 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Salmonella is the only bacterium able to enter a host cell by the two known mechanisms: trigger and zipper. The trigger mechanism relies on the injection of bacterial effectors into the host cell through the Salmonella type III secretion system 1. In the zipper mechanism, mediated by the invasins Rck and PagN, the bacterium takes advantage of a cellular receptor for invasion. This study describes the transcriptomic reprogramming of the IEC-6 intestinal epithelial cell line to Salmonella Typhimurium strains that invaded cells by a trigger, a zipper, or both mechanisms. Using S. Typhimurium strains invalidated for one or other entry mechanism, we have shown that IEC-6 cells could support both entries. Comparison of the gene expression profiles of exposed cells showed that irrespective of the mechanism used for entry, the transcriptomic reprogramming of the cell was nearly the same. On the other hand, when gene expression was compared between cells unexposed or exposed to the bacterium, the transcriptomic reprogramming of exposed cells was significantly different. It is particularly interesting to note the modulation of expression of numerous target genes of the aryl hydrocarbon receptor showing that this transcription factor was activated by S. Typhimurium infection. Numerous genes associated with the extracellular matrix were also modified. This was confirmed at the protein level by western-blotting showing a dramatic modification in some extracellular matrix proteins. Analysis of a selected set of modulated genes showed that the expression of the majority of these genes was modulated during the intracellular life of S. Typhimurium.
Introduction
In salmonellosis, a critical step in pathogenesis is the ability of Salmonella to adhere to and penetrate host cells. The infection process can be divided into four phases: approach, attachment, entry, and intracellular life. To begin its infection cycle, Salmonella needs to reach the cell; the flagellum, a long helical structure, provides the bacterium with a driving force that allows its movement across the extracellular space [Citation1]. Thanks to several adhesins, Salmonella can interact with cellular receptors and with components of the extra cellular matrix (ECM). Numerous pathogens exploit the ECM to reach their target cell. For instance, to be limited to Salmonella, FimH is recognized by human granule membrane glycoprotein (GP2) expressed at the apical pole of M cells [Citation2] while in pigs, FimH can bind the calreticulin [Citation3]. The Lewis X blood group has been shown to be a receptor for PefA [Citation4]; MisL [Citation5], and SdhA [Citation6] binds the fibronectin, and SiiE uses the transmembrane mucin to promote its entry [Citation7]. Even if it has been clearly established that disruption of the ECM is essential to Salmonella entry [Citation8], few data describe the modification induced on ECM after entry.
In non-phagocytic cells, two conceptual frameworks describe the entry of bacteria [Citation9]: the trigger and the zipper entry modes. In Salmonella, two major differences characterize these mechanisms. While the trigger mechanism relies on the type III secretion system 1 (T3SS1) that injects effectors directly into host cell, the zipper mechanism depends on the interaction between a bacterial invasion factor and a cellular receptor, which transduces the internalization signal. In addition, while there is cytoskeleton remodeling leading to entry in both mechanisms, the perturbation of the membrane is less pronounced in the zipper mechanism. However, it has been shown that the T3SS1 can induce both large membrane ruffles and discrete ones [Citation10]. The Salmonella pathogenicity island 1 (SPI1) encodes the majority of the components necessary to build a functional T3SS1 that, from a functional point of view, looks like a syringe. The two invasins PagN [Citation11] and Rck [Citation12] are able to induce invasion through a zipper mechanism. The cellular receptor for PagN has not been clearly identified. However, it has been shown that PagN binds a heparinated proteoglycan to promote entry into the cell [Citation13]. The Rck receptor has been clearly identified as the epidermal growth factor receptor [Citation14]. However, the role of Rck and PagN in the pathogenicity of Salmonella remains somewhat controversial even though few articles have strongly suggested an in vivo role of the invasines PagN [Citation15] and Rck [Citation16].
A large number of studies have established that in vitro and in vivo infection induces a vast reprogramming of gene expression, particularly of those involved in the regulation of the inflammatory response [Citation17]. As the trigger and the zipper mechanisms are based on very different types of interactions with the host cell, the question arose as to how the cell responds to these two entry routes. We analyzed gene expression in the rat epithelial intestinal cell-line IEC-6, which was unexposed or exposed to: 1/S. Typhimurium (STM) expressing constitutively the three invasion factors T3SS1, Rck, and PagN, 2/a STM mutant invalidated for the T3SS1 and expressing constitutively the invasins Rck and PagN, and 3/the STM Rck, PagN double mutant with a functional T3SS1. These three strains can enter the IEC-6 cells through, respectively: 1/both a trigger and a zipper entry, 2/a zipper entry and 3/a trigger entry. For simplicity, hereafter the S. Typhimurium strains will be, respectively, denoted as STM- Z T, STM- Z, STM- T, and STM-3d, which expressed none of these entry factors.
Materials and methods
Bacterial strains and plasmids
To study in-vitro the influence of the zipper entry on cellular gene expression, from the wild type S. Typhimurium (STM ATCC14028) we derived the strain STM- Z invalidated for the T3SS1 by invA deletion and constitutively overexpressing Rck and PagN on a recombinant plasmid (pSUP202:rck-pagN) and strain STM- T invalidated for rck and pagN that expressed the T3SS1 under our growing conditions. In order to compare isogenic strains, we also derived the STM-Z T strain that is the wild type STM ATCC14028TM strain that expresses the T3SS1 genes under our growing conditions and constitutively overexpresses Rck and PagN on a recombinant plasmid (pSUP202:rck-pagN). As a negative control, we used the wild-type strain deleted of invA, rck and pagN named STM-3d [Citation18]. Characteristics of the strains used in this study are given in . Deletion of the invA, rck and pagN was performed according to the Datsenko and Wanner [Citation19] method as described in [Citation18]. To derive the STM- Z and STM- ZT strains, we constructed the plasmid pSUP202: rck - pagN. The plasmid pSUP202: rck, where rck is cloned into the cassette encoding tetracycline resistance [Citation20], was used as the recipient plasmid. The plasmid pSUP202: pagN, containing the pagN gene, cloned into the cassette encoding chloramphenicol resistance [Citation21] was used as the donor plasmid. These two plasmids were digested by the restricted enzymes EcoRI and NcoI. Restricted fragments were separated into agorose gel (0,7%) by electrophoresis. Fragments of interest were purified from the gel with a QIAquick® Gel Extraction Kit (Qiagen) according to the supplier’s recommendations. The purified products were ligated and transformed into E. coli MC1061, verified by sequencing and transformed into desired STM.
Table 1. Characteristics and designation of the strains used in this study.
Cell line
The intestinal epithelial cell-line IEC-6 (ATCC CRL-1592TM) was grown in DMEM (Dulbecco’s modified Eagle’s medium with glucose 4.5 g/L) supplemented with 5% (v/v) fetal calf serum and 0.1 Unit/mL bovine insulin (Sigma). Cells were maintained in a humidified incubator (90% relative humidity) at 37°C under 5% (v/v) CO2.
Invasion assays
At confluence, cells were exposed to a multiplicity of infection of 10 for the indicated time, followed by a measurement of adhesion and a gentamicin protection assay to evaluate the invasion level. These were performed as described before [Citation18]. In brief, the different stages of cell infection were analyzed with cells grown in 24-well tissue culture plates (Falcon) for 5 days to obtain subconfluent monolayers. The cell monolayers were incubated in a culture medium without antibiotics for 24 h, then infected for 1.5 h at 37◦C with 107 CFU in 300 μL of serum-free medium (multiplicity of infection = 10). For the adhesion assays, cell monolayers were gently washed six times with phosphate buffered saline and then disrupted with 1 mL of cold distilled water (4°C). Viable intra- and extracellular bacteria were determined on TSA (Tryptic Soy Agar-Difco). For invasion, after infection for 1.5 h at 37◦C, plates were washed with a cell culture medium and incubated in a medium containing 100 μg of gentamicin per mL. After 1.5 h at 37◦C, cells were washed and lysed with 1 mL cold distilled water. Viable intracellular bacteria were assessed by serial dilutions on TSA. Infection and a gentamicin protection assay were also performed in the presence of the drugs. For these assays, cells were pre-treated with chlorpromazine (Sigma) at 10 µg/mL and amiloride (Sigma) at 1 mM for 30 min in a culture medium. Viability of the bacteria was checked in the presence of the drugs. Six biological replicates were performed for unexposed IEC-6 and for cells treated with the drugs. For the microarray experiments, six biological replicates were performed for unexposed cells, for IEC-6 exposed to STM-Z or STM-ZT and five replicates for the infection with STM-T.
Extracellular matrix protein expression profile by western blotting
Protein expression profiling was carried out using extraction of Non-Infected cells (NI) and cells infected by STM-ZT, STM-Z, STM-T or STM-3d strains. Bacteria were deposited on IEC-6 with a multiplicity of infection of 10, for 1.5 h, followed by gentamicin (100 µg/ml) for 1.5 h. Cells were then resuspended in 100 µL of Laemmli buffer and denaturated 10 min at 100°C. Whole-cell protein samples 25 µL were run on SDS- PAGE (100 V) in a 4–15% Miniprotean TGX Precast Protein gels (Bio-Rad) in a Tris-glycine running buffer (25 mM Tris base, 192 mM glycine, 0.1% [wt/vol] SDS [pH 8.31]) and transferred onto a nitrocellulose membrane with Trans-blot Turbo transfer System (Bio-Rad) in Tris-glycine buffer system 15 min at 25 V and 2.5 mA. The blots were probed with the first antibody (1:1000), rabbit anti-laminin (Invitrogen), rabbit anti-fibronectin (Invitrogen) or mouse anti-tubulin overnight at 4°C and detected by chemiluminescence using goat anti-rabbit secondary antibody (1:25000) (Pierce) or goat anti-mouse secondary antibody (1:5000) (Dako) conjugated to HRP for 1 h. Proteins were revealed using the SuperSignal West Dura Extended Duration Substrate (Thermo Scientific).
RNA labelling and microarray processing
Transcriptional profiling was carried out using all of the 23 samples described above in the invasion assay paragraph. All steps were performed by the @BRIDGe core facility (INRAE Jouy-en-Josas, France, http://abridge.inra.fr). Cyanine-3 (Cy3) labeled cRNA was prepared using 100 ng of total RNA using the One-Color Low Input Quick Amp Labeling kit (Agilent Technologies, Santa Clara, CA, USA) following the recommended protocol. Specific activities and cRNA yields were determined using the NanoDrop ND-1000 (Thermo Fisher Scientific, Waltham, MA, USA).
For each sample, 600 ng of Cy3-labeled cRNA (specific activity >6.0 pmol Cy3/µg of cRNA) were fragmented at 60°C for 30 min in a reaction volume of 25 µl containing 25× Agilent Fragmentation Buffer and 10× Agilent Blocking Agent, following the manufacturer’s instructions. Subsequently, 25 µl of 2× Agilent Hybridization Buffer were added to the fragmentation mixture and hybridized to three SurePrint G3 Rat Gene Expression v2 8 × 60K Microarrays (Agilent Technologies, AMADID: 074036) for 17 h at 65°C in a rotating Agilent hybridization oven (Agilent Technologies). After hybridization, microarrays were washed for 1 min at room temperature with the GE Wash Buffer 1 (Agilent Technologies), for 1 min at 37°C using the GE Wash Buffer 2 (Agilent Technologies) and then dried immediately.
Immediately after washing, the slides were scanned using a G2565CA Scanner System (Agilent Technologies), which used a scan protocol with a resolution of 3 µm and a dynamic range of 20 bit. The resulting tiff images were analyzed with the Feature Extraction Software v10.7.3.1 (Agilent Technologies), using the GE1_107_Sep09 protocol. The microarray data were submitted to the GEO database and received the accession number GSE151881.
Statistical analyses
Microarrays
The expression of each probe was log2-transformed and normalized by median centering for each array after a filtering step. 58,777 probes were kept to differential analysis. A linear model was performed for each probe with R package limma version 3.30.7 [Citation22] to identify differentially expressed (DE) genes between the four biological conditions (IEC-6 cells non-exposed, IEC-6 cells exposed to STM- Z, STM- T, or STM- Z T). The model estimated the differences in expression as fold-change (FC) between two conditions by sharing information between samples. Significance of expression changes were determined using moderated t-statistics. The p-values were adjusted for multiple testing by the Benjamini-Hochberg method [Citation23] to control the False Discovery Rate (FDR). Genes with an adjusted p-value below 0.05 were considered DE.
Cellular tests
A Mann Whitney test was performed to compare invasion levels between the different STM strains, using GraphPad Prism version 6.07 for Windows, GraphPad Software, La Jolla California USA, http://www.graphpad.com. Significance is * p < 0.05, ** p < 0.01, and *** p < 0.001.
RNA extraction, reverse transcription, and purification of cDNA for qPCR analysis
RNA extraction was performed using the NucleoSpin RNA kit (Macherey Nagel, France). Two hundred ng of total RNA, 0.13 µg of Oligo-d(T20) (Eurogentec) and 0.13 µg of random primer (Promega) were denatured at 75°C for 5 min then incubated on ice for 5 min. The reverse transcription reaction was carried out in a final volume of 50 µl containing: dNTP 1 mM, 30 U/µg RNA of AMV reverse transcriptase (Promega) and 1 U/µl of RNasin (Promega) at 42°C for 60 min. After reverse transcription, cDNAs were purified using the QIAquick PCR purification kit (Qiagen); their concentration was measured using the spectrophotometer ND-1000 Nanodrop and concentrations were adjusted to 50 ng/µl.
Primers and high throughput qPCR
Primer pairs used in the study were designed and produced at Fluidigm; their sequences are indicated in supplementary Table S3. Primer stock solutions at 100 µM were kept at -20°C. Prior to specific target amplification (STA), primers were pooled to the final concentration of 500 nM each. BioMark™, a high throughput PCR platform from Fluidigm, was used to perform the qPCR according to manufacturer’s recommendations. Prior to the quantification PCR, the DNA was amplified: sixty ng of purified cDNA was amplified using the pooled primers to a final concentration of 50 nM and a thermal cycling consisting of 5 min at 95°C followed by 18 cycles of 15 s at 95°C and 4 min at 60°C and a final holding step at 40°C. Thereafter, the protocol recommended by the supplier was followed (Fluidigm quick references PN 100–5875 B1, and PN 100–9791 B1). For the quantification of PCR, the following cycling conditions were applied, one Taq activation cycle (95°C, 1 min) and 35 cycles (96°C for 5 sec, 60°C for 20 sec). The software Fluidigm real-time PCR analysis was used to determine the Cq values of each sample/primer pair couple. The Cq was determined by the Auto detector method with a quality control of 0.65 and a linear baseline correction. The fold change (FC) in gene expression was calculated by the method [Citation24]. An unpaired t-test was used to determine the statistically significant differentially expressed (DE) genes. The fold changes (FC) greater than 2 and less than -2, with a p value <0.05 were considered for further analyses.
Bioinformatic analysis
Enriched biological processes were determined and then clustered hierarchically according to Wang’s semantic similarity distance using the Bioconductor R package ViSEAGO [Citation25] and Ingenuity Pathway Analysis (IPA) was used to find the potential transcriptional targets of AhR.
Results
S. Typhimurium can invade IEC-6 cells using both the trigger and the zipper entry processes
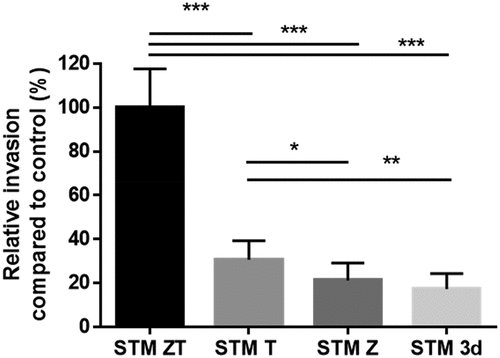
To analyze the cellular response following a trigger or a zipper entry, it was necessary to identify a cell line that was permissive to both entry mechanisms, because in fact S. Typhimurium strains often enter cell lines almost exclusively by only one entry mechanism to the exclusion of the other [Citation18]. Moreover, it was necessary for Salmonella strain to express all known invasion factors in vitro. As Rck and PagN are weakly expressed in standard culture conditions [Citation26], we developed three different modified strains of the wild type STM 14,028: the STM-ZT overexpressing Rck and PagN in vitro and expressing the T3SS1 genes (trigger entry and zipper entry), the STM-Z overexpressing Rck and PagN in vitro and invalidated for the T3SS1 (zipper entry) and the STM-T expressing the T3SS–1 genes and invalidated for Rck and PagN (trigger entry). In the STM-ZT strain, Rck and PagN are cloned on the same plasmid and expressed under the control of two independent promotors. We selected the IEC-6 cell line because the STM-T and STM-Z strains were still able to enter IEC-6 cells at similar levels but, compared to the STM-ZT, with a reduced efficiency of 30% and 20% respectively (). Interestingly, the STM-3d strain, which is unable to express any of these invasion factors has a much lower entry capacity than any of the other strains but retains a small entry capacity, as has been shown previously with other cell lines [Citation18]. This original cellular model allowed us to compare the cellular response to both entry processes in the same cell line.
Figure 1. Invasion of IEC-6 cells by S. Typhimurium strains entering cells by different entry pathways.
Cells exposed to the three Salmonella strains have very similar gene expression profiles
To determine the influence of the entry mechanisms on the modulation of cellular gene expression, we performed whole-genomic expression profiles of IEC-6 exposed to STM strains invalidated or not for factors responsible for the zipper or trigger entry. Gene expression analyses were performed after 1.5 h of interaction between IEC-6 and STM strains and 1.5 h of gentamicin to kill extracellular bacteria. Gene expression was analyzed using microarrays. Differentially expressed (DE) genes, characterized by the fold change (FC), the ratio between exposed cells and non-exposed cells, indicated that very few significant differences were observed between the STM strains, and no DE genes were found between cells exposed to STM-T and STM-ZT strains (). When comparing cells exposed to STM- T and STM- Z strains, the expression of Mlr1, a transcription factor specific to RNA Pol II, Nxf3 involved in mRNA transport, Smpdl3a a phosphodiesterase of nucleoside triphosphate and Nxph4, playing a role in neuronal biology and expressed in cells from colorectal carcinoma [Citation27] was less expressed in STM- T exposed cells. In these latter cells, the expression of Dclk1 and Tgfbr1 was lower. DCLK1 is a marker of the intestinal tuft cells involved in intestinal repair [Citation28], whose expression is correlated with that of TGFBR1 in colon cancer [Citation29]. Finally, the expression of Rhoh, encoding a Rho GTPase recruited at the Salmonella invasion site by SopB, a T3SS1 encoded factor, involved in the activation of Akt [Citation30], was also lower in STM-T exposed cells compared to STM- Z. Smpdl3a, Nxf3, and Rhoh were expressed more in cells exposed to STM- Z than with STM- Z T strain. This low number of DE genes could be related to small differences induced by the entry pathway. Another hypothesis is that the signal is the same and low number of DE genes is related to the slightly different number of intracellular bacteria after the trigger and the zipper entry processes. However, this latter hypothesis is unlikely, as we did not observe any DE genes in the other experiments described below.
Table 2. Paired comparisons of gene expression modification induced in IEC-6 cells exposed to the S. Typhimurium strains STM- Z T, STM- Z, or STM- T.
Cells exposed to the three Salmonella strains have different gene expression profiles compared to unexposed ones
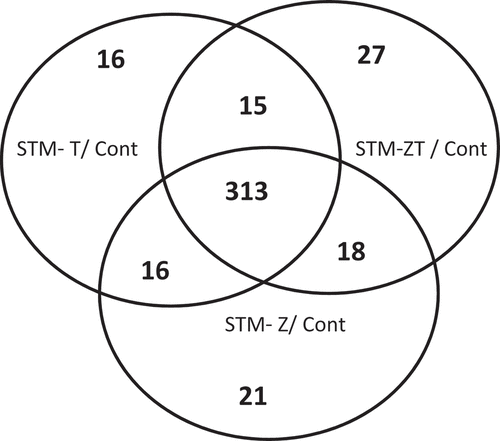
In contrast to the previous observation, and as expected, when IEC-6 cells exposed to one of the three strains were compared to the unexposed cells, numerous DE genes were detected. More than 300 DE genes, with an FC> |2|, were common to the three strains, while around 20 were specific to one strain (). Consistent with observations made when comparing Salmonella strains with each other, the lists of DE genes between control and IEC-6 cells exposed to STM-Z T, STM-Z, or STM-T were very similar, with only occasional minor differences in the magnitude of the FC (data not shown). We therefore decided to perform the subsequent downstream steps of analysis using the DE gene list obtained from the STM-ZT infection. The list of the 372 significantly DE genes with an absolute FC value greater than 2, between cells exposed to STM-ZT and unexposed cells is given in Table S1.
Figure 2. Specific and shared differentially expressed genes between IEC-6 cells exposed to STM-ZT, STM-Z or STM-T strains compared to non-exposed cells (cont).
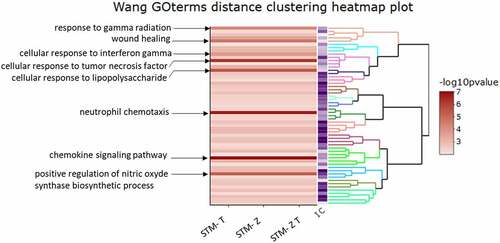
We then used ViSEAGO [Citation25] to determine the biological processes enriched following infection by the mutant strains. We focused on the common functions enriched; for this reason, we performed the ViSEAGO analysis for genes with a FC > |5|. Briefly, enrichment was carried out by aggregating gene ontology (GO) terms using a semantic similarity distance. The resulting functional enrichment analysis is presented in . When compared to unexposed cells, those exposed to STM-ZT, STM-Z or STM-T strains shared identical patterns of enrichment. Immune-related terms such as cellular response to tumor necrosis factor, to interferon gamma and to lipopolysaccharide, as well as neutrophil chemotaxis, chemokine signaling pathway and positive regulation of nitric oxide were among the most enriched. It is noteworthy that wound healing was found to be enriched in cells exposed to the three STM strains. Wound healing consists in a series of interrelated molecular events that work together to restore tissue integrity and cellular function.
Figure 3. Functional enrichment analysis of biological processes.
Expression of numerous ECM-associated, and of aryl hydrocarbon receptor (Ahr) target genes was modulated during salmonella infection
In a bird’s eye view of the list of DE genes, we observed that several of them were linked to the ECM or Ahr. The Extracellular Matrix Interaction Database (MatrixDB: http://matrixdb.univ-lyon1.fr/) lists the molecules that are components or regulators of the ECM mole. Of the 372 DE genes from our study, we used this database to find those that were listed in MatrixDB. We found that 126 of our DE genes were in the database (Table S2); with 113 of them upregulated and 13 downregulated.
A literature search allowed us to assign function to some of the 126 DE genes involved in ECM regulation. only shows the differentially expressed (DE) genes with a p value <0.05, and a fold change (FC) > |2|. These genes are key regulatory genes of the urokinase pathway that control fibrinolysis, like SerpinB2 and Serpine1, which were strongly activated. Genes involved in the ECM formation, synthesis of ECM components or in fibrosis were also upregulated in exposed cells. Conversely, few genes were associated with degradation of ECM ().
Table 3. Differentially expressed genes in the microarray experiment associated with stimulation or inhibition of the ECM.
Analysis of upstream regulators with Ingenuity Pathway Analysis (IPA) indicated that AhR was a significant transcriptional regulator of 101 DE genes from our data (Figure S1). It should be noted that the expression of primary targets of Ahr like Cyp1a1, Cy1b1, and Aldh3a1, was highly increased, while that of Nqo1, Arnt and Tiparp was modestly but significantly increased ().
Table 4. Differentially expressed genes included in the IPA upstream regulator analysis that may be activated by the AHR pathway.
Expression of ECM-associated DE genes by quantitative RT PCR
To focus on the modulation of ECM-associated genes, by means of a Biomark Fluidigm ™ platform, we studied the expression of 40 genes (Table S3) that were found DE in the microarray analysis. Samples of cells exposed or not to STM-ZT, were the same as those used for the microarray experiments. Both techniques gave consistent results for 33 genes (82.5%), showing accordance for both the direction of variation and the order of magnitude of the FC. However, for seven genes (17.5%), the FCs were < I2I or not-significant in the Biomark experiment ().
Table 5. Comparison of gene expression determined by microarray and qRT PC.
Modification of ECM gene expression in IEC-6 cells required Salmonella entry
Next, to determine whether the modification of ECM gene expression occurred when Salmonella had an extra and/or intracellular location, we inhibited the entry of Salmonella and checked the ECM gene expression. As the STM-3d exhibited a small but significant entry, we used a pharmaceutical means to completely prevent Salmonella entry. For this purpose, we incubated IEC-6 cells before and during infection with a cocktail of amiloride and chlorpromazine as previously described [Citation18]. Under these conditions, the entry of STM-ZT was reduced to about 3%, compared to the untreated cells for which the entry was arbitrarily set at 100% (). Similarly, we observed that the cocktail of amiloride and chlorpromazine also significantly inhibited the entry of the STM-3d strain, reinforcing our choice to use drugs to inhibit Salmonella entry. Gene expression measurements in cells with or without intracellular bacteria were performed after 1.5 h of interaction between IEC-6 and Salmonella strains. Gene expression was measured by quantitative RT PCR, using the Biomark Fluidigm™ platform for the same set of 40 genes selected for studying the expression of ECM associated genes. When compared gene expression of cells infected by STM-ZT to un-infected cells expression of 27 genes was significantly increased and three whose expression was decreased (). In contrast, when IEC6 cells, pre-treated with the amiloride/chlorpromazine cocktail, were exposed to the STM-ZT the expression of only 6 genes was modulated after bacteria-cell contact; 2 genes were not found with STM-ZT exposed cells. Four genes (Angptl4, Icam1, Nrg1 and SerpinB2) were common to the two conditions but the magnitude of the FC was lower when invasion was inhibited compared to that observed for the STM-ZT exposed cells. These results showed that the majority of the ECM-related genes were induced when STM was intracellular. These results suggest that under the experimental conditions used here, extracellular bacteria do not profoundly modify the expression of ECM-related genes and that ECM modification does not appear to be necessary for entry. In contrast, the presence of intracellular bacteria is required to profoundly modify the expression of ECM-related genes.
Figure 4. Invasion of the STM-ZT and STM-3d strains impaired for entry by pharmaceutical means.
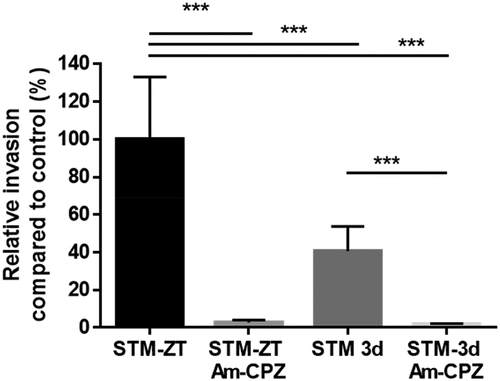
Table 6. Expression profile of selected ECM-associated genes in IEC-6 cells treated or not with chlorpromazine and amiloride before and during infection with the STM-Z T strain.
IEC-6 cell infection by S. Typhimurium modified glycoprotein composition of ECM
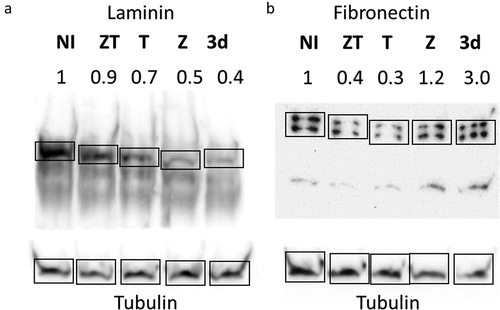
The extracellular matrix is composed of numerous proteins and glycoproteins. It was therefore not possible to carry out a complete screening of the putative modifications induced after cell infection. To confirm that STM-ZT can modify ECM composition, we therefore tested the effect of infection on two major ECM glycoproteins: the fibronectin mainly presents on the apical side and the laminin a major component of the basolateral side. The western blotting presented in clearly shows the dramatic change in ECM composition after cell infection, at least for fibronectin and laminin. However, the fact that the different Salmonella strains did not induce the same modifications on these two glycoproteins and that differences between laminin and fibronectin can be observed, even with the STM-3d strain, will lead us to perform a detailed analysis of the consequences of cellular infection on ECM composition. This protein analysis also showed that similar gene expression profiles could induce different effects on the composition of ECM glycoproteins, probably via post-transcriptional modifications.
Figure 5. Western blotting analysis of IEC6 cells infected by the different S. Typhimurium strains.
Discussion
The first and most extensively studied mechanism of entry of Salmonella is the trigger one. Epithelial intestinal cell lines HT29, Caco-2 and HeLa, that have often been used in these studies, support an overwhelmingly trigger T3SS1-dependent entry. However, T3SS1-independent entry mechanisms have been described [Citation31] like the Rck-dependent entry, which enables S. Enteritidis to enter cells through a zipper mechanism [Citation12,Citation32]. More recently, it has been recognized that in the epithelial cell-line AML12, a wild type S. Typhimurium expressing a functional T3SS1, enters through a zipper mechanism [Citation18]. Thus, Salmonella can use either trigger and zipper entry routes but we were not aware of a cell line that supports both at similar levels. To compare the cellular response induced by bacteria, which entered cells via either the trigger or the zipper entry process, we constructed three genetically modified S. Typhimurium 14,028 strains. The STM-Z is a invA mutant expressing Rck and PagN in-vitro, and can enter cells via the zipper entry; the STM-T is a mutant invalidated for the invasins, Rck and PagN, which can enter cells via the trigger entry; the STM- ZT can use both entry processes and the STM-3d is unable to express any of these three invasion proteins. We found that the STM- Z and STM-T strains were both able to enter the intestinal epithelial cell line IEC-6, but to a lower level compared with the STM- ZT strain. To our knowledge, IEC-6 is, to date, the only cell line that can support both entry routes at similar levels. This peculiar characteristic could be linked to the nature of the cell line used in this study. Indeed, IEC-6 were obtained from a normal rat small intestine by serial subcultures and not from a tumor, like most other intestinal cell lines [Citation33]. Profound cellular modifications accompanying cellular transformation could therefore affect the process of Salmonella entry into cells of non-tumor or tumor origins differentially [Citation32].
Using microarrays, we carried out a transcriptomic profiling of IEC-6 cells exposed to the STM-ZT, STM-Z, and STM-T strains. As suggested by ViSEAGO analysis, compared to unexposed cells, the three strains presented various significantly enriched processes related to immune response as previously shown in different studies [Citation34–36]. In our study, different chemokine-encoding genes like Ccl20, Cxcl2, Ccl2, C×3cl1, or Cxcl3, and also Tnfa, Ptgs2, and the subunits of the NFkB transcription factor Nfkb1, Nfkb2, and Nfkbiz, were among the top upregulated genes with FC higher than 5 and up to 99, indicating that a strong inflammatory response was underway.
Surprisingly, very few differences in gene expression were found when comparing IEC-6 cells exposed to STM-ZT, STM-Z, or STM-T. Modulation of cellular gene expression is a very dynamic process involving different waves of regulation; it is probable that studies at very early time-points could underscore differential responses of the cell to the three STM strains when bacteria enter cells and are not within the cells. On the other hand, during its intracellular life in epithelial cells, Salmonella can occupy two niches: the Salmonella containing vacuole or the cytosol where bacterial replication is very active. Depending on its location in these niches, Salmonella deploys two different transcriptomic programs [Citation37] suggesting that interactions with cellular partners from the two cellular sub-compartments are probably different. As we did not detect any obvious differences in cellular response between IEC-6 exposed to the STM strains, this indicates that in our model, STM-ZT, STM-Z and STM-T occupy the same intracellular niche and therefore the entry pathway does not determine intracellular location.
In cells exposed to the three strains, we found that the expression of more than one hundred genes associated with the ECM was modified compared to unexposed cells. This is in line with another study on organoids exposed to S. Typhimurium which noticed that among the enriched biological processes, in addition to functions associated with immune response, there was “ECM organization” [Citation36]. ECM, a large network of macromolecules, ensures the morphological and mechanical properties of the tissue and is involved in cell signaling across the membrane. Numerous articles have observed that Salmonella interacts with the different proteins of the ECM. Only few have shown that Salmonella infection dramatically modifies the ECM gene expression. Berndt et al. described the reorganization of fibronectin, laminin, and tenascin in chick cells exposed to Salmonella strains with high or low invasiveness capacities [Citation38]. In our data, modulation of the expression of several genes argued for a process of stimulation or stabilization of the ECM and for the establishment of a fibrotic state after 3 h of interaction between the host cell and Salmonella. This hypothesis is strengthened by the modifications in fibronectin and laminin observed by western blotting in our model. The urokinase pathway plays a key role in the homoeostasis of the ECM. By hijacking this pathway, pathogens may enhance their invasiveness [Citation39]. In this way, PtgE [Citation40] and the thin aggregative fimbriae [Citation41] from Salmonella can interfere with the urokinase pathway, which in turn can favour their entry. A key effector of the urokinase pathway is the plasmin that is involved in the degradation of laminin, fibrin, and fibronectin, some major components of the ECM. On the other hand, plasmin also activates the matrix metalloproteinase-9 also known as collagenase. In our data, several genes related to the urokinase pathway were differentially expressed in exposed cells and in particular, the expression of Serpine1 and Serpinb2 was strongly upregulated in exposed cells, suggesting that plasmin production was inhibited. Indeed, these two proteins are potent inhibitors of uPA [Citation42], a protease that drives the activation of plasmin. Similarly, CYR61, which negatively controls plasmin production, by indirectly increasing SERPINE1 production, was modestly but significantly upregulated in exposed cells [Citation43]. This suggested that the urokinase pathway was repressed and could in turn favor ECM restoration. In addition, the expression of genes that positively influence ECM formation or stability was higher in exposed cells. This was the case for Timp1 (inhibitor of matrix metalloproteinases) [Citation44], Plet1 (involved in tissue repair) [Citation45] and Tnfaip6 (ECM stability) [Citation46]. On the other hand, the expression of Lmo7, a negative regulator of ECM deposition [Citation47] was decreased. Furthermore, the expression of genes involved in ECM component synthesis like hyaluronan (Has1), glycosaminoglycans (Ugdh) or N- glycans (Uap1) were upregulated in exposed cells. Finally, modulation of genes with increased expression like Sod2 (protection of heparan from degradation) [Citation48] and Lrp4 (induction of essential components of the ECM) [Citation49] also supported the idea that an ECM restoration process was ongoing. Taken together these observations suggested that, following perturbations induced by the infection, the expression of master components of the urokinase pathway was repressed and that the formation of ECM was consolidated. Activation of ECM formation could correspond to a consolidation phase of the matrix after degradation by the invading Salmonella. However, in different pathological situations, an excess of ECM component deposition induces fibrosis. The latter drives organ damage and most of the time is associated with inflammation. In a model of gut inflammation in mice, it was shown that TNFSF15 (Tl1A) is responsible for the development of fibrosis [Citation50]. In our model, the expression of Tnfsf15 was also increased and could be related to the development of a fibrotic process associated with inflammation. Similarly, other genes like Wnt10A [Citation51], Arel1 [Citation52] and Antxr2 [Citation53] which have been shown to be involved in fibrosis, have an increased expression in exposed cells. Conversely, other genes argued for a process of degradation of the ECM. Expression of Adamts4, which is involved in degradation of aggrecan [Citation54], was upregulated. These apparent contradictory observations most probably reflect the tight and complex regulation exerted on ECM.
As Salmonella can modulate cellular gene expression when they are extra- and intra-cellular, we used drugs (chlorpromazine and amiloride) to completely prevent its entry in order to determine which genes were modulated and at which step. Under our experimental conditions, the expression of far fewer genes was modulated when Salmonella cannot enter cells. The four genes, which were induced by adherent bacteria were involved in angiogenesis, a process that is pivotal to tissue repair. Forbester et al. in their study on organoids exposed to S. Typhimurium found that “positive regulation of angiogenesis” was another enriched biological process [Citation36]. In our study, SerpinB2 [Citation55], Angptl4 [Citation56], Icam1 [Citation57] and Nrg1 [Citation58], whose expressions were upregulated in cells treated with the drugs, were shown to have proangiogenic properties suggesting that an angiogenic process was initiated when Salmonella is extracellular, whereas the ECM modification is mainly driven by intracellular bacteria. The complex interactions between invading pathogens, host tissues, and immune cells occur in the context of the ECM. Several articles demonstrated that Salmonella can modify ECM by a direct interaction in order to improve cell invasion [Citation38]. In our case, we show that Salmonella can modify ECM composition when the bacteria is already within the cells. The modifications of the composition and three-dimensional ultrastructure of ECM should have a profound impact on the specific signals that the ECM conveys to immune cells at the forefront of infection and to epithelial cells and consequently to the outcome of infection. This topic could be addressed with enteroids cultured in the presence of immune cells.
In addition to the high number of ECM-related genes, which were modulated by intracellular Salmonella, we observed that a hundred DE genes were potential transcriptional targets of AhR. At first, the cytosolic ligand-activated transcription factor AhR was shown to be responsible for a response to xenobiotics, but since then its wide pleiotropic functions have been well documented. Activation of AhR can induce both canonical and non-canonical pathways. In the canonical pathway, binding of one of the numerous ligands on AhR induces its translocation to the nucleus where it binds ARNT. The heterodimer AhR/ARNT interacts with an XRE responsive sequence and activates transcription of many target genes [Citation59]. In our study the expression of key genes, regulated by and involved in AhR pathway like Arnt and Ahrr [Citation59] was increased. In addition, the expression of Cyp1a1, usually used as a readout of AhR activation, was strongly increased as well as that of Cyp1B1 and Aldh3a1, two AhR target genes that are important factors in the detoxification process. The role of AhR in a huge number of physiological functions is well recognized and more particularly in intestinal homoeostasis. AhR influences both gut barrier functions and the activity of different intestinal immune cells, particularly by stimulating production of interleukin 22 by group 3 innate lymphocytes [Citation60]. Given the importance of these functions, it is not surprising that AhR is implicated in several pathological processes and more particularly in infections. In this context, the role of AhR in response to pathogens is an emerging theme. Its involvement in viral [Citation61,Citation62], bacterial [Citation63,Citation64] and parasitic infections [Citation65,Citation66] has been documented. Bessede et al [Citation67]. have shown, for example, that in mice: the first challenge with LPS protects them from a second challenge with Salmonella Typhimurium in an AhR-dependant mechanism. More interestingly, AhR activation reduces mortality in a mouse model of systemic Salmonella infection with a concomitant reduced microbicidal capacity of phagocytes and a bacterial burden in surviving mice [Citation68]. Until now, the effect of AhR activation in vivo was assessed via its effect on immune cells [Citation68]. Our study suggests that this effect may also be mediated via enterocytes, directly or indirectly. Activation of AhR during Salmonella infection raises the question: what bacterial factor could bind this cytoplasmic transcription factor and activate the pathway? Moura-Alves suggested that the AhR is not only an important regulator of the immune response but also represents a novel type of pattern recognition receptor (PRR) [Citation69]. However, the plethora of potential microbial AhR ligands, their different affinities and quantities make it difficult to create general concepts.
In conclusion, the IEC-6 cell line supports both a trigger and a zipper entry. Transcriptomic reprogramming associated with both entry mechanisms is very similar, strongly suggesting that the cellular response does not depend on the entry route used by intracellular bacteria. This study also opens up new avenues of research because we have shown that in addition to modification of expression of numerous immune genes, once intracellular, Salmonella modifies in IEC6 cells several factors involved in the structure or regulation of the extracellular matrix and activated the aryl hydrocarbon receptor gene regulation. It is now important to determine the consequences of these modifications on cell invasion and host infection. The ECM plays, indeed, an active role in infection rather than simply providing a scaffold for bacterial adhesion or being a barrier to breach [Citation70]. In addition, it becomes clear that modulation of AhR plays a critical role in the outcome of infections [Citation68]. It is also interesting to note that the modification of both ECM and AhR could have a significant impact on intestinal cell barrier but also on the immune response.
Author contributions
AMC participated in the experimental design, carried out the interpretation of the microarray data, did the quantitative RT PCR on the Biomark Fluidigm™ platform, and drafted the manuscript.
SR participated in the experimental design, performed the cellular experiments, and contributed to the analysis of results.
MM carried out the microarray experiments, and participated in the bioinformatic analysis. CHA participated in the experimental design and did the microarray statistical analysis.
SH participated in the analysis of results, and participated in the discussion.
FK participated in the bioinformatic analysis.
EB participated in the experimental design and performed the cellular experiments.
JT engineered the modified STM strains.
PV conceived the study, participated in its design and the analysis of results, and helped to draft the manuscript.
All authors corrected and approved the final manuscript.
Supplemental Material
Download Zip (1.8 MB)Disclosure statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Data availability statement
The data that support the findings of this study are available in the GEO database with the accession number GSE151881.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21505594.2022.2158663
Additional information
Funding
References
- Berg HC, Huxley, AF, Simmons, RM. Constraints on models for the flagellar rotary motor. Philos Trans R Soc Lond B Biol Sci. 2000;355(1396):491–16.
- Hase K, Kawano K, Nochi T, et al. Uptake through glycoprotein 2 of FimH(+) bacteria by M cells initiates mucosal immune response. Nature. 2009;462(7270):226–230.
- Grzymajlo K, Ugorski M, Suchanski J, et al. The novel type 1 fimbriae FimH receptor calreticulin plays a role in salmonella host specificity. Front Cell Infect Microbiol. 2017;7:326.
- Chessa D, Dorsey CW, Winter M, et al. Binding specificity of salmonella plasmid-encoded fimbriae assessed by glycomics. J Biol Chem. 2008;283(13):8118–8124.
- Dorsey CW, Laarakker MC, Humphries AD, et al. Salmonella enterica serotype typhimurium MisL is an intestinal colonization factor that binds fibronectin. Mol Microbiol. 2005;57(1):196–211.
- Kingsley RA, Santos RL, Keestra AM, et al. Salmonella enterica serotype typhimurium ShdA is an outer membrane fibronectin-binding protein that is expressed in the intestine. Mol Microbiol. 2002;43(4):895–905.
- Li X, Bleumink-Pluym NMC, Luijkx Y, et al. MUC1 is a receptor for the salmonella SiiE adhesin that enables apical invasion into enterocytes. PLOS Pathog. 2019;15(2):e1007566.
- Singh B, Fleury C, Jalalvand F, et al. Human pathogens utilize host extracellular matrix proteins laminin and collagen for adhesion and invasion of the host. FEMS Microbiol Rev. 2012;36(6):61122–61180.
- Velge P, Wiedemann A, Rosselin M, et al. Multiplicity of salmonella entry mechanisms, a new paradigm for salmonella pathogenesis. Microbiologyopen. 2012;1(3):243–258.
- Fattinger SA, Bock D, Di Martino ML, et al. Salmonella typhimurium discreet-invasion of the murine gut absorptive epithelium. PLOS Pathog. 2020;16(5):e1008503.
- Heffernan EJ, Wu L, Louie J, et al. Specificity of the complement resistance and cell association phenotypes encoded by the outer membrane protein genes rck from salmonella typhimurium and ail from yersinia enterocolitica. Infect Immun. 1994;62(11):5183–5186.
- Rosselin M, Virlogeux-Payant I, Roy C, et al. Rck of salmonella enterica, subspecies enterica serovar enteritidis, mediates zipper-like internalization. Cell Res. 2010;20(6):647–664.
- Lambert MA, Smith SG. The PagN protein mediates invasion via interaction with proteoglycan. FEMS Microbiol Lett. 2009;297:209–216.
- Wiedemann A, Mijouin L, Ayoub MA, et al. Identification of the epidermal growth factor receptor as the receptor for salmonella Rck-dependent invasion. FASEB J. 2016;30(12):4180–4191.
- Yang Y, Wan C, Xu H, et al. Identification of an outer membrane protein of salmonella enterica serovar typhimurium as a potential vaccine candidate for salmonellosis in mice. Microbes Infect. 2013;15(5):388–398.
- Dyszel JL, Smith JN, Lucas DE, et al. Salmonella enterica serovar typhimurium can detect acyl homoserine lactone production by yersinia enterocolitica in mice. J Bacteriol. 2010;192(1):29–37.
- Hannemann S, Gao B, Galan JE. Salmonella modulation of host cell gene expression promotes its intracellular growth. PLOS Pathog. 2013;9(10):e1003668.
- Roche SM, Holbert S, Trotereau J, et al. Salmonella typhimurium invalidated for the three currently known invasion factors keeps its ability to invade several cell models. Front Cell Infect Microbiol. 2018;8:273.
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–6645.
- Mambu J, Barilleau E, Fragnet-Trapp L, et al. Rck of salmonella typhimurium delays the host cell cycle to facilitate bacterial invasion. Front Cell Infect Microbiol. 2020;10:586934.
- Barilleau, E, Vedrine M, Koczerka M, et al. Investigation of the invasion mechanism mediated by the outer membrane protein PagN of salmonella typhimurium. BMC Microbiol. 2021;21(1):153.
- Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47.
- Benjamini Y, Hocheberg Y. Controlling the false discovery rate-a practical and powerful approach to multiple testing. J Roy Stat Soc. 1995;57(1):289–300.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–408.
- Brionne A, Juanchich A, Hennequet-Antier C. ViSEAGO: a bioconductor package for clustering biological functions using gene ontology and semantic similarity. BioData Min. 2019;12(1):16.
- Holbert S, Barilleau E, Roche SM, et al. Murine AML12 hepatocytes allow salmonella typhimurium T3SS1-independent invasion and intracellular fate. Sci Rep. 2021;11(1):22803.
- Yusra, Semba S, Yokozaki H. Biological significance of tumor budding at the invasive front of human colorectal carcinoma cells. Int J Oncol. 2012;41:201–210.
- Yi J, Bergstrom K, Fu J, et al. Dclk1 in tuft cells promotes inflammation-driven epithelial restitution and mitigates chronic colitis. Cell Death Differ. 2019;26(9):1656–1669.
- Wu X, Qu D, Weygant N, et al. Cancer stem cell marker DCLK1 correlates with tumorigenic immune infiltrates in the colon and gastric adenocarcinoma microenvironments. Cancers (Basel). 2020;12(2):12.
- Truong D, Boddy KC, Canadien V, et al. Salmonella exploits host Rho GTPase signalling pathways through the phosphatase activity of SopB. Cell Microbiol. 2018;20(10):e12938.
- Radtke AL, Wilson JW, Sarker S, et al. Analysis of interactions of salmonella type three secretion mutants with 3-D intestinal epithelial cells. PLoS ONE. 2010;5(12):e15750.
- Aiastui A, Pucciarelli MG, Garcia-Del Portillo F. Salmonella enterica serovar typhimurium invades fibroblasts by multiple routes differing from the entry into epithelial cells. Infect Immun. 2010;78(6):2700–2713.
- Quaroni A, Wands J, Trelstad RL, et al. Epithelioid cell cultures from rat small intestine. Characterization by morphologic and immunologic criteria. J cell Biol. 1979;80(2):248.
- Eckmann L, Smith JR, Housley MP, et al. Analysis by high density cDNA arrays of altered gene expression in human intestinal epithelial cells in response to infection with the invasive enteric bacteria salmonella. J Biol Chem. 2000;275(19):14084–14094.
- Bruno VM, Hannemann S, Lara-Tejero M, et al. Salmonella typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PLOS Pathog. 2009;5(8):e1000538.
- Forbester JL, Goulding D, Vallier L, et al. Interaction of salmonella enterica serovar typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect Immun. 2015;83(7):2926–2934.
- Powers TR, Haeberle AL, Predeus AV, et al. Intracellular niche-specific profiling reveals transcriptional adaptations required for the cytosolic lifestyle of salmonella enterica. PLOS Pathog. 2021;17(8):e1009280.
- Berndt A, Muller J, Borsi L, et al. Reorganisation of the caecal extracellular matrix upon salmonella infection–relation between bacterial invasiveness and expression of virulence genes. Vet Microbiol. 2009;133(1–2):123–137.
- Bhattacharya S, Ploplis VA, Castellino FJ. Bacterial plasminogen receptors utilize host plasminogen system for effective invasion and dissemination. J Biomed Biotechnol. 2012;2012:482096.
- Haiko J, Laakkonen L, Juuti K, et al. The omptins of yersinia pestis and salmonella enterica cleave the reactive center loop of plasminogen activator inhibitor 1. J Bacteriol. 2010;192(18):4553–4561.
- Sjobring U, Pohl G, Olsen A. Plasminogen, absorbed by Escherichia coli expressing curli or by salmonella enteritidis expressing thin aggregative fimbriae, can be activated by simultaneously captured tissue-type plasminogen activator (t-PA). Mol Microbiol. 1994;14(3):443–452.
- Tang L, Han X. The urokinase plasminogen activator system in breast cancer invasion and metastasis. Biomed Pharmacother. 2013;67(2):179–182.
- Chen CC, Mo FE, Lau LF. The angiogenic factor Cyr61 activates a genetic program for wound healing in human skin fibroblasts. J Biol Chem. 2001;276(50):47329–47337.
- Brew K, Dinakarpandian D, Nagase H. Tissue inhibitors of metalloproteinases: evolution, structure and function. Biochim Biophys Acta. 2000;1477(1–2):267–283.
- Zepp JA, Zhao J, Liu C, et al. IL-17A-induced PLET1 expression contributes to tissue repair and colon tumorigenesis. J Immunol. 2017;199(11):3849–3857.
- Lauer ME, Cheng G, Swaidani S, et al. Tumor necrosis factor-stimulated gene-6 (TSG-6) amplifies hyaluronan synthesis by airway smooth muscle cells. J Biol Chem. 2013;288(1):423–431.
- Xie Y, Ostriker AC, Jin Y, et al. LMO7 is a negative feedback regulator of transforming growth factor beta signaling and fibrosis. Circulation. 2019;139(5):679–693.
- Petersen SV, Oury TD, Ostergaard L, et al. Extracellular superoxide dismutase (EC-SOD) binds to type I collagen and protects against oxidative fragmentation. J Biol Chem. 2004;279(14):13705–13710.
- Asai N, Ohkawara B, Ito M, et al. LRP4 induces extracellular matrix productions and facilitates chondrocyte differentiation. Biochem Biophys Res Commun. 2014;451(2):302–307.
- Barrett R, Zhang X, Koon HW, et al. Constitutive TL1A expression under colitogenic conditions modulates the severity and location of gut mucosal inflammation and induces fibrostenosis. Am J Pathol. 2012;180(2):636–649.
- Oda K, Yatera K, Izumi H, et al. Profibrotic role of WNT10A via TGF-beta signaling in idiopathic pulmonary fibrosis. Respir Res. 2016;17(1):39.
- Lear T, McKelvey AC, Rajbhandari S, et al. Ubiquitin E3 ligase FIEL1 regulates fibrotic lung injury through SUMO-E3 ligase PIAS4. J Exp Med. 2016;213(6):1029–1046.
- Burgi J, Kunz B, Abrami L, et al. CMG2/ANTXR2 regulates extracellular collagen VI which accumulates in hyaline fibromatosis syndrome. Nat Commun. 2017;8(1):15861.
- Westling J, Fosang AJ, Last K, et al. ADAMTS4 cleaves at the aggrecanase site (Glu(373)-Ala(374)) and secondarily at the matrix metalloproteinase site (Asn(341)-Phe(342)) in the aggrecan interglobular domain. J Biol Chem. 2002;277(18):16059–16066.
- Isogai C, Laug WE, Shimada H, et al. Plasminogen activator inhibitor-1 promotes angiogenesis by stimulating endothelial cell migration toward fibronectin. Cancer Res. 2001;61(14):5587–5594.
- La Paglia L, Listi A, Caruso S, et al. Potential role of ANGPTL4 in the cross talk between metabolism and cancer through PPAR signaling pathway. PPAR Res. 2017;2017:8187235.
- Gho YS, Kleinman HK, Sosne G. Angiogenic activity of human soluble intercellular adhesion molecule-1. Cancer Res. 1999;59(20):5128–5132.
- Hedhli N, Russell KS. Cytostatic drugs, neuregulin activation of erbB receptors, and angiogenesis. Curr Hypertens Rep. 2010;12(6):411–417.
- Larigot L, Juricek L, Dairou J, et al. AhR signaling pathways and regulatory functions. Biochim Open. 2018;7:1–9.
- Qiu J, Heller JJ, Guo X, et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36(1):92–104.
- Torti MF, Giovannoni F, Quintana FJ, et al. The aryl hydrocarbon receptor as a modulator of anti-viral immunity. Front Immunol. 2021;12:624293.
- Giovannoni F, Li Z, Remes-Lenicov F, et al. AHR signaling is induced by infection with coronaviruses. Nat Commun. 2021;12(1):5148.
- He R, Zhao L, Xu X, et al. Aryl hydrocarbon receptor is required for immune response in epinephelus coioides and danio rerio infected by pseudomonas plecoglossicida. Fish Shellfish Immunol. 2020;97:564–570.
- Rademacher F, Simanski M, Hesse B, et al. Staphylococcus epidermidis activates aryl hydrocarbon receptor signaling in human keratinocytes: implications for cutaneous defense. J Innate Immun. 2019;11(2):125–135.
- Munck NA, Roth J, Sunderkotter C, et al. Aryl hydrocarbon receptor-signaling regulates early leishmania major-induced cytokine expression. Front Immunol. 2019;10:2442.
- Ambrosio LF, Insfran C, Volpini X, et al. Role of aryl hydrocarbon receptor (AhR) in the regulation of immunity and immunopathology during trypanosoma cruzi infection. Front Immunol. 2019;10:631.
- Bessede A, Gargaro M, Pallotta MT, et al. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature. 2014;511(7508):184–190.
- Fueldner C, Riemschneider S, Haupt J, et al. Aryl hydrocarbon receptor activation by benzo[a]pyrene prevents development of septic shock and fatal outcome in a mouse model of systemic salmonella enterica infection. Cells. 2022;11(4):737.
- Moura-Alves P, Fae K, Houthuys E, et al. AhR sensing of bacterial pigments regulates antibacterial defence. Nature. 2014;512(7515):387–392.
- Tomlin H, and AM Piccinini. A complex interplay between the extracellular matrix and the innate immune response to microbial pathogens. Immunology. 2018;155(2):186–201.
