ABSTRACT
The length of stay (LOS) in hospital varied considerably in different patients with COVID-19 caused by SARS-CoV-2 Omicron variant. The study aimed to explore the clinical characteristics of Omicron patients, identify prognostic factors, and develop a prognostic model to predict the LOS of Omicron patients. This was a single center retrospective study in a secondary medical institution in China. A total of 384 Omicron patients in China were enrolled. According to the analyzed data, we employed LASSO to select the primitive predictors. The predictive model was constructed by fitting a linear regression model using the predictors selected by LASSO. Bootstrap validation was used to test performance and eventually we obtained the actual model. Among these patients, 222 (57.8%) were female, the median age of patients was 18 years and 349 (90.9%) completed two doses of vaccination. Patients on admission diagnosed as mild were 363 (94.5%). Five variables were selected by LASSO and a linear model, and those with P < 0.05 were integrated into the analysis. It shows that if Omicron patients receive immunotherapy or heparin, the LOS increases by 36% or 16.1%. If Omicron patients developed rhinorrhea or occur familial cluster, the LOS increased by 10.4% or 12.3%, respectively. Moreover, if Omicron patients’ APTT increased by one unit, the LOS increased by 0.38%. Five variables were identified, including immunotherapy, heparin, familial cluster, rhinorrhea, and APTT. A simple model was developed and evaluated to predict the LOS of Omicron patients. The formula is as follows: Predictive LOS = exp(1*2.66263 + 0.30778*Immunotherapy + 0.1158*Familiar cluster + 0.1496*Heparin + 0.0989*Rhinorrhea + 0.0036*APTT).
Introduction
In recent years, the coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2 had become one of the most public-threatening infectious diseases [Citation1]. The original wild-type of SARS-CoV-2 caused severe disease and mutations of the virus will coexist with humans, possibly forever [Citation2]. Since the discovery of the Omicron variant in November 2021, it has continued to sweep the globe and mutate. Currently, Omicron variants are characterized by enhanced transmissibility, immune escape, and breakthrough reinfection including BA.1, BA.2, BA.3, BA.4, BA.5, and descendant lineages [Citation3–6]. As of 22 September 2022, more than 610 million laboratory-confirmed cases of infection with SARS-CoV-2 were reported globally, and these numbers continue to rise rapidly [Citation7].
There are many reports on the clinical characteristics, outcomes, and prognostic factors of patients infected with the Omicron variant [Citation8–10]. However, no predictive models have been developed for the length of hospital stay in Omicron patients. Outbreaks of Omicron Variant have led to a substantial increase in the demand for hospital beds and a shortage of medical equipment. Doctors can alleviate the shortage of medical resources by predicting LOS based on the clinical condition of patients. Two previous studies were used to predict LOS in SARS-CoV-2 patients. However, in one study, the variables of the model contained “Wuhan travelling history” [Citation11], and in another the virus was not Omicron variant [Citation12]. Therefore, it is an urgent need to develop a validated predictive model to predict LOS in Omicron patients, which can optimize the management of patients to alleviate the strain on medical resources.
In the present study, we screened a total of 384 Omicron patients from January 8 to February 13 in The Fifth People’s Hospital of Anyang City, which was initially and completely taken over by The First Affiliated Hospital of Zhengzhou University. We explored the clinical characteristics of 384 patients and found that cough, sputum, fever, and nasal congestion were the most common on admission. Using a screening of two models, we subsequently identified five variables, and developed and evaluated a simple model to predict LOS in hospitals for patients infected with the Omicron variant. These results can be used to provide enhanced and targeted management of Omicron patients and also to provide clinical decision-making support for doctors.
Materials and methods
Study population and data source
From January 8 to 13 February 2022, the data of 384 patients infected with the SARS-CoV-2 Omicron Variant were collected at the Fifth People’s Hospital of Anyang. Cases confirmed that SARS-CoV-2 Omicron Variant infection fulfilled the guidelines of “Diagnosis and Treatment Protocol for COVID-19.” First, we stratified the patients into the following three groups based on LOS: the lowest quartile (25th quartile; 15 days; n = 85, 22.1%), the median (25th–75th quartiles; n = 212, 55.2%), and the highest quartile (75th quartile; 75 days; n = 87, 22.7%). Respectively, the age ranges for the three groups were 9–14 years, 15–25 years, and 26–39 years. The baseline characteristics were summarized in . Missing values were imputed with the median of each corresponding variable. Finally, this study was approved by the Institutional Review Board of the First Affiliated Hospital of Zhengzhou University (L2021-Y429-002). The study was implemented under the Helsinki Declaration and Rules of Good Clinical Practice.
Table 1. General characteristics of the 384 patients on admission.
Feature extraction
The LOS was skewed and thus was transformed using the natural log for model training and testing. The final predicted LOS was then converted back to the standard LOS within days. Using all the sample and candidate predictors, we employed LASSO to select the primitive predictors. The final predictive model was constructed by fitting a linear regression model using the predictors selected by the LASSO method. Due to the small sample size, bootstrap validation was used to test the model performance, and a total of 2000 bootstrap samples were drawn with replacement of the sample size as the original sample. Prediction models were developed for each bootstrap sample and then tested for the original sample. The differences in performance between the bootstrap sample and the original sample indicated optimism, the mean of which was subtracted from the apparent performance of the original model to get the actual performance of the model. We calculated four indicators to evaluate the performance of the predictive model, that is, the explained variation R^2, and three accuracy indicators calculated by comparing the predicted LOS with the actual LOS. Predictors with missing values were imputed using multiple imputations with 10 imputations. The final imputed value was an average of 10 imputations. Rates of missing ranged from 0.26% (Neu-count) to 14.06% (ESR). The outcome was LOS.
Information collection
Detailed patients’ information was collected from electronic medical records. Clinically meaningful data (which were reliable and easily collected) were selected as candidate variables. Initial candidate variables included epidemiological data, clinical (COVID-19 related) and laboratory data. Treatments during hospitalization, including traditional Chinese medicine (TCM), immunotherapy, heparin, interferon inhalation, antibiotic therapy, and others, were also collected and used as candidate variables. Variables with more than 20% missing and less than 1% frequency were excluded.
Definition of terms
Immunotherapy mainly referred to treatment with interleukin 2 (IL2). TCM treatment is based on the principle of “one person, one treatment” improved the treatment protocol. Patients could be discharged when their temperature was normal for three consecutive days, respiratory symptoms and chest CT images improved, and two consecutive novel coronavirus nucleic acid tests’ were Ct values ≥35 for both the N and ORF genes (fluorescent quantitative PCR method with a cut-off value of 40 and a sampling interval of at least 24 h). LOS refers to the length of stay in Omicron patients.
Primary outcome measure
In the study, the primary outcome was LOS. Patients who reappeared in positive nasopharyngeal swab collection within 2 weeks of meeting the discharge criteria would be excluded.
Statistical analysis
The analysis was conducted using SAS statistical software 9.4 (SAS institute Inc.). The study followed the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) reporting guidelines. Each of the 22 items of the TRIPOD statement was addressed.
Results
Patients’ characteristics
As shown in , a total of 384 patients was enrolled in the study. The baseline characteristics of these patients are summarized in . Among the 384 patients, 222 (57.8%) were female, the mean age of patients was 19 years and 349 (90.9%) completed two doses of vaccination. Patients on admission diagnosed as mild were 363 (94.5%). Common symptoms on admission were cough, expectoration, fever, and nasal congestion. In the group of LOS <15 days, 24 (28.2%) patients were associated with familial cluster, which was notably different from other two groups (100 [47.2%] vs. 42 [48.3%], p = 0.0084). Then, the median value of aspartate aminotransferase (18 vs. 20 vs. 18 U/L, P = 0.0205) was statistically significant among the three groups (Table S1).
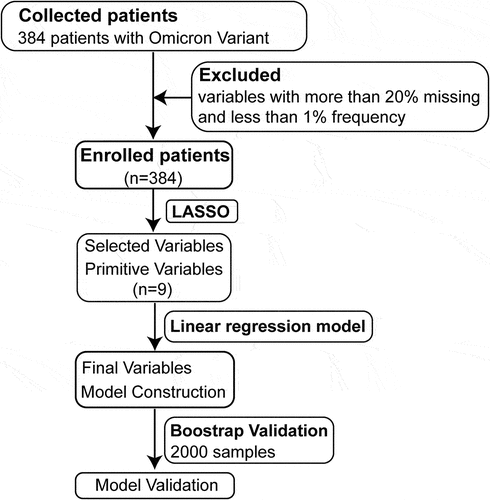
Figure 1. Flow diagram of the study. A total of 384 patients infected with Omicron variant from January 8 to February 13 in 2022 were enrolled. The final predictive model was constructed by fitting a linear regression model using the predictors selected by the LASSO method. Due to small sample size, bootstrap validation was used to test model performance. A total of 2,000 bootstrap samples were taken and replaced with the same sample size as the original, resulting in the final model.
Management of 384 patients during hospitalization is shown in . In this cohort, no patients had experienced complications. Overall, 381 (99.2%) patients received traditional Chinese medicine (TCM) treatment, 143 (37.2%) patients underwent immunotherapy, 20 (5.2%) received heparin and 22 (5.7%) underwent interferon inhalation. Treatment among the three groups differed in terms of immunotherapy, heparin, and interferon inhalation.
Table 2. Treatments and outcomes for 384 patients.
Feature selection
presents a flow diagram depicting the sampling strategy for this study. Because of the skewed distribution of hospital LOS, a logarithmic function transformation with base e was made to obtain a distribution of LOS (). We then used LASSO regression simulations to select nine candidate variables, with non-zero coefficients taking a penalty parameter 0.395477 (; ). On this basis, variables were again selected using a linear model, and those with P < 0.05 were integrated into the analysis, including immunotherapy, familial clustering, heparin, rhinorrhea, activated partial thromboplastin time (APTT; ). The factors affecting the LOS and the model are shown in . It showed that if the Omicron patients developed rhinorrhea or occur familial cluster, the LOS increased by 10.4% (95%CI: 1.1%–20.6%) or 12.3% (95%CI: 6.4%–18.5%), respectively. If the Omicron patients received immunotherapy or heparin, the LOS increased by 36% (95% CI: 28.6%–43.9%)or 16.1% (95%CI: 2.8%–31.3%). Moreover, if the Omicron patients’ APTT increased by one unit, the LOS increased by 0.38%(95%CI: 0.02%–0.70%).
Figure 2. Screening process and analysis of results. (a) Histogram of length of hospital stay. Because the patients’ LOS showed a skewed state, log transformation with base e was made to it. (b) Using all the sample and candidate predictors, we employ LASSO to select the primitive predictors. Optimal lambda is 0.395477. (c) Box plot of real LOS and its predicted value. (d) Scatterplot of real LOS and its predicted value.
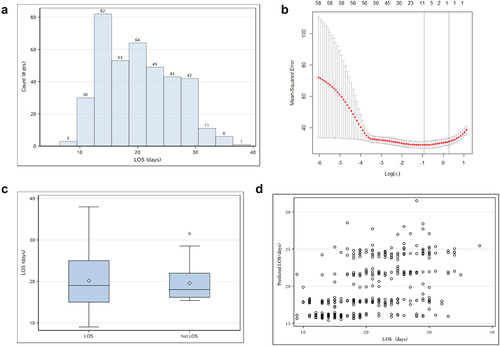
Table 3. Candidate variables selected by the LASSO simulation.
Table 4. Candidate variables again selected by a linear regression model.
Table 5. A prognostic model to predict the length of stay of Omicron patients.
Predictive model
Specific points on variables are shown in . The formula for predicting LOS is as follows: Predictive LOS = exp(1 × 2.66263 + 0.30778*Immunotherapy+0.1158*Familiar cluster+0.1496*Heparin+0.0989*Rhinorrhea+0.0036*APTT). The effect of each variable on LOS was obtained by multiplying the value of each factor by the corresponding point. The sum was then converted to an exponential function with base e. The distribution of LOS and its predictive value is plotted in .
Performance of hospital LOS prediction model
We test the predictive efficiency of the model by using the R-squared, which measured how well the predicted value fits the true value, with an R-squared of 0.3021. There were 59 (15.4%) cases where the prediction error was less than or equal to 1 day, 119 (30%) where it was less than or equal to 2 days and 171 (44.5%) where it was less than or equal to 3 days.
Testing and calibration of models
In order to accurately evaluate the predictive ability of the model, bootstrap with put-back resampling was used 2000 times with the same sample size as the original sample size. The model was built with each bootstrap sample separately, validated with the original sample, and the ratio of prediction error less than or equal to 1 day, less than or equal to 2 days, less than or equal to 3 days, and R-square were calculated for each of the two samples. Finally, the difference between the means of these four indicators in the resampled sample and the original sample was calculated and used to adjust the values of the four evaluation indicators obtained from the original sample. After the calibration, R-squared was 0.2864. There were 15.2% cases where the prediction error was less than or equal to 1 day, 29.6% where it was less than or equal to 2 days and 43.8% where it was less than or equal to 3 days.
Disscussion
Previous studies have been carried out to determine the LOS associated with Omicron patients, but the results varied from study to study, which identified five variables including age, alanine aminotransferase, pneumonia, platelet count, and PF ratio (PaO2/FiO2) [Citation12]. In our study, we conducted a single-centre retrospective study of 384 Omicron patients in Anyang City, Henan Province. We analysed the clinical characteristics, identified five variables, and developed and evaluated a predictive model to predict LOS in hospitalized Omicron patients. It was beneficial to optimize the use efficiency of hospital beds and medical resources and alleviate medical resource shortages [Citation13].
We found that cough, expectoration, fever, and nasal congestion were common clinical manifestations in Omicron patients, in accordance with previous studies [Citation14]. Previous evidence indicated that the incidence of fatalities from COVID-19 depends crucially on the infected age groups [Citation15]. As most of the patients were young, they had few underlying diseases and were mildly ill.
The results of previous studies differed from ours [Citation16]. Our study showed that immunotherapy, familial clustering, heparin, rhinorrhea, and APTT were independent factors that influenced the LOS in hospitalized patients, and they were included in the final formula. In addition, the prediction model had a low percentage of errors in predicting the LOS within 1, 2, and 3 d. These results confirmed that the formula accurately predicted the LOS of Omicron patients.
Adaptive immune responses play key roles in SARS-CoV-2 clearance and protection of reinfection [Citation17]. Weakened cell-mediated immunity and humoral immune function can prolong LOS. The data showed that D-dimer levels were prognostically significant for mortality in COVID-19 [Citation18,Citation19]. Improvements in D-dimer levels by heparin might indicate clinical improvement. Moreover, rhinorrhea was less common in COVID-19 and SARS than in influenza and common cold.
There are several limitations to this study. First was a single-centre study involving only Chinese populations, which limits the validity of the results. Second, only 83 variables were included in this study, which led to less-than-expected predictive test results. Third, the study lacked external validity. Finally, more studies are needed to verify the generality of the results.
In summary, in this study, we analysed the clinical characteristics and outcomes of Chinese patients with a single centre Omicron variant. Common symptoms on admission were cough, expectoration, fever, and nasal congestion. The use of immunotherapy was the main reason for the LOS of patients. Five variables were identified, including immunotherapy, heparin, familial cluster, rhinorrhea, and APTT. A simple model was developed and evaluated to predict the LOS of Omicron patients. The formula is as follows: Predictive LOS = exp(1 × 2.66263 + 0.30778*Immunotherapy+0.1158*Familiar cluster+0.1496*Heparin+0.0989*Rhinorrhea+0.0036*APTT). These findings help doctors to optimize the management of patients to alleviate the strain on medical resources caused by Omicron Variant.
Author contributions
RZG and YZJ designed the study. ZJX, HXB, ZDH, LJ, LYH, and HP diagnosed and treated COVID-19 patients; ZJX, HXB and LL collected clinical data. LL and RZG analyzed clinical data. LL and RZG wrote the manuscript. All authors reviewed and approved the manuscript.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board from the First Affiliated Hospital of Zhengzhou University (L2021-Y429-002). The study was performed in accordance with the Helsinki Declaration and Rules of Good Clinical Practice. The requirement for informed consent was waived due to the urgent need to collect data on this emerging pathogen.
Patient and Public Involvement
Patients or the public were not involved in the design, conduct, reporting, or dissemination plans of our research.
Supplemental Material
Download MS Word (18 KB)Acknowledgements
We thank all the generous volunteer subjects who participated in the study. We sincerely thank all the doctors, nurses, and medical technicians who bravely fight the COVID-19 pandemic.
Data availability statement
The data that support the findings of this study are available from the corresponding author [RZG], upon reasonable request.
Disclosure statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21505594.2023.2196177.
Additional information
Funding
References
- Meganck RM, Baric RS. Developing therapeutic approaches for twenty-first-century emerging infectious viral diseases. Nat Med. 2021;27(3):401–7.
- The People’s Republic of China. Press Conference of the Joint Prevention and Control Mechanism of the State Council. 2022 [cited 2022 Sep 23]. Available from: http://www.gov.cn/xinwen/gwylflkjz206/index.htm
- WHO. Coronavirus (COVID-19) Dashboard. [cited 2022 Sep 23. Available from: https://covid19.who.int/
- Wu L, Zhou L, Mo M, et al. SARS-CoV-2 Omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2. Signal Transduct Target Ther. 2022;7(1):8.
- Zhang X, Wu S, Wu B, et al. SARS-CoV-2 Omicron strain exhibits potent capabilities for immune evasion and viral entrance. Signal Transduct Target Ther. 2021;6(1):430.
- He X, Hong W, Pan X, et al. SARS-CoV-2 Omicron variant: characteristics and prevention. MedComm. 2021;2:838–845.
- WHO. Currently circulating variants of concern. [cited 2022 Sep 23]. Available from: https://www.who.int/activities/tracking-SARS-CoV-2-variants
- Tian D, Sun Y, Xu H, et al. The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 Omicron variant. J Med Virol. 2022 Jun;94(6):2376–2383.
- Maslo C, Friedland R, Toubkin M, et al. Characteristics and Outcomes of Hospitalized Patients in South Africa During the COVID-19 Omicron Wave Compared with Previous Waves. JAMA. 2022 Feb 8;327(6):583–584.
- Qiu W, Shi Q, Chen F, et al. The derived neutrophil to lymphocyte ratio can be the predictor of prognosis for COVID-19 Omicron BA.2 infected patients. Front Immunol. 2022 Nov 2;13:1065345.
- Hong Y, Wu X, Qu J, et al. Clinical characteristics of Coronavirus Disease 2019 and development of a prediction model for prolonged hospital length of stay. Ann Transl Med. 2020 Apr;8(7):443.
- Li K, Zhang C, Qin L, et al. A Nomogram Prediction of Length of Hospital Stay in Patients with COVID-19 Pneumonia: A Retrospective Cohort Study. Dis Markers. 2021;2021:5598824.
- Leclerc QJ, Fuller NM, Keogh RH, et al. Importance of patient bed pathways and length of stay differences in predicting COVID-19 hospital bed occupancy in England. BMC Health Serv Res. 2021;21(1):566.
- Li Q, Liu X, Li L, et al. Comparison of clinical characteristics between SARS-CoV-2 Omicron variant and Delta variant infections in China. Front Med (Lausanne). 2022;9:944909.
- Levin AT, Hanage WP, Owusu-Boaitey N, et al. Assessing the age specificity of infection fatality rates for COVID-19: systematic review, meta-analysis, and public policy implications. Eur J Epidemiol. 2020;35(12):1123–1138.
- Ebell MH, Hamadani R, Kieber-Emmons A. Development and Validation of Simple Risk Scores to Predict Hospitalization in Outpatients with COVID-19 Including the Omicron Variant. J Am Board Fam Med. 2022;35:1058–1064.
- Castro Dopico X, Ols S, Lore K, et al. Immunity to SARS-CoV-2 induced by infection or vaccination. J Intern Med. 2022;291(1):32–50.
- Middeldorp S, Coppens M, van Haaps TF, et al. Incidence of venous thromboembolism in hospitalized patients with COVID-19. J Thromb Haemost. 2020;18(8):1995–2002.
- Helms J, Tacquard C, Severac F, et al. High risk of thrombosis in patients with severe SARS-CoV-2 infection: a multicenter prospective cohort study. Intensive care Med. 2020;46(6):1089–1098.
