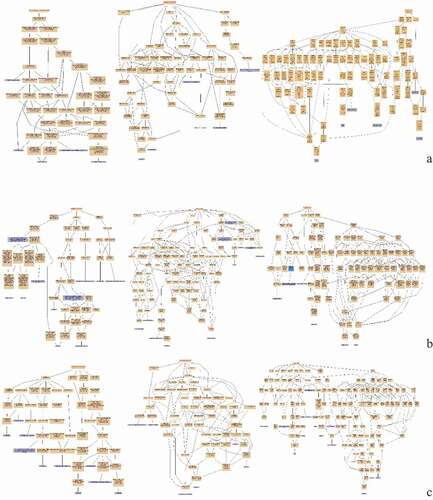
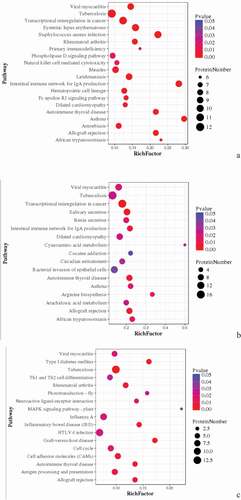
Figure 11. Pathway enrichment analyses of DEPs of each comparison group. A: 15A-VS-15P; B: 15A-VS-15 T; C: 15P-VS-15 T. The enrichment factor is the number of DEPs annotated to the pathway divided by all of the identified proteins annotated to the pathway. The higher the value, the higher the proportion of differentially expressed proteins annotated to this pathway. The dot size in the figure represented the number of DEPs annotated to this pathway.
The CSTF2, FIP1L1, SMNDC1, PCF11, and other DEPs were the center of the interaction network of Avian broilers and Pengxian yellow chickens after ALV-J infection, and there was positive regulation between DEPs, such as NUP153, which had positive regulation on NUP35, and STAT1, which had positive regulation on IRF4 and CD274 ( (A)). The interaction network of DEPs of Avian broilers and Tibetan chickens after ALV-J infection was complex, mainly centered on RAD51, ZNRF2, CBLB, OASL, CWC25, RBM5, etc., and there was positive or negative regulation among DEPs, such as DDB2 with positive regulation of STMN1 and EPB41L2 with negative regulation of RNF7 ( (B)). The DEPs of Pengxian yellow chicken and Tibetan chicken after ALV-J infection were formed, respectively, by OASL, RPL19, RPL7L1 as the center of the interaction network, CBLB, UBR2, FBXL20 as the center of the interaction network, ARID4A, KDM5A, CREBBP as the center of the interaction network, and there was positive or negative regulation among DEPs, such as RBL2 positively regulated SMC5, CBLB negatively regulated OASL ( (C)).
(A) shows the GO enrichments of DEPs of Avian broiler and Pengxian yellow chickens after ALV-J infection. DEPs were enriched in voltage-gated potassium channel complex, HOPS complex, and protein-DNA complex (cellular component); voltage-gated potassium channel activity, delayed rectifier potassium channel activity, and ligand-gated sodium channel activity (molecular function); negative regulation of endothelial cell differentiation, positive regulation by host of viral transcription and negative regulation of vascular endothelial growth factor signaling pathway (biochemical process).
(B) shows the GO enrichments of DEPs of Avian broilers and Tibetan chickens after ALV-J infection. DEPs were enriched in low-density lipoprotein particle, proteinaceous extracellular matrix, and extracellular vesicular exosome (cellular component); DNA replication origin binding, adenylate cyclase activity, and guanylate cyclase activity (molecular function); response to lipopolysaccharide, defense response to bacterium, defense response to fungus, positive regulation by host of viral transcription and negative regulation by host of viral transcription (biochemical process).
(C) shows the GO enrichments of DEPs of Pengxian yellow chickens and Tibetan chickens after ALV-J infection. DEPs were enriched in MHC class I protein complex, MHC class II protein complex, and AP-1adaptor complex (cellular component); DNA replication origin binding, stem cell factor receptor activity, and ATPase binding (molecular function); immunoglobulin mediated immune response, immune system process, innate immune response, interleukin-12-mediated signaling pathway and defense response to bacterium (biochemical process).
KEGG pathway enrichment was conducted for DEPs in each comparison group. DEPs in Avian broilers and Pengxian yellow chickens after ALV-J infection were involved in viral myocarditis, transcriptional misregulation in cancer, primary immunodeficiency, natural killer cell-mediated cytotoxicity, and intestinal immune network for IgA production such as ZYX, SNRNP40, and BLB () ( (A)). DEPs in Avian broilers and Tibetan chickens after ALV-J infection were involved in viral myocarditis, transcriptional misregulation in cancer, intestinal immune network for IgA production, B arterials invasion of epithelial cells and autoimmune thyroid disease such as ANKRD27, HSPG, RELA, and ZNF148 () ( (B)). DEPs in Pengxian yellow chickens and Tibetan chickens after ALV-J infection were involved in viral myocarditis, Th1 and Th2 cell differentiation, MAPK signaling pathway, inflammatory bowel disease, HTLV-I infection, graft-versus-host disease, autoimmune thyroid disease, and antigen processing and presentation such as BLBII, BF2, CREBBP, and NCOR1 () ( (C)).
The DEPs detected by MRM
MRM technology was used to scan the partial DEPs of pairwise analysis results. Beta-galactosidase was used as the control protein and the data was naturalized with the intention of reducing the experimental error of MRM nonstandard quantification [Citation19–Citation21]. The experiment verified the difference in proteomics according to the scanning results, and the MRM results are shown in . As shown by the results, the MRM verification results were basically consistent with the results from the DIA results.
Table 6. Comparison of differential proteins from MRM and DIA.
Discussion
Genetic selection is considered a feasible and reliable method for improving immunity in chickens. China’s local chicken genetic resources are rich and provide a lot of material for breeding disease resistance. In order to select chicken breeds with higher disease resistance, it is necessary to first understand the relevant molecular mechanisms, so as to explore the selection of markers related to disease resistance.
The Pengxian yellow chicken has a round and medium-sized body; the Tibetan chicken has a light, small, long, low, symmetrical, and compact body; and the Avian broiler has full, heavy, wide, deep body. ALV-J is an RNA retrovirus, which has a complex pathogenic mechanism and high variability. The effective methods to improve the resistance of breeds to the virus is the genetic breeding. According to the disease statistics, the Tibetan chicken has the strongest resistance to ALV-J, followed by the Pengxian yellow chicken and then the Avian broiler, which is the most sensitive, which showed that local chickens have great potential as candidates for disease resistance breeding.
Protein function is a dynamic biological process. Proteomics is the macroscopic detection of changes in protein expression through omics technology, which is conducive to the in-depth study of biological processes. Proteomics analysis is a powerful and relatively new technique for studying biomarkers of protein response to viral infection and has been used in chickens [Citation22]. Chickens with different genetic backgrounds have different levels of proteins during development. In the current study, the phenotype of the two local breeds (Pengxian yellow chicken and Tibetan chicken) were confirmed to be different from the Avian broiler after ALV-J infection, which might be related to the different development of immune organs in the three breeds. DIA technology was used to analyze proteomics in the spleens of 15-day-old healthy hens from three different breeds and the specific proteins of each breed were detected, such as PCNT, DBI, CATHL1, MHCI, VCAM1, and IGF2BP1 of Tibetan chicken, PCNT, RBM14, HNMT, TFEB, CD247, and DDB2 of Avian broilers, ZNF622, TAF10, TPN, SNX13, TIE1, ASPR, and HINT2 of Pengxian yellow chickens. These DEPs were involved in tissue components (lysosomes, cell receptor complexes, and transcription factor complexes), metabolism (fatty acid transport, arachidonic acid secretion, and nucleotide biosynthesis), signal transmission (acetylation and groups of protein tyrosine kinase activity), and immune response (Notch signal pathway, lipopolysaccharide reaction, and defense). The biological functions enriched with different proteins led to the differences in spleen development among the three breeds which specific effect on virus resistance still needs further study of virus infection.
AL molecular markers are difficult to study due to the complex mechanisms involved in ALV-J infection and the complicated immune mechanisms in chickens. Only a few studies have used proteomics to study molecular markers in AL [Citation12–Citation14]. Chickens are susceptible to ALV-J and the spleen plays an important role in anti-infection and immune response function in viral infection. In this study, the index of spleens of Tibetan chickens infected with ALV-J was no significantly different from control groups, while the index of spleens of Pengxian yellow chicken and Avian broilers infected with ALV-J was significantly higher than control groups of each breed. Proteomics analysis of spleens was conducted in each breed infected with ALV-J groups and control groups. Using proteomics techniques, there were no common DEPs in three breeds of infected with ALV-J groups and control groups, but MKI67, RAB44, ELP4, HINT2, ASCC3, MRPL50, and XRN1 were both detected in Pengxian yellow chickens and Avian broilers infected with ALV-J groups and control groups, and BLB, TPN, PRMT7, NSUN5, DDX20, TRAF3, NFATC1, and NFIX were detected in Tibetan chicken infected with ALV-J groups and control groups. These DEPs were enriched in inflammatory bowel disease, antigen processing and presentation, Th17 cell differentiation, toll-like receptor signaling pathway, TNF signaling pathway, and other signaling pathways related to the immune response. The differences in proteins among the three breeds showed that the infection mechanism of ALV-J is complex and is associated with different genetic backgrounds. These DEPs may be potential markers of spleen immune response after ALV-J infection in chickens.
The application of proteomics technology in ALV-J infection provides a new way to explore the mechanism of ALV-J infection and the immune capacity of the body at a macro level. After taking into account normal genetic background differences among the three breeds, DEPs were analyzed between three breeds after ALV-J infection. The proteins RFX1, VCAM1, PRMT7, TCF, GNMT, CATHL1, MHCI, TRMT11, and CD48 of Tibetan chickens were differently expressed with Pengxian yellow chickens and Avian broilers. The proteins TAF10, KDM5A, SFN, OSBPL7, CNOT11, and PNPLA6 of Pengxian yellow chicken were differently expressed with Tibetan chicken and Avian broilers. The proteins VH1, IRF4, MHCII, LBP, CD247, PIAS1, ERG, CREB, YTHDF3, YTHDF2, BCR, and STAT1 of Avian broilers were differently expressed with Tibetan chicken and Pengxian yellow chickens. These DEPs are involved in cancer transcriptional dysregulation, IgA production of intestinal immune network, TNF signaling pathway, IL-17 signaling pathway, toll-like receptor signaling pathway.
Vascular cell adhesion molecule 1 (VCAM1, also known as CD106) belongs to the immunoglobulin (Ig) superfamily of cell surface proteins [Citation23,Citation24]. VCAM1 is highly expressed in acute myeloid leukemia (AML) cells [Citation25]. AML is one of the diseases of avian leukemia caused by ALV-J [Citation2]. The up-regulated expression of VCAM1 in Tibetan chickens infected with ALV-J revealed that VCAM1 plays an important role in resistance to ALV-J infection in chickens; thus, VCAM1 could be a potential immune marker. Transcription factor X1 (RFX1) is a widely expressed dual active transcription factor that can activate and inhibit target genes. RFX1 is important in regulating the epigenetic state of T cells [Citation26]. Deficiency of RFX1 promotes CD4 + T cells to differentiate into Th17 cells, which can be reversed by forcing the expression of RFX1 [Citation27]. ALV-J induced immunosuppression is related to T cell differentiation [Citation2]. The high expression of RFX1 in Tibetan chicken indicates that RFX1 may be related to the immunosuppression caused by ALV-J. RFX1 can play an anti-cancer role by down-regulating the original oncogene c-MYC [Citation28]. The pathogenic mechanism caused by the insertion of ALV-J nucleic acid into host DNA is related to c-MYC [Citation29]. Avian leukemia caused by ALV-J is a neoplastic disease. According to the function of RFX1 in cancer tumors, it could be that RFX1 may have an anti-virus function in ALV-J infection. The high expression of RFX1 in Tibetan chickens infected with ALV-J indicates that RFX1 plays a role in inhibiting the virus in ALV-J infection, which is consistent with the results found in other studies of RFX1. RFX1 could be used as a candidate marker molecule for poultry resistance to ALV-J infection. T cytokine 3 (TCF3, also known as E2A) is closely related to human acute lymphoid leukemia [Citation30]. TCF3 protein is highly expressed in Tibetan chicken, suggesting that TCF3 may play the same role as RFX1 in suppressing the immune function of the virus. A host defense peptide (CATHL1) is an important component of innate immunity and can activate innate and adaptive immunity [Citation31]. ALV-J induces immune response of the body [Citation32–Citation34]. The high expression of CATHL1 in Tibetan chicken indicates that CATHL1 plays an important role in anti-virus. Previous studies have shown the great potential of CATHL1 as an antiviral infectant [Citation35], which is consistent with the results of this study. Major histocompatibility class I molecules (MHCI) are essential for host–pathogen interactions and deliver both their own and external antigenic peptides to T lymphocytes [Citation36]. Studies on MHCI in poultry mainly focus on the relationship between its haplotype and resistance [Citation37–Citation39]. In this study, the expression of MHCI protein in Tibetan chickens was lower than in Avian broilers and Pengxian yellow chickens, indicating that MHCI protein may not play a major role in anti-virus. B cell antigen receptor (BCR) regulates B cell development by mediating the selection of functional and self-tolerant of B cells, thus ensuring immune protection and avoiding autoimmunity [Citation40,Citation41]. In chronic lymphocytic leukemia, BCR signal is in an abnormal state of activation [Citation42]. In the current study, Avian broilers were sensitive to ALV-J and BCR was up-regulated, indicating that BCR could be used as a potential molecular marker of ALV-J infection. MHCII is a class II major histocompatibility complex involved in antigen presentation, restricted recognition between immune cells, T cell differentiation, and genetic control of the immune response [Citation43–Citation45]. The expression of MHCII protein was up-regulated in Avian broilers, down-regulated in Pengxian yellow chickens, and no significant difference in Tibetan chickens. This may be related to the different genetic background of Avian broilers, Pengxian yellow chickens, and Tibetan chickens. MHCII protein can be used as a potential marker molecule of ALV-J infection, but it is not suitable to be used as an evaluation factor of interbreed immunity. Lipopolysaccharide binding protein (LBP) is an acute protein synthesized in the liver. LBP and LPS complexes bind to CD14 expressed by macrophages and neutrophils and mediate signal transduction, including activation of NFκB via TLR4, which activates innate and adaptive inflammatory responses [Citation46,Citation47]. The interaction between LBP and CD14/TLR4 is directly involved in the activation of LPS-mediated function and the expression of innate immune genes in chicken [Citation48]. The expression of LBP protein in Avian broilers was up-regulated; therefore, LBP protein could be used as one of the potential immune marker factors after ALV-J infection. Transcription regulator cAMP response element-binding protein (CREB) is a factor involved in the regulation of various cellular processes. IL-10 is the most important anti-inflammatory cytokine and CREB stimulates the transcription of IL-10 [Citation49]. The expression of CREB protein in Avian broilers was down-regulated compared with the two other breeds, indicating that CREB protein could be used as a potential immune marker molecule after ALV-J infection.
Conclusion
In this study, DIA technology was used to detect the DEPs of three breeds of chicken according to different comparison to investigate the potential markers. Special DEPs for spleen development of each breed were detected, such as PCNT, DDB2, and ZNF62. These DEPs were involved in intestinal immune network used in the production of IgA signaling pathways and related to immune response which can be used as potential markers for spleen development in different breeds. The DEPs such as RAB44 and TPN involved in viral myocarditis, transcriptional misregulation in cancer, and tuberculosis can be used as potential markers of spleen immune response after ALV-J infection in chickens. Pair-wise analysis was performed for the three breeds after the infection of ALV-J. The proteins such as RFX1, TAF10 and VH1 were differently expressed between three breeds. These DEPs involved in antigen processing and expression, acute myelogenous leukemia, and viral carcinogenesis can be used as potential immune markers after ALV-J infection of different genetic backgrounds.
Author contributions statements
Conceived and designed the experiments: FY, YW, HL, and QZ; Performed the experiments: FY, QJH, ZSW, EYM, SLZ, and HLY; Analyzed the data: FY; Contributed reagents/materials/analysis tools: HYX, DYL, and XLZ; Wrote the paper: FY; Revised the manuscript: YW, HL, and QZ. All authors read and approved the final manuscript. We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Acknowledgments
The Sequencing work was financially supported by the China Agriculture Research System (CARS-41), National Natural Science Foundation of China (No: 31601936), the Engineering Centre of Chicken Breeding in Guangdong Province (2017-1649), and Guangdong Provincial Key Laboratory of Animal Molecular Design and Precise Breeding (2019B030301010).
Disclosure statement
The authors declare no conflicts of interest.
Additional information
Funding
References
- Kim Y, Brown TP, Pantin-Jackwood MJ. The effects of cyclophosphamide treatment on the pathogenesis of subgroup J avian leukosis virus (ALV-J) infection in broiler chickens with Marek’s disease virus exposure. J Vet Sci. 2004;5:49–58.
- Payne L, Nair V. The long view: 40 years of avian leukosis research. Avian Pathol. 2012;41:11–19.
- Guo H, Cui Z, Sun S, et al. Influence of REV and ALV-J infection on cellular immunity responses in commercial broiler chickens. Chin J Vet Sci. 2007;27:632–639.
- Pan W, Gao Y, Qin L, et al. Genetic diversity and phylogenetic analysis of glycoprotein GP85 of ALV-J isolates from Mainland China between 1999 and 2010: coexistence of two extremely different subgroups in layers. Vet Microbiol. 2012;156:205–212.
- Dong X, Zhao P, Li W, et al. Diagnosis and sequence analysis of avian leukosis virus subgroup J isolated from Chinese Partridge Shank chickens. Poult Sci. 2015;94:668–672.
- Zhang L, Cai D, Zhao X, et al. Liposomes containing recombinant gp85 protein vaccine against ALV-J in chickens. Vaccine. 2014;32:2452–2456.
- Chen B, Pan W, Zhang L, et al. NHE1 gene associated with avian leukosis virus subgroup J infection in chicken. Mol Biol Rep. 2014;41:6519–6524.
- Li L, Feng W, Cheng Z, et al. TRIM62-mediated restriction of avian leukosis virus subgroup J replication is dependent on the SPRY domain. Poult Sci. 2019;98:6019–6025.
- Dai M, Feng M, Liao M, et al. Inhibition of ERK/MAPK suppresses avian leukosis virus subgroup A and B replication. Microb Pathog. 2017;102:29–35.
- Feng M, Zhang N, Xie T, et al. Chichen type III interferon produced by silkworm bioreactor induces ISG expression and restricts ALV-J infection in vitro. Appl Microbiol Biotechnol. 2019;103:1–11.
- Collins BC, Hunter CL, Liu Y, et al. Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat Commun. 2017;8:291.
- Fan Z, Hu X, Zhang Y, et al. Proteomics of DF-1 cells infected with avian leukosis virus subgroup J. Virus Res. 2012;167:314–321.
- Li X, Wang Q, Gao Y, et al. Quantitative iTRAQ LC-MS/MS proteomics reveals the proteome profiles of DF-1 cells after infection with subgroup J Avian leukosis virus. Biomed Res Int. 2015;2015:395307.
- Ye F, He Q, Wang Y, et al. Data-independent acquisition of the proteomics of spleens from chickens infected by avian leukosis virus. 3 Biotech. 2019;9:332.
- F M, X L, J F, Y G, L Z, G X, et al. Avian leukosis virusGenomic diversity of the subgroup J gene in different organs of an infected chicken. J Vet Sci. 2016;17:497–503.
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372.
- Bruderer R, Bernhardt O, Gandhi T, et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol Cell Proteomics. 2015;14:1400–1410.
- Choi M, Chang C, Clough T, et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 2014;30:2524–2526.
- Tang H-Y, Beer LA, Barnhart KT, et al. Rapid verification of candidate serological biomarkers using gel-based, label-free multiple reaction monitoring. J Proteome Res. 2011;10:4005–4017.
- Martins-de-Souza D, Alsaif M, Ernst A, et al. The application of selective reaction monitoring confirms dysregulation of glycolysis in a preclinical model of schizophrenia. BMC Res Notes. 2012;5:146.
- Martínez-Aguilar J, Molloy MP. Label-free selected reaction monitoring enables multiplexed quantitation of S100 protein isoforms in cancer cells. J Proteome Res. 2013;12:3679–3688.
- Korte J, Fröhlich T, Kohn M, et al. 2D DIGE analysis of the bursa of Fabricius reveals characteristic proteome profiles for different stages of chicken B-cell development. Proteomics. 2013;13:119–133.
- Osborn L, Hession C, Tizard R, et al. Direct expression cloning of vascular cell adhesion molecule 1, a cytokine-induced endothelial protein that binds to lymphocytes. Cell. 1989;59:1203–1211.
- Pepinsky B, Hession C, Chen -L-L, et al. Structure/function studies on vascular cell adhesion molecule-1. J Biol Chem. 1992;267:17820–17826.
- Pinho S, Qiaozhi W, Maryanovich M, et al. Vcam1 Is a” Don’t-Eat-Me” Signal on Healthy Hematopoietic and Leukemic Stem Cells. Am Soc Hematol. 2016;128:565-565.
- Zhao M, Sun Y, Gao F, et al. Epigenetics and SLE: RFX1 downregulation causes CD11a and CD70 overexpression by altering epigenetic modifications in lupus CD4+ T cells. J Autoimmun. 2010;35:58–69.
- Zhao M, Tan Y, Peng Q, et al. IL-6/STAT3 pathway induced deficiency of RFX1 contributes to Th17-dependent autoimmune diseases via epigenetic regulation. Nat Commun. 2018;9:583.
- Chen L, Smith L, Johnson MR, et al. Activation of protein kinase C induces nuclear translocation of RFX1 and down-regulates c-myc via an intron 1 X box in undifferentiated leukemia HL-60 cells. J Biol Chem. 2000;275:32227–32233.
- Li Y, Liu X, Yang Z, et al. The MYC, TERT, and ZIC1 genes are common targets of viral integration and transcriptional deregulation in avian leukosis virus subgroup J-induced myeloid leukosis. J Virol. 2014;88:3182–3191.
- Lin A, Cheng FW, Chiang AK, et al. Excellent outcome of acute lymphoblastic leukaemia with TCF3-PBX1 rearrangement in Hong Kong. Pediatr Blood Cancer. 2018;65:e27346.
- Bommineni YR, Pham GH, Sunkara LT, et al. Immune regulatory activities of fowlicidin-1, a cathelicidin host defense peptide. Mol Immunol. 2014;59:55–63.
- Li J, Meng F, Li W, et al. Characterization of avian leukosis virus subgroup J isolated between 1999 and 2013 in China. Poult Sci. 2018;97:3532–3539.
- Venugopal K. Avian leukosis virus subgroup J: a rapidly evolving group of oncogenic retroviruses. Res Vet Sci. 1999;67:113–119.
- Feng M, Zhang X. Immunity to Avian Leukosis virus: where are we now and what should we do? Front Immunol. 2016;7:624.
- Xiao Y, Dai H, Bommineni YR, et al. Structure-activity relationships of fowlicidin-1, a cathelicidin antimicrobial peptide in chicken. Febs J. 2006;273:2581–2593.
- Van KL, Parekh VV, Postoak JL, et al. Role of autophagy in MHC class I-restricted antigen presentation. Mol Immunol. 2019;113:2–5.
- JK M, LD B, AR P, et al. Response of white leghorn chickens of various B haplotypes to infection at hatch with subgroup J avian leukosis virus. Avian Dis. 2005;49:214–219.
- Gao C, Han L, Han J, et al. Establishment of six homozygous MHC-B haplotype populations associated with susceptibility to Marek’s disease in Chinese specific pathogen-free BWEL chickens. Infect Genet Evol. 2015;29:15–25.
- Gaston JH, Life PF, van der Zee R, et al. Epitope specificity and MHC restriction of rheumatoid arthritis synovial T cell clones which recognize a mycobacterial 65 kDa heat shock protein. Int Immunol. 1991;3:965–972.
- Shlomchik MJ. Sites and stages of autoreactive B cell activation and regulation. Immunity. 2008;28:18–28.
- Pelanda R, Torres RM. Central B-cell tolerance: where selection begins. Cold Spring Harb Perspect Biol. 2012;4:a007146.
- Gururajan M, Jennings CD, Bondada S. Cutting edge: constitutive B cell receptor signaling is critical for basal growth of B lymphoma. J Immunol. 2006;176:5715–5719.
- Motta JV, Crittenden LB, Pollard WO. The inheritance of resistance to subgroup C leukosis-sarcoma viruses in New Hampshire chickens. Poult Sci. 1973;52:578–586.
- Garboczi DN, Ghosh P, Utz U, et al. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134.
- Hare BJ, Wyss DF, Osburne MS, et al. Structure, specificity and CDR mobility of a class II restricted single-chain T-cell receptor. Nat Struct Mol Biol. 1999;6:574.
- Gubern C, Lopez-Bermejo A, Biarnés J, et al. Natural antibiotics and insulin sensitivity: the role of bactericidal/permeability-increasing protein. Diabetes. 2006;55:216–224.
- Liu X, Lu L, Yao P, et al. Lipopolysaccharide binding protein, obesity status and incidence of metabolic syndrome: a prospective study among middle-aged and older Chinese. Diabetologia. 2014;57:1834–1841.
- Kogut M, He H, Kaiser P. Lipopolysaccharide binding protein/CD14/TLR4-dependent recognition of Salmonella LPS induces the functional activation of chicken heterophils and up-regulation of pro-inflammatory cytokine and chemokine gene expression in these cells. Anim Biotechnol. 2005;16:165–181.
- Ananieva O, Darragh J, Johansen C, et al. The kinases MSK1 and MSK2 act as negative regulators of Toll-like receptor signaling. Nat Immunol. 2008;9:1028.
