ABSTRACT
In this study, we examined the occurrence of acquired and transmitted drug resistance to integrase strand transfer inhibitor (INSTI) in HIV-1 strains in Chongqing (China) for guiding for the routine testing of INSTI-associated HIV-1 genotype resistance. Plasma samples were obtained from HIV-1 patients at Chongqing Public Health Medical Center from July 2019 to August 2022. Besides, amplification, sequence, and analysis of the portion of the HIV-1 pol gene that encodes the integrase protein were implemented to identify INSTI resistance. Integrase sequence data was harvested for a comprehensive cohort of 1032 patients infected with HIV-1. This cohort consisted of 564 ART-naive patients, 465 ART-treated patients, and 3 patients with an unknown treatment history. Within the study group, we identified INSTI resistance in 21 patients (2.03%, 21/1032), including 17 ART-treated patients (3.66%, 17/465). Among the ART-treated patients, 12 were INSTI-treated (11.76%, 12/102), 5 were INSTI-naive (1.38%, 5/363), and 4 were ART-ineffective patients (0.71%, 4/564). The prevalent major resistance mutation was Q148R (0.48%, 5/1032), while the most prevalent accessory resistance mutation was E157Q (1.65%, 17/1032). In light of the above, it is recommended that the incidence of accessory genotype analysis should be considered before starting any future INSTI-based therapy, especially in patients with drug resistance to NRTIs and NNRTIs and the reduction of INSTI sensitivity should be carefully monitored and investigated. Regular monitoring for resistance should be implemented after the use of INSTIs, and, importantly, ongoing monitoring of the decreasing susceptibility to INSTIs is crucial following the initiation of treatment with INSTIs.
Introduction
Acquired immunodeficiency syndrome (AIDS), caused by the infection of the human immunodeficiency virus (HIV), is a highly prevalent global health concern that continues to pose a significant threat to human well-being worldwide. By the end of 2020, approximately 1,053,000 individuals in China had been infected with HIV-1, resulting in 351,000 AIDS-related deaths [Citation1]. Highly active antiretroviral therapy (HAART) has proven to be efficacious in suppressing HIV replication and mitigating HIV-related morbidity and mortality [Citation2]. Nevertheless, the wide utilization of antiretroviral drugs in clinical practice has given rise to HIV drug resistance, which significantly impacts the effectiveness of ART in both treatment-naive and experienced patients [Citation3–5].
Integrase strand transfer inhibitors (INSTIs) represent a contemporary category of antiretroviral drugs that inhibit strand transfer by competitively binding to the enzyme-binding site of the target DNA. They achieve this effect not only by replacing the 3’ end of the viral DNA from the active site but also by chelating divalent ions (Mg2+ or Mn2+) essential for integrase enzyme activity, thus blocking the integration of proviral DNA into the host chromosomal DNA [Citation6].
Notably, integrase inhibitors have emerged as preferred treatment options for both treatment-naive and experienced patients due to their outstanding tolerability, minimal toxicity, high efficacy, and user-friendly administration [Citation7]. Consequently, these inhibitors are now characterized as the first-line treatment regimens for patients infected with HIV-1.
Acquisition of resistance to INSTIs is dependent upon multiple factors, such as drug efficacy and viral heterogeneity. Singleton drug usage such as raltegravir (RAL) and elvitegravir (EVG) with low genetic barrier results in rapid emergence of resistance and such drug resistance is generally associated with accumulation of primary substitutions along with accessory substitutions in the integrase gene. The second-generation INSTIs have shown excellent efficacy and a high genetic barrier to drug resistance. However, the growing application of INSTIs in clinical practice has resulted in the inevitable emergence of resistance. Previously published studies have reported significant INSTIs resistance mutations among HIV-1 patients in various regions, including the United States [Citation8], Canada [Citation9], Europe [Citation10], and Spain [Citation5]. Likewise, major INSTIs resistance mutations have also been identified in several locations within China, such as Shenyang [Citation11], Henan [Citation4], Yunnan [Citation12], and Jiangsu [Citation13]. Thus, early monitoring of resistance to INSTIs is crucial for informing clinical decisions regarding HIV medication and facilitating timely adjustment of dosing regimens. Chongqing, the largest municipality in China and a key economic centre of the southwest region, ranked sixth in terms of the number of people living with HIV (PLWH) by the end of October 2021, with 58,000 individuals affected and 19,000 AIDS-related deaths [Citation14]. However, there is currently no existing literature regarding the prevalence of drug resistance to INSTIs in Chongqing. Therefore, this study conducted a preliminary analysis of mutations in the integrase gene and its associated drug resistance in Chongqing, aiming to offer valuable insights for preventing and treating HIV-1-infected individuals in China.
Materials and methods
Ethics statement
This study was approved by the ethics committee of Chongqing Public Health Medical Center and conducted in accordance with the Declaration of Helsinki (2008). All data were analysed anonymously, and the requirement for written or oral informed consent was waived.
Study population
HIV-1-infected inpatients and outpatients who visited Chongqing Public Health Medical Center between July 2019 and August 2022 were enrolled in this study. Plasma samples were collected from patients with HIV-1 viral load (VL) greater than 1,000 copies/ml and subjected to a subsequent polymerase chain reaction (PCR) to amplify the IN gene. Data on the patient’s general condition, transmission route, VL, CD4 + T-cell count, and ART history were collected. Blood samples were drawn into EDTA-containing tubes, and subsequently, plasma samples were obtained following centrifugation. The plasma samples were then analysed for HIV-1 RNA VL employing the Roche HIV-1 nucleic Acid Quantitative Detection System (HiScript®Ampli Prep/COBAS®Taq COBAS®HIV-1 Test, 2.0).
Genotypic resistance testing
Whole blood samples were harvested and centrifuged at 3000×g for 15 minutes to remove leucocytes, and then plasma (supernatant) was harvested for RNA extraction. HIV-RNA was extracted utilizing a viral nucleic acid extraction kit (Jiangsu Shuoshi), followed by amplification of the target fragment by nested PCR. The HIV-1 IN gene was amplified by a validated In-house method. The first round of PCR was completed utilizing the HiScript®II one-step RT-PCR kit (Nanjing Vazyme, China). The second round was implemented with the help of an Ace Taq kit (Nanjing Vazyme, China) for nested PCR amplification of target sequences (1157–1213 bp) with two primer sets (4007F and 5219 R for the first round of PCR and 4063F and 5219 R for the second round of PCR) (). Then, sequences were subjected to the regularly updated Stanford HIV-1 Drug Resistance Database (http://hivdb.stanford.edu/) to analyse INSTI drug resistance mutations and antiretroviral susceptibility. Accordingly, drug resistance is categorized into five levels: S (susceptible), P (potential low-level), L (low-level), I (intermediate), and H (high-level). Strains exhibiting the latter three levels were considered drug-resistant strains.
Table 1. Primers for amplification and sequence analysis of HIV-1 integrase (IN) region.
Phylogenetic analysis
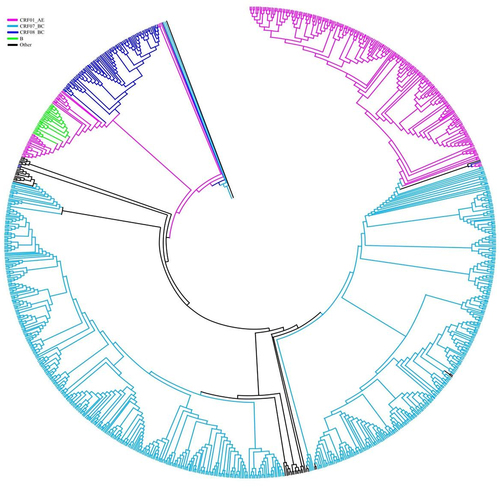
The pol region is widely recognized as a reliable region for HIV-1 subtyping analysis. For subtype analysis, HIV-1 genotypes were estimated employing the REGA HIV-1 Subtype Analysis Tool version 3.0 (http://dbpartners.stanford.edu:8080/RegaSubtyping/stanford-hiv/typingtool/) and substantiated through additional analysis. The newly characterized partial pol gene sequences (IN) were aligned with the subtype reference sequences retrieved from the Los Alamos HIV sequence database (http://www.hiv.lanl.gov/) utilizing the CLUSTAL W alignment tool. For phylogenetic analysis, 21 reference sequences or all sequences for each isoform were chosen. A phylogenetic tree was prepared with the help of 1000 bootstrap repeats using Molecular Evolutionary Genetic Analysis (MEGA) software version 6 (https://www.megasoftware.net/home), employing the neighbour-joining method and the Tamura-Nei model. A Kimura two-parameter model with a transition-transformation rate of 2.0 was prepared in our study, and scale bars represented a 2% genetic distance.
Results
Characteristics of the patients
Samples were harvested from 1080 patients, and successful amplification of the IN gene was achieved in 1032 samples (). The demographic characteristics of all enrolled patients (n = 1032, median age of 52 years, ranging from 12 to 89 years) are depicted in . Among those patients, 79.26% were male with a median age of 50 years (range 12 to 89 years), and 20.73% were female with a median age of 54 years (range 14 to 80 years). In addition, 98.16% of the patients identified as Han ethnicity. Additionally, heterosexual activity and men having sex with other men were identified as the two main routes of transmission among the included patients. Considering the clinical information, 564 patients were confirmed as patients on ART-naive, representing 54.65% of the study population (564/1032), while the treatment status of three patients remained unknown. The remaining patients on ART (45.06%, 465/1032) were treated with dolutegravir, rilpivirine, lamivudine, tenofovir, crizaline and efavirenz, among others.
Figure 1. Flow chart of our study.(INSTI integrase strand transfer inhibitor;ART antiretroviral therapy;DR drug resistance).
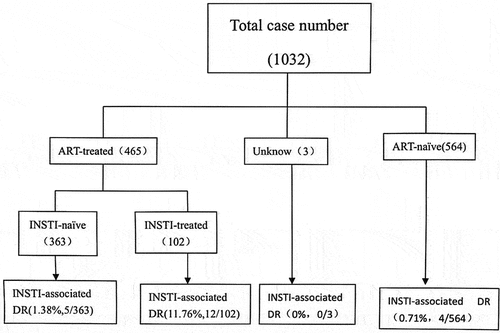
Table 2. Baseline demographic and viral characteristics.
Resistance mutations to INSTIs
Out of the total of 1032 patients, 4.17% (43/1032) exhibited major and/or accessory INSTI resistance mutations, with major mutations present in 1.45% (15/1032) of cases and accessory mutations present in 2.71% (28/1032) of cases. A total of 21 patients showed mutations associated with INSTI resistance (). Information on the INSTI resistance mutations associated with specific drugs is exhibited in . Three cases exhibited mutations associated with high-level resistance to bictegravir(BIC), eight cases showed high-level resistance to cabotegravir(CAB), four had high-level resistance to dolutegravir(DTG), thirteen had high-level resistance to elvitegravir(EVG), and ten had high-level resistance to raltegravir(RAL). Mutations leading to medium-level resistance to BIC, CAB, DTG, EVG, and RAL were observed in five, one, four, one, and four cases, respectively. Mutations leading to low-level resistance to BIC, CAB, DTG, EVG, and RAL were discovered in one, eleven, seven, seven, and seven cases, respectively.
Table 3. INSTI-associated mutation in ART-treated and naive patients.
Table 4. Analysis of INSTI drug resistance mutations.
The Q148R mutation was the most commonly observed major INSTI resistance mutation, with a resistance rate of 0.48% (5/1032). The E138K/A mutation occurred more frequently (0.39%, 4/1032). The G118R mutation is a nonpolymorphic mutation with a mutation frequency of 0.39% (4/1032). The E92Q mutation is a common non-polymorphic mutation with a mutation frequency of 0.29% (3/1032). The E157Q mutation was the most frequently observed accessory INSTI resistance mutation, with a frequency of 1.65% (17/1032). The S230R mutation leads to a resistance rate of 0.58% (6/1032) ().
Table 5. Prevalence of drug-specific INSTI mutation in ART-treated and naive patients.
The study participants were then categorized into ART-naive and ART-treated groups. We primarily observed major and/or accessory mutations, which resulted in resistance to INSTIs, within the group of ART-treated patients (3.66%, 17/465). The patients in the ART-treated group were sub-divided into INSTI-treated and INSTI-naive groups. Among the seventeen patients with INSTI resistance, twelve were INSTI-treated (11.76%, 12/102), and five were INSTI-naive (1.38%, 5/363). Among patients treated with INSTIs, varying combinations of major and/or accessory resistance mutations resulted in diverse degrees of drug resistance to INSTIs. In one patient, the major resistance mutations E138K plus the major resistance mutations G118R resulted in high-level resistance to all INSTIs (H1718 in ). In another patient, the major resistance mutations E138K, S147G, and Q148R plus the accessory mutation A128T caused high-level resistance to all INSTIs (H4778 in ). In the remaining patients, major resistance mutations E138K, S147G, Q148R, and R263K without accessory mutations resulted in high-level resistance to all INSTIs (H3944 in ). In ART-naive patients, accessory resistance mutations (S230R) (0.71%, 4/564) () in four patients resulted in low-level resistance to RAL, EVG, CAB, and DTG.
Primarily, INSTI resistance was identified in the CRF07_BC subtype of the virus. A phylogenetic analysis of the IN gene demonstrated that the sequenced strains could be categorized into 13 different subtypes (). The prevalence of each subtype was: A (0.39%, 4/1032), B (1.16%, 12/1032), C (2.42%, 25/1032), CRF01_AE (19.86%, 205/1032), CRF07_BC (57.66%, 595/1032), CRF08_BC (14.24%, 147/1032), CRF55_01B (2.42%, 25/1032), CRF52_01B (0.78%, 8/1032), CRF59_01B (0.19%, 2/1032), CRF67_01B (0.09%, 1/1032), CRF77_cpx (0.09%, 1/1032), CRF78_cpx (0.09%, 1/1032), and CRF85_BC (0.58%, 6/1032). Circulating recombinant forms accounted for 95.63% of all subtypes (987/1032), with CRF07_BC and CRF01_AE being the major recombinant strains ().
Table 6. Distribution and prevalence of INSTI DR among HIV-1 subtype.
Discussion
According to the Chinese Guidelines for Diagnosis and Treatment of HIV/AIDS (2021) in China, authored by the AIDS and Hepatitis C Professional Group (2021), ART treatment plans that include INSTIs have been established as the standard care for patients infected with HIV-1 [Citation15]. However, with the widespread use of INSTIs, INSTI-related drug resistance mutations have gradually emerged. This study aimed to assess the prevalence of HIV INSTI-associated drug resistance mutations in Chongqing from July 2019 to August 2022, making it the first investigation to examine the status of drug resistance related to INSTIs in Chongqing, China. In the current study, we identified four cases of transmitted resistance to INSTIs, resulting in a prevalence of INSTI resistance of 0.71% (4/564) in ART-naïve patients. Among these four patients, no major drug resistance mutations were detected, except for a single accessory mutation (S230R) associated with low levels of resistance to CAB, DTG, EVG, and RAL.
INSTI resistance rates among primary ART patients were higher than in Henan (0.63%) [Citation4] and Beijing (0.34%) [Citation16] but lower than in Jiangsu (0.76%) [Citation13] and Yunnan [Citation12] (5.7%), suggesting a low transmission rate of pre-treatment INSTI resistant variants.
Importantly, prolonged use of first-generation INSTIs RAL and EVG can lead to rapid development of drug resistance. This study identified moderate-to-high-level resistance to RAL and EVG in 12 of 102 INSTI-treated patients, highlighting a resistance rate of 11.76%. This proportion closely aligns with the resistance rate reported in Henan Province (14.63%) [Citation4]. It is well-known that many patients who were prescribed RAL or EVG did not undergo INSTI resistance testing prior to the initiation of their treatment. In fact, almost all of these patients treated with INSTIs in Chongqing have HIV and exhibit resistance to NNRTIs or NRTIs. Resistance to NNRTIs and/or NRTIs before treatment has been validated to increase the risk of developing resistance to INSTIs. Specifically, the five ART-treated patients in our study who had resistance to INSTIs (regimens without INSTIs) were all resistant to NNRTIs and/or NRTIs. The highest mutation frequency of 41.67% (5/12) was found in Q148R. Q148H/K/R are non-polymorphic mutations reported in persons receiving RAL, EVG, CAB, and DTG. These mutations typically occur in combination with G140A/S or E138K. In this context, these mutations share a linkage with near complete resistance to RAL and EVG, high levels of reduction in CAB susceptibility, and low-to-intermediate reductions in DTG and BIC susceptibility. The G118R mutation had a high prevalence of 33.33% (4/12). This mutation is the major resistance mutation commonly responsible for DTG and CAB resistance. Three patients showed high resistance to all five INSTIs. All three patients with high resistance to each of the five INSTIs had the E138K resistance mutation. Two of them had concomitant Q148R mutations, and two of them had mutations in S147G, which reduced sensitivity to EVG. Besides, E138K/A are non-polymorphic mutations that often develop in patients taking RAL, EVG, and DTG. Typically, these mutations co-occur with Q148 mutations. On their own, they do not reduce INSTI susceptibility. Nevertheless, when they occur in tandem with Q148 mutations, they result in high-level resistance to RAL and EVG, as well as intermediate reductions in susceptibility to DTG and BIC.
Our study has several limitations, including the exclusive utilization of the IN gene in the phylogenetic analysis, which could potentially limit the accuracy of subtyping and the recognition of CRF. Additionally, Sanger dideoxy sequencing was employed for the identification of the clinically significant known drug resistance mutations. Even though Sanger sequencing is confirmed as the gold standard approach for HIV drug resistance testing, it is incapable of identifying variants within the viral population that exhibit less than 20% drug resistance. The method may underestimate the true prevalence of drug-resistant mutations in treatment-naive patients.
Conclusion
Although no major resistance mutations were identified in ART-naïve patients, ART experience patients in Chongqing had an incidence of 1.45% for major resistance mutations related to INSTI. In patients who had used INSTIs, the incidence of major resistance mutations was as high as 11.76%. Those patients with potential low-level or low-level drug resistance to INSTIs can be treated with INSTIs in combination with other classes of drugs while those with intermediate- or high-level drug resistance, particularly against second-generation INSTIs, should be fully evaluated as such patients are generally associated with high-level resistance to other classes of drugs. Additionally, patients resistant to NNRTIs or NRTIs will experience increased resistance to INSTIs. Therefore, it is essential to consider conducting accessory genotype analysis before starting any future INSTI-based therapy, especially in patients with drug resistance to NRTIs and NNRTIs. Additionally, it is crucial to continually monitor and investigate any reduction in INSTI sensitivity. Resistance should be monitored more regularly after INSTIs have been used. More importantly, the decreasing susceptibility to INSTIs should be monitored regularly following the initiation of treatment with INSTIs.
Author contributions
ZHZ and LJG designed the study. LM and LJG obtained the sequences. WP, ZHZ, LM, and LJG analysed and interpreted the data. ZHZ and LM wrote the manuscript. WP and LJG reviewed the manuscript. All authors read and agreed to the published version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data Availability statement
The data in this study are available from the first author (HZ.Z) on reasonable request.
Additional information
Funding
References
- Liu Z, Tang X, Liu Y, et al. HIV prevention and health poverty alleviation - liangshan prefecture, Sichuan Province, China, 2017-2020. Chin CDC Weekly. 2021 Nov 26;3(48):1031–8. doi: 10.46234/ccdcw2021.250
- Bandera A, Gori A, Clerici M, et al. Phylogenies in ART: HIV reservoirs, HIV latency and drug resistance. Curr Opin Pharmacol. 2019 Oct;48:24–32.
- Mbisa JL, Ledesma J, Kirwan P, et al. Surveillance of HIV-1 transmitted integrase strand transfer inhibitor resistance in the UK. J Antimicrob Chemother. 2020 Nov 1;75(11):3311–3318. doi: 10.1093/jac/dkaa309
- Yang Z, Yang X, Deng X, et al. Prevalence of integrase strand transfer inhibitor (INSTIs) resistance mutations in Henan Province, China (2018-2020). Infection. 2021 Dec;49(6):1195–1202. doi: 10.1007/s15010-021-01668-9
- Casadella M, Santos JR, Noguera-Julian M, et al. Primary resistance to integrase strand transfer inhibitors in Spain using ultrasensitive HIV-1 genotyping. J Antimicrob Chemother. 2020 Dec 1;75(12):3517–3524. doi: 10.1093/jac/dkaa349
- Pommier Y, Johnson AA, Marchand C. Integrase inhibitors to treat HIV/AIDS. Nat Rev Drug Discov. 2005 Mar;4(3):236–248. doi: 10.1038/nrd1660
- Chen Q, Cheng X, Wei D, et al. Molecular dynamics simulation studies of the wild type and E92Q/N155H mutant of elvitegravir-resistance HIV-1 integrase. Interdiscip Sci. 2015 Mar;7(1):36–42. doi: 10.1007/s12539-014-0235-8
- Stekler JD, McKernan J, Milne R, et al. Lack of resistance to integrase inhibitors among antiretroviral-naive subjects with primary HIV-1 infection, 2007-2013. Antivir Ther. 2015;20(1):77–80. doi: 10.3851/IMP2780
- Ji H, Patterson A, Taylor T, et al. Prevalence of primary drug resistance against HIV-1 integrase inhibitors in Canada. J Acquir Immune Defic Syndr. 2018 May 1;78(1):e1–e3. doi: 10.1097/QAI.0000000000001649
- Antiretroviral Therapy Cohort C, May MT, Vehreschild J-J. Survival of HIV-positive patients starting antiretroviral therapy between 1996 and 2013: a collaborative analysis of cohort studies. Lancet HIV. 2017 Aug;4(8):e349–e356. doi: 10.1016/S2352-3018(17)30066-8
- Huang Xiaotong SZ, Minghui A. Primary drug resistance to integrase inhibitors among newly diagnosed HIV infected patients in Shenyang city. Chin J Lab. 2019;37(10):5.
- Deng XM, Liu JF, Zhang ME. Mutations of primary integrase gene resistance of HIV/AIDS patients in Yunan province. J AIDS Clin Res. 2019;25(4):327–330+341.
- Yin YQ, Lu J, Zhou Y, et al. Drug resistance to HIV-1 integrase inhibitors among treatment-naive patients in Jiangsu, China. Biomed Environ Sci. 2021 May 20;34(5):400–403. doi: 10.3967/bes2021.053
- Upstream News. By the end of October 2021, Chongqing ranked sixth in China, in the number of people living with HIV (PLWH; 58,000), with 19,000 deaths related to AIDS . 2021.
- Group AaHCP. The Chinese guidelines for diagnosis and treatment of HIV/AIDS. Chin J AIDS STD. 2021;27(11):1182–1201.
- Yu F, Li Q, Wang L, et al. Drug resistance to HIV-1 integrase inhibitors among treatment-naive patients in Beijing, China. Pharmgenomics Pers Med. 2022;15:195–203. doi: 10.2147/PGPM.S345797