ABSTRACT
Rab proteins are the major regulators of vesicular trafficking in eukaryotic cells. Their activity can be tightly controlled within cells: Regulated by guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs), they switch between an active GTP-bound state and an inactive GDP-bound state, interacting with downstream effector proteins only in the active state. Additionally, they can bind to membranes via C-terminal prenylated cysteine residues and they can be solubilized and shuttled between membranes by chaperone-like molecules called GDP dissociation inhibitors (GDIs). In this review we give an overview of Rab proteins with a focus on the current understanding of their regulation by GEFs, GAPs and GDI.
Introduction
Rabs are the largest branch of the superfamily of small GTPases with more than 60 members in humans and 11 members in budding yeast.Citation1,2 Since their discovery in the 1980s,Citation3-5 their major role in regulation of vesicular trafficking has been well established and many regulating factors have been identified, some of which will be discussed in detail in the following. In their physiologic role, Rab proteins can attach reversibly to membranes and bind to the nucleotides guanosine-5′-di- (GDP) or -triphosphate (GTP).Citation2
As a prerequisite for their function and to be able to localize to internal membranes within eukaryotic cells, Rab proteins need to become prenylated at C-terminal cysteine residues.Citation6 In order for this to happen, the newly synthesized Rab protein binds to the Rab escort protein (REP, termed Mrs6 in yeast) and is only then prenylated by Rab geranylgeranyltransferase (RabGGTase or GGTase-II, ).Citation7-9 After prenylation, the Rab protein can be delivered to a target membrane, where it is activated by a guanine nucleotide exchange factor (GEF) and bound GDP is replaced by the approximately 10-fold more abundant GTP.Citation1,10 It should be noted that GEFs catalyze nucleotide exchange in both directions and directionality is only a result of the higher concentration of GTP compared with GDP. In this active state, Rabs interact with a variety of different effector proteins which help to select cargo, form and bud vesicles from donor membranes, transport vesicles along cytoskeletal tracks and finally attach and fuse vesicles with the target membrane (reviewed in ref. Citation11). Finally, the Rab interacts with GTPase activating proteins (GAPs) and becomes inactivated by hydrolysis of GTP to GDP. It can then be extracted from the membrane by the GDP dissociation inhibitor (GDI), which solubilizes the inactive prenylated Rab protein to provide a pool of inactive Rab in the cytosol ready for the next round of vesicular transportCitation12 ().
Figure 1. Rab proteins as molecular switches. (a) After being synthesized, Rab proteins bind to the Rab escort protein (REP) and become prenylated by RabGGTase at C-terminal cysteine residues. The prenylated protein can be solubilized in the cytosol by the GDP dissociation inhibitor (GDI), which shields the hydrophobic geranylgeranyl groups from the hydrophilic environment (1). Guanine nucleotide exchange factors (GEFs) catalyze nucleotide exchange and help to recruit Rabs to certain membranes within the cell (2). In their GTP-bound active state, Rabs interact with effectors and regulate different steps in vesicular trafficking (budding, vesicular transport along the cytoskeleton, tethering and fusion with a target membrane; 3). Finally, inactivation takes place via interaction with GTPase activating proteins (GAPs) to yield the GDP-bound Rab (4). (b) Comparison of Rab proteins bound to GppNHp (left; Rab5 (1HUQ), Rab8 (4LHW), Rab9 (2OCB), Rab11 (2F9M), Sec 4 (1G17) and Ypt7 (1KY2)) and bound to GDP (right; Rab5 (3CLV), Rab8 (4LHV), Rab9 (1WMS), Rab11 (2F9L), Sec 4 (1G16) and Ypt7 (1KY3)). The nucleotide (sticks) and Mg2+ (green sphere), switch I (red), switch II (blue) and the P-loop (magenta) are highlighted (N: N-terminus, C: C-terminus). Note the switch regions that adopt an ordered and defined conformation in the active state, whereas more flexible and less well defined conformations can be seen in the inactive Rabs. (c) A sequence alignment of different representatives of Rab proteins from different Rab families (according to Klöpper et al.Citation134) is shown highlighting important features within the sequences. The P-loop (magenta), switch I (red) and switch II (blue), the nucleotide-binding G-motifs (green, G1-G5) as well as the C-terminal cysteine residues used for prenylation (brown) are highlighted. Additionally, the position of Rab-family (RabF, gray) and Rab-subfamily (RabSF, orange) motifs that play an important role in recognition of general Rab interacting proteins or Rab family specific interactions, respectively, are indicated. The secondary structure (α-helices 1–5 and β-sheets 1–6) is shown below the aligned sequences.
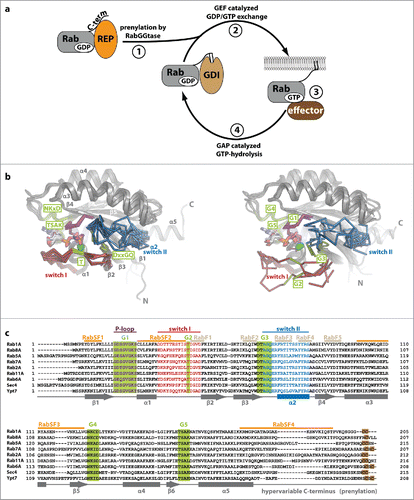
The G-domain common to small GTPases and Rab proteins shows a globular fold with a 6-stranded β-sheet surrounded by 5 α-helices (). The differences between the inactive and the active state of small GTPases were first resolved on a structural level in the late 1980s and the early 1990s,Citation13,14 and are well understood today: Upon exchange of the nucleotide, the major changes of conformation take place within regions termed switch I and switch II, which adopt an ordered conformation in the triphosphate state and a less well defined conformation in the diphosphate state (reviewed in ref. Citation15 see ). The high affinity binding and the specific recognition and discrimination of guanine over other nucleotides takes place via interactions between the nucleotide and specific motifs termed G-motifs (G1-G5).Citation16 The G1-motif (P-loop, GxxxxGK[S/T]) is present in many nucleotide binding proteins and binds the β-phosphate and a Mg2+-ion,Citation17 the G2-motif (T, part of switch I) makes contacts with the γ-phosphate and the Mg2+-ion,Citation15 the G3-motif (DxxGQ at the beginning of switch II) contains the Gln-residue necessary for GTP-hydrolysisCitation18 and the G4- (NKxD) and G5-motifs (TSAK) contain essential residues that form specific contacts with the guanine base to distinguish guanine from other nucleotides ( and ).Citation15,19 Besides these G-motifs, Rab proteins also contain short stretches of amino acids that have been termed RabF and RabSF motifs (). These distinguish Rabs from other small GTPases and provide the specificity toward general Rab interaction partners such as REP/GDI (RabF motifs) or toward specific interaction partners that only bind a subset of or single Rabs, typically effector proteins (RabSF motifs).Citation20,21
Here we present an overview of different regulatory factors of Rabs (GEFs, GAPs and GDI), with a focus on their discoveries and their molecular mechanisms of action and function within cells.
Guanine nucleotide exchange factors (GEFs)
Families and numbers
Guanine nucleotide exchange factors play an important role in activating Rab proteins in a spatiotemporally controlled manner.Citation22 In the past, a variety of different Rab GEFs have been identified (summarized in ), including the Vps9Citation23 and DENN domainCitation24 families of Rab GEFs as well as several other unrelated proteins. The low homology between Rab GEFs is probably the reason that for many Rabs, the corresponding GEFs are yet to be identified.Citation25
Table 1. Rab proteins and their known GEFs and GAPs.
The first large family of Rab GEFs identified were Vps9-domain containing proteins (at least 9 members in humans) that are specific toward Rab5 family members and act in early endocytic trafficking.Citation26-28 The second large family of RabGEFs, with 18 members in humans, are DENN domain GEFs.Citation29 Compared to the Rab5-family specific Vps9 domains, this family has a broader substrate spectrum and DENN domain containing proteins act on several different Rab proteins (see ). The domain architecture of these proteins shows 3 distinct regions, the upstream (uDENN), the central (DENN) and the downstream (dDENN) segments that are separated by linkers of different length within the primary sequence,Citation30 but form a 2-domain closely packed tertiary structure with one longin domain (a domain found in several GEFsCitation31) and a C-terminal lobe.Citation32 Besides these 2 major families of related Rab GEFs, several other proteins have been shown to possess GEF activity, including multi-subunit complexes such as the TRAPP I and II complexes (GEFs for Ypt1/Rab1 and Ypt31/32, respectively),Citation33 the Mon1/Ccz1 complex (GEF for Rab7/Ypt7),Citation34 Ric1-Rgp1 (GEF for Rab6/Ypt6)Citation35 and BLOC-3 (GEF for Rab32 and Rab38).Citation36 Interestingly, some of these contain longin-fold domains (Mon1/Ccz1, BLOC-3 and the TRAPP complexCitation31), however they seem to fulfill different functions within these proteins compared with DENN domains.Citation22 The related Sec2/GRAB/Rabin-8 GEFs that act on Rab8 or the yeast Rab8 homolog Sec4 all consist of a long parallel coiled-coil that catalyzes nucleotide exchange.Citation37,38 Very recently, the C. elegans protein REI-1 and its human homolog SH3BP5 were found to possess GEF activity toward Rab11.Citation39
As a mechanism of fine-tuning their activity, several GEFs of small GTPases have been reported to be controlled by auto-inhibition and release of this autoinhibition by regulatory proteins.Citation22 One such example is the Vps9 domain of Rabex-5, which shows basal activity toward Rab5, but is strongly activated in a positive feedback loop only upon interaction with a Rab5 effector protein, Rabaptin-5.Citation40 Furthermore, several GEFs have been implicated in Rab cascades, where they are recruited by one Rab to activate another Rab protein acting further down the pathway.Citation41 One example of this is the yeast Rab protein Ypt32p that recruits Sec2p, the GEF of downstream acting Sec4 (the yeast Rab8 homolog).Citation42 Additionally it has been shown that GEFs (together with other factors) play a major role in recruitment of Rabs to certain sites within cells.Citation43,44 Therefore, untangling the role of Rab-GEF networks including further interacting proteins and the identification of as yet unknown (possibly multi-subunit) GEFs will be a major challenge to fully understand their precise roles in the regulation of vesicular trafficking.
The possibility of manipulating vesicular transport for their own survival is exploited by several intracellular surviving pathogens.Citation45 One example is the pathogenic bacterium Legionella pneumophila that provides a plethora of proteins for manipulation of vesicular transport and other physiologic processes.Citation46 The protein DrrA (defect in Rab recruitment protein A) is among these and it was shown that Legionella injects this protein into infected host cells and uses its GEF activity to recruit and mislocalize Rab1 to the intracellular vacuole where Legionella resides.Citation47,48 Similarly, the Salmonella protein SopE was reported to act as a GEF for and recruit Rab5 to the intracellular Salmonella containing phagosomes, thereby promoting fusion with endosomes.Citation49
Structures and mechanisms
The first structure of a Rab:GEF complex, published in 2006, was that of Rab8:Mss4, even though structural analysis showed some peculiarities compared with other known GTPase:GEF structures and indicated a function of Mss4 as a chaperone rather than a GEF.Citation50 In the following years, a variety of different GEFs in complex with their cognate Rabs were successfully crystallized and their structures solved (Sec4:Sec2,Citation51,52 Rab21:Rabex-5,Citation53 Ypt1p:TRAPP,Citation54 Rab35:DennD1B,Citation32 Rab8:GRAB/Rabin855 and Rab5:Rabex-5,Citation56 ), allowing a structural view of intermediates of the nucleotide exchange reaction. Since the general mechanism of action of GEFs is the stabilization of the intermediate nucleotide-free state of small GTPases, GEFs bind these nucleotide-free GTPases with high affinities. Accordingly, most structures of GTPase:GEF complexes have been obtained in the absence of nucleotides (), with the exception of the recently published structures of Rab5:Vps957 and Rab8:Rabin855 that could be crystallized in the presence of different nucleotides.
Figure 2. Conformational changes during GEF-catalyzed nucleotide exchange. (a) Structures of Rab1b:GppNHp (pdb id 3NKV) and the Rab:GEF complexes Rab21:Rabex-5 (pdb id 2OT3), Ypt1:TRAPP (pdb id 3CUE), Rab35:DennD1 (pdb id 3TW8) and Rab1b:DrrA (pdb id 3JZA) Sec 4:Sec 2 (pdb id 2EQB and 2OCY). The relative distance of residues of the different Rab proteins (Cα-positions) within the P-loop, switch I and switch II compared with Rab1b:GppNHp is shown below each structure, highlighting the structural changes during GEF-catalyzed nucleotide exchange. Note the structures of Sec 4:Sec 2 in the presence of a PO42−-ion (pdb id 2EQB) and Sec 2:Sec 4 in the absence of a PO42−-ion (pdb id 2OCY), showing the collapsed state of the P-loop due to missing interactions with a negatively charged ion. (b) A comparison of the P-loop conformation of myosin bound to a nucleotide analogon (ADP-Metavanadate; pdb id 3MJX) and of the P-loop conformation of myosin in the absence of any nucleotide (pdb id 2AKA) indicates that the collapsed state of the P-loop is solely caused by missing interactions with a negatively charged phosphate group, not by binding of an exchange factor.
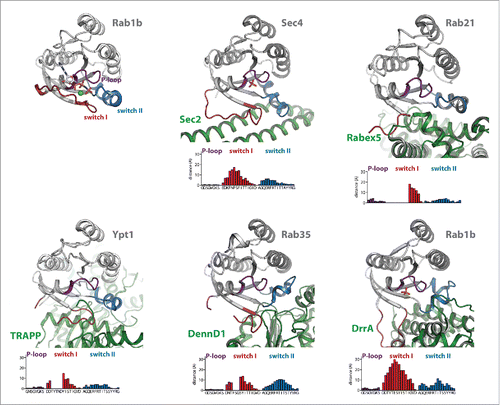
The general exchange mechanism of all GEFs must follow a common route with a first low-affinity encounter complex with the nucleotide-bound GTPase and subsequent release of the nucleotide. In all cases, GEFs bind to the switch I/switch II and interswitch regions of the small GTPases, inducing structural rearrangements within these regions that are incompatible with high affinity nucleotide binding (). The largest conformational changes (compared with the active GTP-bound GTPases) generally take place in switch I, where interactions occur mostly between the GEF and the C-terminal half of switch I (where the γ-phosphate and Mg2+ binding G2-motif Thr is located) to pull the switch I into an open conformation. This movement also leads to displacement of a highly conserved aromatic Phe or Tyr residue in Rab proteins (Tyr33 in Rab1b) from an edge-to-face interaction with the guanine base,Citation32 thus lowering the nucleotide affinity. In many cases, the residues surrounding this aromatic residue in switch I move approximately 10–30 Angstrom away from their position in the GTP-bound GTPase and often adopt a disordered state. For this reason, amino acids within this region often cannot be traced in the electron density of Rab:GEF structures (), similar to the situation in GDP-bound GTPases. In contrast, switch II adopts an ordered conformation in Rab:GEF structures that is more similar to the GTP- than the GDP-bound state of Rabs and the overall conformational change of switch II shown in is less dramatic than that of switch I. Since GEFs catalyze nucleotide exchange in both directions (i.e. from GDP- to GTP-bound state or vice versa), these results indicate that, depending on the educt of the GEF catalyzed reaction (i.e., Rab:GDP or Rab:GTP), the initial recognition of the GTPase by the GEF might be either by binding to switch I (GDP-bound state) or switch II (GTP-bound state).
In contrast to the switch regions, the conformational change of the P-loop is usually less dramatic and changes in the P-loop are mostly secondary effects due to a collapse of this loop into the nucleotide-binding pocket in the absence of interactions with negatively charged phosphates.Citation51,53 In fact, PO42− or SO42− ions were found to be bound at a position resembling the β-phosphate of the nucleotides in some structures (e.g. Rab1b:DrrA and Rab8:Rabin8) and seem to stabilize the P-loop in a conformation similar to the nucleotide-bound form. In this respect, comparison of Sec2:Sec4 structures with (pdb 2EQB) and without (pdb 2OCY) a bound PO42− ion nicely illustrate this effect with a collapsed conformation of the P-loop only in the PO42−-free structure ().Citation51,52 Furthermore, a similar collapsed state of the P-loop can also be observed in the related P-loop NTPase myosin in the absence of both an exchange factor and a bound nucleotide (). However, in some GEF:Rab complexes (Ypt1:TRAPP, Rab21:Rabex-5), a negatively charged residue from the GEF is projected into the vicinity of the highly conserved P-loop lysine, thus substituting for the missing negative charge.Citation58
In addition to binding to the switch and interswitch regions and opening of the nucleotide binding pocket, binding of the GEF in many cases involves projection of residues toward and steric hindrance of Mg2+ binding (e.g., Sec2, Vps9).Citation51,53 as well as electrostatic repulsion between acidic residues of GEFs or the Rab itself pointing toward the phosphate groups (e.g., Vps9, TRAPP)Citation53,54 These effects additionally lower the nucleotide affinity and thereby further accelerate nucleotide dissociation from the open nucleotide binding pocket.
GTPase activating proteins (GAPs)
Families and numbers
Small GTPases have a low intrinsic nucleotide hydrolysis activity with the half-life of the active state being of the order of 30 min to several hours.Citation59-62 Therefore, to be turned off in a physiologically meaningful timeframe, additional proteins termed GTPase activating proteins need to bind to the small GTPases and assist in hydrolysis. Whereas Rab GEFs are highly diverse in primary sequence and structure, Rab GAPs consist of one major family, the TBC- (Tre-2/Bub2/Cdc16) domain GAPs with more than 40 different members present in humans.Citation63 They were first described in yeast in the early 1990s where they are called Gyps (“GAP for Ypt proteins”).Citation64 Only one GAP in humans is known that does not contain this conserved TBC domain: The Rab3GAP complex, consisting of 2 different proteins and acts on members of the Rab3 family.Citation65
As can be seen in , for many Rabs, no corresponding GAP has been identified. However, many GAPs have been found to be promiscuous toward RabsCitation63 and possibly evolved low specificity to ensure that different active Rabs that reach a certain destination/organelle in a cell are inactivated by the GAP(s) present at this site.Citation66 Additionally, the boundaries between different Rabs are regulated by cascades similar to the case of GEFs described above. Thus, besides recruiting the downstream acting GEF Sec2, Ypt32 has also been described to recruit Gyp1, the GAP acting on Ypt1, as well as Gyp6, the GAP for Ypt6, thus helping to establish a sharp transition between active Ypt1/Ypt6 and Ypt32 on intracellular membranes.Citation41,67,68
Similar to DrrA as a GEF, Legionella pneumophila also harbors a Rab GAP in its arsenal of translocated proteins for the manipulation of host vesicular transport, the GAP protein LepB.Citation69 LepB also binds to the Legionella containing vacuole and appears to inactivate Rab1 (and possibly other Rab proteins) to allow their removal at a later stage in the reproduction cycle.Citation70
Structures and mechanisms
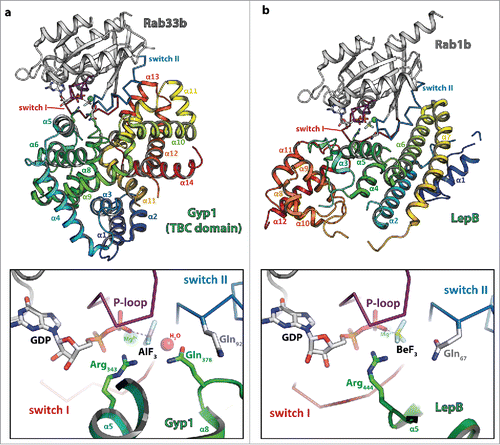
The first structure of a TBC domain GAP was that of Gyp1 in 2000,Citation71 followed by the structure of the complex between Gyp1 and Rab33b in 2006.Citation72 In this paper, Lambright and colleagues made the surprising discovery that, in contrast to other known GAPs of small GTPases, TBC-domain GAPs use a slightly different mechanism for catalysis. Whereas other GAPs were shown to provide an Arg finger to assist hydrolysis together with the Gln within the G3-motif of the small GTPases, Gyp1 provides an Arg- and an additional Gln-finger in trans that substitutes the G3-motif Gln that is used by many other GTPases in their GAP activated mechanism. TBC-domain GAPs were thus shown to act via a dual-finger mechanism ().Citation72 In contrast, the Legionella pneumophila GAP LepB uses a single Arg-finger comparable to other GAPs of small GTPases in conjunction with the cis-Gln provided by the Rab protein ().Citation70 The general role of the Gln (whether provided by the GAP or the Rab itself) is to align a water molecule for an in-line attack of the γ-phosphate, whereas the Arg finger (generally provided by the GAP) neutralizes developing negative charges during the hydrolysis reaction.Citation22 Since TBC-domain GAPs are not dependent on the Gln provided by the small GTPase, they have also been shown to still be able to catalyze GTP-hydrolysis in Rab proteins carrying a mutation often considered to stabilize GTPases in the active state (e.g., the G3-Gln mutated to Leu or Ala), thus possibly leading to misinterpretations obtained with Rabs carrying these mutations in in vivo experiments.Citation70,72 On the other hand, some TBC-domain GAPs lack the Arg, the Gln or both and might thus be either inactive or act via a different mechanism.Citation66
Figure 3. Dual- and single-finger mechanism used by RabGAPs. The structures of Gyp1:Rab33 (a) and LepB:Rab1 (b) are shown (left; Rab: gray cartoon with the switch I and II in red and blue, respectively and the P-loop in magenta (the switch regions and the P-loop are shown as ribbon), the GAP is shown as rainbow colored cartoon) and a zoom into the active site (right; residues provided by the GAP are colored in green, residues of the Rab protein in gray). Both structures were obtained in the presence of GDP and BeF3 or AlF3 as transition state mimetics of the hydrolysis reaction. (a) Gyp1 uses a dual-finger mechanism to catalyze GTP-hydrolysis and provides both Arg343 and Gln378 (green) in trans. The G3-motif Gln92 (gray) of Rab33 is repositioned by interactions with backbone atoms of Gyp1 and does not participate in catalysis. (b) In contrast to TBC-domain GAPs, the Legionella pneumophila GAP LepB uses a single-finger mechanism and provides only one residue (Arg444, green). Gln67 of Rab1b remains in its usual conformation to position the attacking water for the in-line attack of the γ-phosphate.
Prenylation of Rab proteins and membrane binding
Besides cycling between an active and an inactive state regulated by GEFs and GAPs, Rab proteins also cycle between a cytosolic and a membrane bound form and these cycles are tightly interconnected. As a necessary requirement for the reversible membrane attachment, Rab proteins need to become irreversibly prenylated at C-terminal cysteine residues.Citation73
Prenylation as a posttranslational modification was first discovered in the late 1970s,Citation74 but it took another decade until C-terminal prenylation was also shown for RasCitation75-77 and Rabs.Citation78,79 Prenylation is catalyzed by 3 different enzymes using either farnesylpyrophosphate (FPP) or geranylgeranylpyrophosphate (GGPP) for modification of target proteins. Farnesyltransferase (FTase) and geranylgeranyltransferase I (GGTase I) both recognize C-terminal CaaX-motifs (C - Cys, a - aliphatic amino acid, X - any amino acid) and attach a single farnesyl- or geranylgeranyl-group to the cysteine via a thioether linkage.Citation80,81 In contrast, geranylgeranyltransferase II (GGTase II, also termed RabGGTase) recognizes C-terminal cysteine residues within different motifs (e.g., CC, CXC, see ) and usually modifies 2 cysteine residues at the C-terminus of Rab proteins.Citation82 Rab proteins ending on CXC, but not CC, are then further processed by carboxymethylation.Citation83
Prenylation machinery and REP
After ribosomal synthesis, Rab proteins first need to bind to the Rab escort protein (REP), which only then presents the Rab proteins to RabGGTase for prenylation.Citation7,9 The double prenylation observed in most Rab proteins proceeds without intermediate release of the mono-prenylated Rab protein.Citation84 After binding of a new molecule of GGPP to the active site of the RabGGTase, the doubly prenylated Rab in complex with REP dissociates from RabGGTase and can deliver the Rab protein to a target membrane.Citation85,86
The X-ray structures of mono-prenylated Rab7 in complex with REPCitation87 as well as that of the RabGGTase:REP complexCitation88 have been solved (). REP, which exists in 2 different isoforms in humans and one isoform (termed Mrs6) in yeast,Citation89,90 consists of 2 different domains. Whereas domain I recognizes and binds the globular G-domain and the extended C-terminus of the Rab protein via regions referred to as the Rab binding platform (RBP) and the C-terminus binding region (CBR), respectively, the prenyl-groups bind between helices D and E of domain II ().Citation87 These 2 helices within domain II of REP additionally form the binding platform for the heterodimeric RabGGTase which consists of one large (60kDa) α-subunit and a smaller (38kDa) β-subunit (termed Bet4 and Bet2, respectively, in yeast).Citation88,89 The large subunit consists of 3 domains, an immunoglobulin- (Ig-) like domain, a leucine-rich repeat- (LRR-) domain and a solely α-helical domain which provides the binding interface with REP. The interaction is formed by the α-helices 8, 10 and 12 within this domain interacting with helices D and E of REP-1 ( and ). The active site, containing a Zn2+-ion for catalysis of prenylation, is located within the β-subunit.
Figure 4. The interaction of Rab proteins with REP, RabGGTase and GDI. (a) Model of the ternary complex between Rab7:GDP (gray, switch I – red, switch II – blue, P-loop – magenta, Mg2+ - green sphere, GDP - sticks), REP (cyan) and RabGGTase (α-subunit – green, β-subunit – blue, Zn2+ - orange sphere). The ternary complex was modeled from the structures of Rab7:REP (pdb id 1VG0) and REP:RabGGTase (pdb id 1LTX). (b) Model of doubly prenylated Ypt1:GDP (colors as above) in complex with GDI (yellow, pdb id 2BCG). Note the structural similarity between Rab7:REP and Ypt1:GDI. (c) Close-up view of helices D, E and H of the lipid-binding domain II of REP in complex with RabGGTase (left) or in complex with one geranylgeranyl-group (middle) and of the corresponding domain II of GDI in complex with 2 geranylgeranyl-groups (right). A conformational change within this domain upon binding of the C-terminally linked geranylgeranyl-groups of the Rab is presumably the cause of dissociation of RabGGTase from REP subsequent to prenylation.
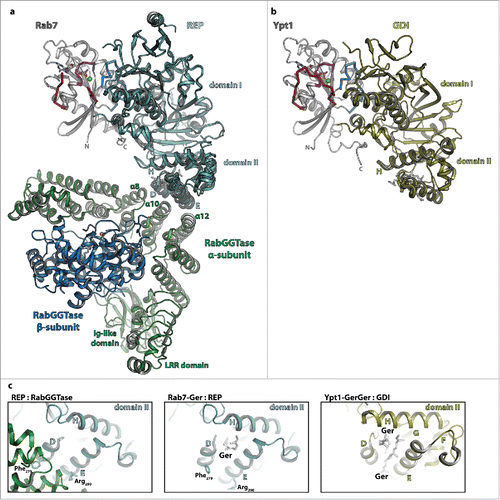
Based on the available structures of Rab7:REP and REP:RabGGTase, modeling of the ternary complex was also possible which in turn allowed proposal of a model of the catalytic mechanism87: Whereas the Rab-protein is bound to domain I of REP distal from the RabGGTase, the flexible C-terminus can extend toward and bind within the active site of the β-subunit of RabGGTase to become prenylated. As discussed before, binding of another GGPP leads to displacement of the already prenylated C-terminus from the active site which then migrates toward and binds to helices D and E of domain II of REP. This in turn leads to a conformational change that interferes with the interaction of Phe279 (α-helix D) and Arg290 (α-helix E) with RabGGTase ( and ), so that the Rab:REP complex finally dissociates from RabGGTase and the prenylated Rab can be delivered to a target membrane.Citation87
GDI function and structures
To be recycled after one round of vesicular trafficking, Rab proteins need to be extracted from membranes and solubilized in the cytosol. The protein responsible for this was originally identified as a Rab3 interacting protein that inhibited dissociation of GDP and was therefore named GDP dissociation inhibitor (GDI).Citation91 In contrast to REP, GDI only binds prenylated and inactive GDP-bound Rab proteins with high affinity,Citation91,92 thus ensuring extraction of the Rab proteins only after their trafficking cycle is complete and they have been inactivated by a GAP. Whereas GDI displays similar affinities for mono- and diprenylated Rabs (KD = 1.5 nM and 5.2 nM, respectively), but does not bind to unprenylated Rab, REP has a higher affinity for the mono-prenylated form (KD = 61 pM) compared with the unprenylated or diprenylated Rab (KD = 1 nM and 1.3 nM, respectively).Citation92,93 These observations provide an explanation for the thermodynamic driving force of extraction of Rab proteins from membranes by GDI, but not REP: Whereas GDI binds only weakly to the G-domain of a Rab, the affinity increases approximately 3 orders of magnitude upon additional binding of the prenylated C-terminus. On the other hand, REP binds with similar affinities to the prenylated and unprenylated Rab, thus allowing binding and presentation of the unprenylated Rab to RabGGTase.Citation92
The first structure of GDI was solved in 1996, displaying 2 separate domains termed domain I and II.Citation94 A first structure of GDI in complex with a prenylated cysteineCitation95 initially suggested the lipid binding site to be located in domain I. However some years later, the structures of mono-Citation96 and diprenylatedCitation97 Rab proteins bound to GDI showed that the lipid binding site was actually located in domain II, whereas the globular G-domain and the extended C-terminus both bind to domain I, with an overall fold, binding regions and relative orientation of Rab and GDI similar to the situation in the Rab:REP complex (see comparison in and ).
Targeting and retrieval of Rab proteins to and from intracellular membranes
One major issue that still remains to be fully resolved is the mechanism of specific targeting of Rab proteins to certain membranes within a cell. Initially, the hypervariable C-terminus was thought to contain the information encoded in the amino acid sequence for targeting to certain membranes,Citation98,99 but it was later shown that this does not apply to all Rabs and that sequences important for the intracellular localization are distributed throughout the primary sequence of Rabs.Citation100 Further work implied membrane localized GDI dissociation factors (GDFs) that are able to specifically dissociate Rab:GDI complexes and thereby contribute to specific localization.Citation101,102 However, Pra1 and the yeast homolog Yip3 remain the only proteins with proven GDF activities that have been identified. Since GDIs bind with high affinity to Rabs in their inactive, but not their active state,Citation103 a role of specific membrane targeting was also suggested for GEFs, since they can catalyze activation and therefore stabilize Rabs at a membrane.Citation43,48 A present consensus view is that targeting of Rabs to certain membranes is most probably a process involving many different factors, including the C-terminus of Rabs, further putative GDFs, GEFs, GAPs and effectors.Citation44 On the other hand, it is currently not clear whether extraction of Rabs by GDI is actively regulated and possibly accelerated beyond the presumably slow spontaneous dissociation rate of Rabs from membranes. The precise mechanisms of delivery and extraction of Rabs to and from intracellular membranes and additional factors that might catalyze these processes and contribute to spatial specificity therefore need to be established in future research.
PTMs and their role in regulation
Besides prenylation of the C-terminus, several other posttranslational modifications (PTMs) at different positions within the primary sequence of Rabs have been reported, including for example phosphorylation and serotonylation.Citation104-109 p34cdc2 kinase for example phosphorylates Ser204 within the C-terminus of Rab4 close to the prenylateable cysteines,Citation105 Parkinson's disease kinase LRRK2 phosphorylates several Rabs including Rab10 at position Thr73 within switch IICitation109 and a yet to be identified kinase phosphorylates Rab8 at Ser111 within the RabSF3 motif and close to the switch II region in the tertiary structure.Citation108 The latter was also shown to negatively influence the activation of Rab8 by its cognate GEF Rabin8.Citation108 In a very recent study, TGF-β activated kinase 1 mediated phosphorylation of Rab1a at Thr75 in switch II was shown to disrupt interaction with GDI.Citation110 Serotonlyation (the attachment of serotonin via an amide bond to the catalytic glutamine side chain by transglutaminases) on the other hand has been shown to cause constitutive activation of the modified GTPase.Citation106 Additionally, not only Rabs can be posttranslationally modified, but also their regulatory proteins including for example GEFs, GAPs and GDIs.Citation111-115
Again, much has been learned from the pathogenic bacterium Legionella pneumophila concerning the potential of PTMs for manipulation of Rabs. The Legionella proteins DrrA/SidDCitation116-119 and AnkX/Lem3Citation120-122 were shown to reversibly modify a Tyr and a Ser/Thr residue (Tyr77 and Ser76 in Rab1b within switch II) with AMP or phosphocholine, respectively, having an impact on the interaction of these Rab proteins with effectors, GAPs, GEFs and GDI. The potential role of PTMs was therefore discussed as means of modulating Rab interactions, including an effective displacement of Rab:GDI complexes and stabilizing Rabs at a certain membrane.Citation114,123
Even though several examples have been published, a universal regulatory role or the impact of the different PTMs on Rab regulated membrane trafficking is still uncertain for many PTMs and will be subject to further interesting investigations in the future. However, the positions of the posttranslational modification within functionally important regions of Rab proteins indicate that regulatory roles are highly probable.
Genetic disorders involving regulatory proteins
Besides being targeted by pathogenic bacteria, Rabs and associated regulatory proteins are also mutated in several genetic disorders.Citation45,124 Mutations of the GAPs Rab3GAP or TBC1D20 for instance are known to cause the severe Warburg Micro syndrome with neurologic and ocular disorders as well as microgenitalia.Citation125,126 Another mutation in Rab3GAP causes cataracts, mental retardation and hypogonadism in Martsolf syndrome.Citation127 The GEF BLOC-3 is mutated in Hermansky-Pudlak syndrome, causing albinism and bleeding disorders.Citation36 A very recent discovery shows that the product of the C90rf72 gene, which harbors expanded GGGCCC repeats in non-coding regions in many cases of amyotrophic lateral sclerosis (ALS) leading to underexpression of the gene product, is a GEF for Rab8 and Rab39. These Rabs are involved in autophagy, which is defective in ALS.Citation128,129
Many other mutations are known or can be assumed to effect prenylation, delivery and/or retrieval of Rabs from membranes. For instance, Hermansky-Pudlak syndrome is also caused by a mutation of RabGGTase presumably causing deficient prenylation.Citation130 Several mutations within the gene coding for REP-1 cause Choroideremia, an X-chromosomal disease causing degeneration of the retina and resulting night blindness as well as loss of peripheral vision,Citation131,132 and another X-chromosomal mutation within the gene for GDI-1 is known to cause X-linked non-specific mental retardation.Citation133 Together, these severe impacts on human physiology illustrate the importance of regulated membrane cycling of Rab proteins.
Outlook
Even though further GEFs and GAPs still will (and need to be) identified, many have been found and biochemically and structurally characterized in vitro. Similarly, the structures of GDI and REP in complex with Rabs have been reported as discussed above. Together, these insights give a rough scheme of the regulation of vesicular transport and factors involved. However, comparatively little is still known about the situation in vivo, especially regarding the localization and the specificity of these regulatory proteins toward Rabs as well as regarding their spatiotemporal regulation by additional proteins and posttranslational modifications. Our knowledge of the underlying protein interaction networks therefore still needs to be expanded to finally understand the fine details of the different Rab regulated vesicular transport processes in physiological and pathological states.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to ask for understanding in all instances where reviews were cited instead of the original work.
Funding
The Max Planck Society and the SFB642 (project A4) are acknowledged for financial support.
References
- Barr F, Lambright DG. Rab GEFs and GAPs. Curr Opin Cell Biol 2010; 22(4):461-70; PMID:20466531; http://dx.doi.org/10.1016/j.ceb.2010.04.007
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 2009; 10(8):513-25; PMID:19603039; http://dx.doi.org/10.1038/nrm2728
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 1980; 21(1):205-15; PMID:6996832; http://dx.doi.org/10.1016/0092-8674(80)90128-2
- Salminen A, Novick PJ. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell 1987; 49(4):527-38; PMID:3552249; http://dx.doi.org/10.1016/0092-8674(87)90455-7
- Schmitt HD, Wagner P, Pfaff E, Gallwitz D. The ras-related YPT1 gene product in yeast: a GTP-binding protein that might be involved in microtubule organization. Cell 1986; 47(3):401-12; PMID:3094963; http://dx.doi.org/10.1016/0092-8674(86)90597-0
- Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem 1996; 65:241-69; PMID:8811180; http://dx.doi.org/10.1146/annurev.bi.65.070196.001325
- Andres DA, Seabra MC, Brown MS, Armstrong SA, Smeland TE, Cremers FP, Goldstein JL. cDNA cloning of component A of Rab geranylgeranyl transferase and demonstration of its role as a Rab escort protein. Cell 1993; 73(6):1091-9; PMID:8513495; http://dx.doi.org/10.1016/0092-8674(93)90639-8
- Alexandrov K, Simon I, Yurchenko V, Iakovenko A, Rostkova E, Scheidig AJ, Goody RS. Characterization of the ternary complex between Rab7, REP-1 and Rab geranylgeranyl transferase. Eur J Biochem 1999; 265(1):160-70; PMID:10491170; http://dx.doi.org/10.1046/j.1432-1327.1999.00699.x
- Alexandrov K, Horiuchi H, Steele-Mortimer O, Seabra MC, Zerial M. Rab escort protein-1 is a multifunctional protein that accompanies newly prenylated rab proteins to their target membranes. EMBO J 1994; 13(22):5262-73; PMID:7957092
- Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem 1994; 140(1):1-22; PMID:7877593; http://dx.doi.org/10.1007/BF00928361
- Oesterlin LK, Pylypenko O, Goud B. Effectors of Rab GTPases: Rab Binding Specificity and Their Role in Coordination of Rab Function and Localization, in Ras Superfamily Small G Proteins: Biology and Mechanisms 2, A. Wittinghofer, Editor. 2014, Springer International Publishing, 39-66. http://dx.doi.org/10.1007/978-3-319-07761-1
- Pfeffer S, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol 2004; 5(11):886-96; PMID:15520808; http://dx.doi.org/10.1038/nrm1500
- Milburn MV, Tong L, deVos AM, Brünger A, Yamaizumi Z, Nishimura S, Kim SH. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science 1990; 247(4945):939-45; PMID:2406906; http://dx.doi.org/10.1126/science.2406906
- Pai EF, Kabsch W, Krengel U, Holmes KC, John J, Wittinghofer A. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature 1989; 341(6239):209-14; PMID:2476675; http://dx.doi.org/10.1038/341209a0
- Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science 2001; 294(5545):1299-304; PMID:11701921; http://dx.doi.org/10.1126/science.1062023
- Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature 1991; 349(6305):117-27; PMID:1898771; http://dx.doi.org/10.1038/349117a0
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1982; 1(8):945-51; PMID:6329717
- Prive GG, Milburn MV, Tong L, de Vos AM, Yamaizumi Z, Nishimura S, Kim SH. X-ray crystal structures of transforming p21 ras mutants suggest a transition-state stabilization mechanism for GTP hydrolysis. Proc Natl Acad Sci U S A 1992; 89(8):3649-53; PMID:1565661; http://dx.doi.org/10.1073/pnas.89.8.3649
- Rensland H, John J, Linke R, Simon I, Schlichting I, Wittinghofer A, Goody RS. Substrate and product structural requirements for binding of nucleotides to H-ras p21: the mechanism of discrimination between guanosine and adenosine nucleotides. Biochemistry 1995; 34(2):593-9; PMID:7819254; http://dx.doi.org/10.1021/bi00002a026
- Pereira-Leal JB, Seabra MC. The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J Mol Biol 2000; 301(4):1077-87; PMID:10966806; http://dx.doi.org/10.1006/jmbi.2000.4010
- Pereira-Leal JB, Seabra MC. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol 2001; 313(4):889-901; PMID:11697911; http://dx.doi.org/10.1006/jmbi.2001.5072
- Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 2013; 93(1):269-309; PMID:23303910; http://dx.doi.org/10.1152/physrev.00003.2012
- Hama H, Tall GG, Horazdovsky BF. Vps9p is a guanine nucleotide exchange factor involved in vesicle-mediated vacuolar protein transport. J Biol Chem 1999; 274(21):15284-91; PMID:10329739; http://dx.doi.org/10.1074/jbc.274.21.15284
- Allaire PD, Marat AL, Dall'Armi C, Di Paolo G, McPherson PS, Ritter B. The Connecdenn DENN domain: a GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol Cell 2010; 37(3):370-82; PMID:20159556; http://dx.doi.org/10.1016/j.molcel.2009.12.037
- Koch D, et al. A pull-down procedure for the identification of unknown GEFs for small GTPases. Small GTPases 2016:1-14. PMID: 26918858; http://dx.doi.org/10.1080/21541248.2016.1156803
- Carney DS, Davies BA, Horazdovsky BF. Vps9 domain-containing proteins: activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol 2006; 16(1):27-35; PMID:16330212; http://dx.doi.org/10.1016/j.tcb.2005.11.001
- Burd CG, Mustol PA, Schu PV, Emr SD. A yeast protein related to a mammalian Ras-binding protein, Vps9p, is required for localization of vacuolar proteins. Mol Cell Biol 1996; 16(5):2369-77; PMID:8628304; http://dx.doi.org/10.1128/MCB.16.5.2369
- Horiuchi H, Lippé R, McBride HM, Rubino M, Woodman P, Stenmark H, Rybin V, Wilm M, Ashman K, Mann M. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell 1997; 90(6):1149-59; PMID:9323142; http://dx.doi.org/10.1016/S0092-8674(00)80380-3
- Marat AL, Dokainish H, McPherson PS. DENN domain proteins: regulators of Rab GTPases. J Biol Chem 2011; 286(16):13791-800; PMID:21330364; http://dx.doi.org/10.1074/jbc.R110.217067
- Levivier E, et al. uDENN, DENN, and dDENN: Indissociable domains in Rab and MAP kinases signaling pathways. Biochem Biophys Res Commun 2001; 287(3):688-695; PMID:11563850; http://dx.doi.org/10.1006/bbrc.2001.5652
- Levine TP, Goud B, Souchet M, Calmels TP, Mornon JP, Callebaut I. Discovery of new Longin and Roadblock domains that form platforms for small GTPases in Ragulator and TRAPP-II. Small GTPases 2013; 4(2):62-9; PMID:23511850; http://dx.doi.org/10.4161/sgtp.24262
- Wu X, Bradley MJ, Cai Y, Kümmel D, De La Cruz EM, Barr FA, Reinisch KM. Insights regarding guanine nucleotide exchange from the structure of a DENN-domain protein complexed with its Rab GTPase substrate. Proc Natl Acad Sci U S A 2011; 108(46):18672-7; PMID:22065758; http://dx.doi.org/10.1073/pnas.1110415108
- Jones S, Newman C, Liu F, Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell 2000; 11(12):4403-4411; PMID:11102533; http://dx.doi.org/10.1091/mbc.11.12.4403
- Nordmann M, Cabrera M, Perz A, Bröcker C, Ostrowicz C, Engelbrecht-Vandré S, Ungermann C. The mon1-Ccz1 complex is the GEF of the late endosomal rab7 homolog Ypt7. Curr Biol 2010; 20(18):1654-1659; PMID:20797862; http://dx.doi.org/10.1016/j.cub.2010.08.002
- Pusapati GV, Luchetti G, Pfeffer SR. Ric1-Rgp1 complex is a guanine nucleotide exchange factor for the late golgi Rab6A GTPase and an effector of the medial golgi Rab33B GTPase. J Biol Chem 2012; 287(50):42129-37; PMID:23091056; http://dx.doi.org/10.1074/jbc.M112.414565
- Gerondopoulos A, Langemeyer L, Liang JR, Linford A, Barr FA. BLOC-3 mutated in hermansky-pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr Biol 2012; 22(22):2135-9; PMID:23084991; http://dx.doi.org/10.1016/j.cub.2012.09.020
- Hattula K, Furuhjelm J, Arffman A, Peränen J. A Rab8-specific GDP/GTP exchange factor is involved in actin remodeling and polarized membrane transport. Mol Biol Cell 2002; 13(9):3268-80; PMID:12221131; http://dx.doi.org/10.1091/mbc.E02-03-0143
- WalchSolimena C, Collins RN, Novick PJ. Sec 2p mediates nucleotide exchange on Sec 4p and is involved in polarized delivery of post-Golgi vesicles. J Cell Biol 1997; 137(7):1495-509; PMID:9199166; http://dx.doi.org/10.1083/jcb.137.7.1495
- Sakaguchi A, Sato M, Sato K, Gengyo-Ando K, Yorimitsu T, Nakai J, Hara T, Sato K. Sato K5. REI-1 is a guanine nucleotide exchange factor regulating RAB-11 localization and function in C. elegans Embryos. Dev Cell 2015; 35(2):211-21; PMID:26506309; http://dx.doi.org/10.1016/j.devcel.2015.09.013
- Lippe R, Miaczynska M, Rybin V, Runge A, Zerial M. Functional synergy between Rab5 effector Rabaptin-5 and exchange factor Rabex-5 when physically associated in a complex. Mol Biol Cell 2001; 12(7):2219-28; PMID:11452015; http://dx.doi.org/10.1091/mbc.12.7.2219
- Nottingham RM, Pfeffer SR. Defining the boundaries: Rab GEFs and GAPs. Proc Natl Acad Sci U S A 2009; 106(34):14185-6; PMID:19706500; http://dx.doi.org/10.1073/pnas.0907725106
- Ortiz D, Medkova M, Walch-Solimena C, Novick P. Ypt32 recruits the Sec 4p guanine nucleotide exchange factor, Sec 2p, to secretory vesicles; evidence for a Rab cascade in yeast. J Cell Biol 2002; 157(6):1005-15; PMID:12045183; http://dx.doi.org/10.1083/jcb.200201003
- Blumer J, Rey J, Dehmelt L, Mazel T, Wu YW, Bastiaens P, Goody RS, Itzen A. RabGEFs are a major determinant for specific Rab membrane targeting. J Cell Biol 2013; 200(3):287-300; PMID:23382462; http://dx.doi.org/10.1083/jcb.201209113
- Li F, Yi L, Zhao L, Itzen A, Goody RS, Wu YW. The role of the hypervariable C-terminal domain in Rab GTPases membrane targeting. Proc Natl Acad Sci U S A 2014; 111(7):2572-7; PMID:24550285; http://dx.doi.org/10.1073/pnas.1313655111
- Stein MP, Müller MP, Wandinger-Ness A. Bacterial pathogens commandeer Rab GTPases to establish intracellular niches. Traffic 2012; 13(12):1565-88; PMID:22901006; http://dx.doi.org/10.1111/tra.12000
- Heidtman M, Chen EJ, Moy MY, Isberg RR. Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cell Microbiol 2009; 11(2):230-48; PMID:19016775; http://dx.doi.org/10.1111/j.1462-5822.2008.01249.x
- Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol 2006; 8(9):971-7; PMID:16906144; http://dx.doi.org/10.1038/ncb1463
- Schoebel S, Oesterlin LK, Blankenfeldt W, Goody RS, Itzen A. RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity. Mol Cell 2009; 36(6):1060-72; PMID:20064470; http://dx.doi.org/10.1016/j.molcel.2009.11.014
- Mukherjee K, Parashuraman S, Raje M, Mukhopadhyay A. SopE acts as an Rab5-specific nucleotide exchange factor and recruits non-prenylated Rab5 on Salmonella-containing phagosomes to promote fusion with early endosomes. J Biol Chem 2001; 276(26):23607-15; PMID:11316807; http://dx.doi.org/10.1074/jbc.M101034200
- Itzen A, Pylypenko O, Goody RS, Alexandrov K, Rak A. Nucleotide exchange via local protein unfolding–structure of Rab8 in complex with MSS4. EMBO J 2006; 25(7):1445-55; PMID:16541104; http://dx.doi.org/10.1038/sj.emboj.7601044
- Dong G, Medkova M, Novick P, Reinisch KM. A catalytic coiled coil: structural insights into the activation of the Rab GTPase Sec 4p by Sec 2p. Mol Cell 2007; 25(3):455-62; PMID:17289591; http://dx.doi.org/10.1016/j.molcel.2007.01.013
- Sato Y, Fukai S, Ishitani R, Nureki O. Crystal structure of the Sec 4p.Sec 2p complex in the nucleotide exchanging intermediate state. Proc Natl Acad Sci U S A 2007; 104(20):8305-10; PMID:17488829; http://dx.doi.org/10.1073/pnas.0701550104
- Delprato A, Lambright DG. Structural basis for Rab GTPase activation by VPS9 domain exchange factors. Nat Struct Mol Biol 2007; 14(5):406-12; PMID:17450153; http://dx.doi.org/10.1038/nsmb1232
- Cai Y, Chin HF, Lazarova D, Menon S, Fu C, Cai H, Sclafani A, Rodgers DW, De La Cruz EM, Ferro-Novick S. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell 2008; 133(7):1202-13; PMID:18585354; http://dx.doi.org/10.1016/j.cell.2008.04.049
- Guo Z, Hou X, Goody RS, Itzen A. Intermediates in the guanine nucleotide exchange reaction of Rab8 protein catalyzed by guanine nucleotide exchange factors Rabin8 and GRAB. J Biol Chem 2013; 288(45):32466-74; PMID:24072714; http://dx.doi.org/10.1074/jbc.M113.498329
- Zhang Z, Zhang T, Wang S, Gong Z, Tang C, Chen J, Ding J. Molecular mechanism for Rabex-5 GEF activation by Rabaptin-5. Elife 2014; 3. PMID:24957337; http://dx.doi.org/10.7554/eLife.02687
- Uejima T, Ihara K, Goh T, Ito E, Sunada M, Ueda T, Nakano A, Wakatsuki S. GDP-bound and nucleotide-free intermediates of the guanine nucleotide exchange in the Rab5.Vps9 system. J Biol Chem 2010; 285(47):36689-97; PMID:20833725; http://dx.doi.org/10.1074/jbc.M110.152132
- Langemeyer L, Nunes Bastos R, Cai Y, Itzen A, Reinisch KM, Barr FA. Diversity and plasticity in Rab GTPase nucleotide release mechanism has consequences for Rab activation and inactivation. Elife 2014; 3:e01623; PMID:24520163; http://dx.doi.org/10.7554/eLife.01623
- Esters H, Alexandrov K, Constantinescu AT, Goody RS, Scheidig AJ. High-resolution crystal structure of S. cerevisiae Ypt51(DeltaC15)-GppNHp, a small GTP-binding protein involved in regulation of endocytosis. J Mol Biol 2000; 298(1):111-21; PMID:10756108; http://dx.doi.org/10.1006/jmbi.2000.3645
- Huber SK, Scheidig AJ. High resolution crystal structures of human Rab4a in its active and inactive conformations. FEBS Lett 2005; 579(13):2821-9; PMID:15907487; http://dx.doi.org/10.1016/j.febslet.2005.04.020
- Rensland H, Lautwein A, Wittinghofer A, Goody RS. Is There a Rate-Limiting Step before Gtp Cleavage by H-Ras P21. Biochemistry 1991; 30(46):11181-85; PMID:1932038; http://dx.doi.org/10.1021/bi00110a023
- Simon I, Zerial M, Goody RS. Kinetics of interaction of Rab5 and Rab7 with nucleotides and magnesium ions. J Biol Chem 1996; 271(34):20470-8; PMID:8702787; http://dx.doi.org/10.1074/jbc.271.34.20470
- Fukuda M. TBC proteins: GAPs for mammalian small GTPase Rab? Biosci Rep 2011; 31(3):159-68; PMID:21250943; http://dx.doi.org/10.1042/BSR20100112
- Strom M, Vollmer P, Tan TJ, Gallwitz D. A yeast GTPase-activating protein that interacts specifically with a member of the Ypt/Rab family. Nature 1993; 361(6414):736-9; PMID:8441469; http://dx.doi.org/10.1038/361736a0
- Nagano F, Sasaki T, Fukui K, Asakura T, Imazumi K, Takai Y. Molecular cloning and characterization of the noncatalytic subunit of the Rab3 subfamily-specific GTPase-activating protein. J Biol Chem 1998; 273(38):24781-5; PMID:9733780; http://dx.doi.org/10.1074/jbc.273.38.24781
- Frasa MA, Koessmeier KT, Ahmadian MR, Braga VM. Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat Rev Mol Cell Biol 2012; 13(2):67-73; PMID:22251903
- Rivera-Molina FE, Novick PJ. A Rab GAP cascade defines the boundary between two Rab GTPases on the secretory pathway. Proc Natl Acad Sci U S A 2009; 106(34):14408-13; PMID:19666511; http://dx.doi.org/10.1073/pnas.0906536106
- Suda Y, Kurokawa K, Hirata R, Nakano A. Rab GAP cascade regulates dynamics of Ypt6 in the Golgi traffic. Proc Natl Acad Sci U S A 2013; 110(47):18976-81; PMID:24194547; http://dx.doi.org/10.1073/pnas.1308627110
- Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 2007; 450(7168):365-9; PMID:17952054; http://dx.doi.org/10.1038/nature06336
- Mihai Gazdag E, Streller A, Haneburger I, Hilbi H, Vetter IR, Goody RS, Itzen A. Mechanism of Rab1b deactivation by the Legionella pneumophila GAP LepB. EMBO Rep 2013; 14(2):199-205; http://dx.doi.org/10.1038/embor.2012.211
- Rak A, Fedorov R, Alexandrov K, Albert S, Goody RS, Gallwitz D, Scheidig AJ. Crystal structure of the GAP domain of Gyp1p: first insights into interaction with Ypt/Rab proteins. Embo J 2000; 19(19):5105-13; http://dx.doi.org/10.1093/emboj/19.19.5105
- Pan XJ, Eathiraj S, Munson M, Lambright DG. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature 2006; 442(7100):303-6; PMID:16855591; http://dx.doi.org/10.1038/nature04847
- Goody RS, Rak A, Alexandrov K. The structural and mechanistic basis for recycling of Rab proteins between membrane compartments. Cell Mol Life Sci 2005; 62(15):1657-70; PMID:15924270; http://dx.doi.org/10.1007/s00018-005-4486-8
- Kamiya Y, Sakurai A, Tamura S, Takahashi N. Structure of rhodotorucine A, a novel lipopeptide, inducing mating tube formation in Rhodosporidium toruloides. Biochem Biophys Res Commun 1978; 83(3):1077-83; PMID:708426; http://dx.doi.org/10.1016/0006-291X(78)91505-X
- Casey PJ, Solski PA, Der CJ, Buss JE. P21ras is modified by a farnesyl isoprenoid. Proc Natl Acad Sci U S A 1989; 86(21):8323-7; PMID:2682646; http://dx.doi.org/10.1073/pnas.86.21.8323
- Hancock JF, Magee AI, Childs JE, Marshall CJ. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell 1989; 57(7):1167-77; PMID:2661017; http://dx.doi.org/10.1016/0092-8674(89)90054-8
- Schafer WR, Kim R, Sterne R, Thorner J, Kim SH, Rine J. Genetic and Pharmacological Suppression of Oncogenic Mutations in Ras Genes of Yeast and Humans. Science 1989; 245(4916):379-85; PMID:2569235; http://dx.doi.org/10.1126/science.2569235
- Farnsworth CC, Kawata M, Yoshida Y, Takai Y, Gelb MH, Glomset JA. C terminus of the small GTP-binding protein smg p25A contains two geranylgeranylated cysteine residues and a methyl ester. Proc Natl Acad Sci U S A 1991; 88(14):6196-200; PMID:1906176; http://dx.doi.org/10.1073/pnas.88.14.6196
- Khosravi-Far R, Clark GJ, Abe K, Cox AD, McLain T, Lutz RJ, Sinensky M, Der CJ. Ras (CXXX) and Rab (CC/CXC) prenylation signal sequences are unique and functionally distinct. J Biol Chem 1992; 267(34):24363-8; PMID:1332953
- Reiss Y, Goldstein JL, Seabra MC, Casey PJ, Brown MS. Inhibition of purified P21ras farnesyl - protein transferase by Cys-Aax tetrapeptides. Cell 1990; 62(1):81-88; PMID:2194674; http://dx.doi.org/10.1016/0092-8674(90)90242-7
- Seabra MC, Reiss Y, Casey PJ, Brown MS, Goldstein JL. Protein farnesyltransferase and geranylgeranyltransferase share a common alpha-subunit. Cell 1991; 65(3):429-34; PMID:2018975; http://dx.doi.org/10.1016/0092-8674(91)90460-G
- Seabra MC, Goldstein JL, Südhof TC, Brown MS. Rab geranylgeranyl transferase. A multisubunit enzyme that prenylates GTP-binding proteins terminating in Cys-X-Cys or Cys-Cys. J Biol Chem 1992; 267(20):14497-503; PMID:1321151
- Smeland TE, Seabra MC, Goldstein JL, Brown MS. Geranylgeranylated rab proteins terminating in Cys-Ala-Cys, but not Cys-Cys, are carboxyl-methylated by bovine brain membranes in-vitro. Proc Natl Acad Sci U S A 1994; 91(22):10712-6; PMID:7938016; http://dx.doi.org/10.1073/pnas.91.22.10712
- Thoma NH, Niculae A, Goody RS, Alexandrov K. Double prenylation by RabGGTase can proceed without dissociation of the mono-prenylated intermediate. J Biol Chem 2001; 276(52):48631-6; PMID:11591706; http://dx.doi.org/10.1074/jbc.M106470200
- Thoma NH, Iakovenko A, Goody RS, Alexandrov K. Phosphoisoprenoids modulate association of Rab geranylgeranyltransferase with REP-1. J Biol Chem 2001; 276(52):48637-43; PMID:11675392; http://dx.doi.org/10.1074/jbc.M108241200
- Thoma NH, Iakovenko A, Kalinin A, Waldmann H, Goody RS, Alexandrov K. Allosteric regulation of substrate binding and product release in geranylgeranyltransferase type II. Biochemistry 2001; 40(1):268-274; PMID:11141079; http://dx.doi.org/10.1021/bi002034p
- Rak A, Pylypenko O, Niculae A, Pyatkov K, Goody RS, Alexandrov K. Structure of the Rab7 : REP-1 complex: Insights into the mechanism of rab prenylation and choroideremia disease. Cell 2004; 117(6):749-60; PMID:15186776; http://dx.doi.org/10.1016/j.cell.2004.05.017
- Pylypenko O, Rak A, Reents R, Niculae A, Sidorovitch V, Cioaca MD, Bessolitsyna E, Thomä NH, Waldmann H, Schlichting I. Structure of Rab escort protein-1 in complex with Rab geranylgeranyltransferase. Mol Cell 2003; 11(2):483-94; PMID:12620235; http://dx.doi.org/10.1016/S1097-2765(03)00044-3
- Leung KF, Baron R, Seabra MC. Thematic review series: lipid posttranslational modifications. geranylgeranylation of Rab GTPases. J Lipid Res 2006; 47(3):467-75; PMID:16401880; http://dx.doi.org/10.1194/jlr.R500017-JLR200
- Waldherr M, Ragnini A, Schweyer RJ, Boguski MS. MRS6–yeast homologue of the choroideraemia gene. Nat Genet 1993; 3(3):193-4; PMID:8387377; http://dx.doi.org/10.1038/ng0393-193
- Sasaki T, Kikuchi A, Araki S, Hata Y, Isomura M, Kuroda S, Takai Y. Purification and characterization from bovine brain cytosol of a protein that inhibits the dissociation of GDP from and the subsequent binding of GTP to smg p25A, a ras p21-like GTP-binding protein. J Biol Chem 1990; 265(4):2333-7; PMID:2105320
- Wu YW, Tan KT, Waldmann H, Goody RS, Alexandrov K. Interaction analysis of prenylated Rab GTPase with Rab escort protein and GDP dissociation inhibitor explains the need for both regulators. Proc Natl Acad Sci U S A 2007; 104(30):12294-9; PMID:17640890; http://dx.doi.org/10.1073/pnas.0701817104
- Alexandrov K, Simon I, Iakovenko A, Holz B, Goody RS, Scheidig AJ. Moderate discrimination of REP-1 between Rab7-GDP and Rab7-GTP arises from a difference of an order of magnitude in dissociation rates. Febs Lett 1998; 425(3):460-4; PMID:9563513; http://dx.doi.org/10.1016/S0014-5793(98)00290-7
- Schalk I, Zeng K, Wu SK, Stura EA, Matteson J, Huang M, Tandon A, Wilson IA, Balch WE. Structure and mutational analysis of Rab GDP-dissociation inhibitor. Nature 1996; 381(6577):42-8; PMID:8609986; http://dx.doi.org/10.1038/381042a0
- An Y, Shao Y, Alory C, Matteson J, Sakisaka T, Chen W, Gibbs RA, Wilson IA, Balch WE. Geranylgeranyl switching regulates GDI-Rab GTPase recycling. Structure 2003; 11(3):347-57; PMID:12623022; http://dx.doi.org/10.1016/S0969-2126(03)00034-0
- Rak A, Pylypenko O, Durek T, Watzke A, Kushnir S, Brunsveld L, Waldmann H, Goody RS, Alexandrov K. Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase. Science 2003; 302(5645):646-50; PMID:14576435; http://dx.doi.org/10.1126/science.1087761
- Pylypenko O, Rak A, Durek T, Kushnir S, Dursina BE, Thomae NH, Constantinescu AT, Brunsveld L, Watzke A, Waldmann H. Structure of doubly prenylated Ypt1:GDI complex and the mechanism of GDI-mediated Rab recycling. EMBO J 2006; 25(1):13-23; PMID:16395334; http://dx.doi.org/10.1038/sj.emboj.7600921
- Chavrier P, Gorvel JP, Stelzer E, Simons K, Gruenberg J, Zerial M. Hypervariable C-terminal domain of Rab proteins acts as a targeting signal. Nature 1991; 353(6346):769-72; PMID:1944536; http://dx.doi.org/10.1038/353769a0
- Stenmark H, Valencia A, Martinez O, Ullrich O, Goud B, Zerial M. Distinct structural elements of Rab5 define its functional specificity. Embo J 1994; 13(3):575-83; PMID:8313902
- Ali BR, Wasmeier C, Lamoreux L, Strom M, Seabra MC. Multiple regions contribute to membrane targeting of Rab GTPases. J Cell Sci 2004; 117(26):6401-12; PMID:15561774; http://dx.doi.org/10.1242/jcs.01542
- DiracSvejstrup AB, Sumizawa T, Pfeffer SR. Identification of a GDI displacement factor that releases endosomal Rab GTPases from Rab-GDI. Embo J 1997; 16(3):465-72; PMID:9034329; http://dx.doi.org/10.1093/emboj/16.3.465
- Sivars U, Aivazian D, Pfeffer SR. Yip3 catalyses the dissociation of endosomal Rab-GDI complexes. Nature 2003; 425(6960):856-9; PMID:14574414; http://dx.doi.org/10.1038/nature02057
- Wu YW, Oesterlin LK, Tan KT, Waldmann H, Alexandrov K, Goody RS. Membrane targeting mechanism of Rab GTPases elucidated by semisynthetic protein probes. Nat Chem Biol 2010; 6(7):534-40; PMID:20512138; http://dx.doi.org/10.1038/nchembio.386
- Bailly E, McCaffrey M, Touchot N, Zahraoui A, Goud B, Bornens M. Phosphorylation of two small GTP-binding proteins of the Rab family by p34cdc2. Nature 1991; 350(6320):715-8; PMID:1902553; http://dx.doi.org/10.1038/350715a0
- Vandersluijs P, Hull M, Huber LA, Mâle P, Goud B, Mellman I. Reversible phosphorylation dephosphorylation determines the localization of Rab4 during the cell-cycle. Embo J 1992; 11(12):4379-89; PMID:1425574
- Walther DJ, Peter JU, Winter S, Höltje M, Paulmann N, Grohmann M, Vowinckel J, Alamo-Bethencourt V, Wilhelm CS, Ahnert-Hilger G, et al. Serotonylation of small GTPases is a signal transduction pathway that triggers platelet alpha-granule release. Cell 2003; 115(7):851-62; PMID:14697203; http://dx.doi.org/10.1016/S0092-8674(03)01014-6
- Paulmann N, Grohmann M, Voigt JP, Bert B, Vowinckel J, Bader M, Skelin M, Jevsek M, Fink H, Rupnik M, et al. Intracellular serotonin modulates insulin secretion from pancreatic beta-cells by protein serotonylation. PLoS Biol 2009; 7(10):e1000229; PMID:19859528; http://dx.doi.org/10.1371/journal.pbio.1000229
- Lai YC, Kondapalli C, Lehneck R, Procter JB, Dill BD, Woodroof HI, Gourlay R, Peggie M, Macartney TJ, Corti O, et al. Phosphoproteomic screening identifies Rab GTPases as novel downstream targets of PINK1. EMBO J 2015; 34(22):2840-61; PMID:26471730; http://dx.doi.org/10.15252/embj.201591593
- Steger M, Tonelli F, Ito G, Davies P, Trost M, Vetter M, Wachter S, Lorentzen E, Duddy G, Wilson S, et al. Phosphoproteomics reveals that Parkinson's disease kinase LRRK2 regulates a subset of Rab GTPases. Elife 2016; 5:e12813; PMID:26824392; http://dx.doi.org/10.7554/eLife.12813
- Levin RS, Hertz NT, Burlingame AL, Shokat KM, Mukherjee S. Innate immunity kinase TAK1 phosphorylates Rab1 on a hotspot for posttranslational modifications by host and pathogen. Proc Natl Acad Sci U S A 2016; 113:E4776-83; PMID:27482120
- Peck GR, Chavez JA, Roach WG, Budnik BA, Lane WS, Karlsson HK, Zierath JR, Lienhard GE. Insulin-stimulated phosphorylation of the Rab GTPase-activating protein TBC1D1 regulates GLUT4 translocation. J Biol Chem 2009; 284(44):30016-23; PMID:19740738; http://dx.doi.org/10.1074/jbc.M109.035568
- Sano H, Kane S, Sano E, Mîinea CP, Asara JM, Lane WS, Garner CW, Lienhard GE. Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J Biol Chem 2003; 278(17):14599-602; PMID:12637568; http://dx.doi.org/10.1074/jbc.C300063200
- Liu W, Yuen EY, Yan Z. The stress hormone corticosterone increases synaptic alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors via serum- and glucocorticoid-inducible kinase (SGK) regulation of the GDI-Rab4 complex. J Biol Chem 2010; 285(9):6101-8; PMID:20051515; http://dx.doi.org/10.1074/jbc.M109.050229
- Steele-Mortimer O, Gruenberg J, Clague MJ. Phosphorylation of GDI and membrane cycling of rab proteins. FEBS Lett 1993; 329(3):313-8; PMID:8365473; http://dx.doi.org/10.1016/0014-5793(93)80244-O
- Kulasekaran G, Nossova N, Marat AL, Lund I, Cremer C, Ioannou MS, McPherson PS. Phosphorylation-dependent regulation of connecdenn/DENND1 guanine nucleotide exchange factors. J Biol Chem 2015; 290(29):17999-8008; PMID:26055712; http://dx.doi.org/10.1074/jbc.M115.636712
- Muller MP, Peters H, Blümer J, Blankenfeldt W, Goody RS, Itzen A. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 2010; 329(5994):946-9; PMID:20651120; http://dx.doi.org/10.1126/science.1192276
- Neunuebel MR, Chen Y, Gaspar AH, Backlund PS Jr, Yergey A, Machner MP. De-AMPylation of the small GTPase Rab1 by the pathogen legionella pneumophila. Science 2011; 333(6041):453-6; PMID:21680813; http://dx.doi.org/10.1126/science.1207193
- Tan YH, Luo ZQ. Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature 2011; 475(7357):506-U102; PMID:21734656; http://dx.doi.org/10.1038/nature10307
- Müller MP, Shkumatov AV, Oesterlin LK, Schoebel S, Goody PR, Goody RS, Itzen A. Characterization of enzymes from Legionella pneumophila involved in reversible adenylylation of Rab1 protein. J Biol Chem 2012; 287(42):35036-46; PMID:22872634; http://dx.doi.org/10.1074/jbc.M112.396861
- Goody PR, Heller K, Oesterlin LK, Müller MP, Itzen A, Goody RS. Reversible phosphocholination of Rab proteins by Legionella pneumophila effector proteins. EMBO J 2012; 31(7):1774-84; PMID:22307087; http://dx.doi.org/10.1038/emboj.2012.16
- Mukherjee S, Liu X, Arasaki K, McDonough J, Galán JE, Roy CR. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 2011; 477(7362):103-6; PMID:21822290; http://dx.doi.org/10.1038/nature10335
- Tan Y, Arnold RJ, Luo ZQ. Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc Natl Acad Sci U S A 2011; 108(52):21212-7; PMID:22158903; http://dx.doi.org/10.1073/pnas.1114023109
- Oesterlin LK, Goody RS, Itzen A. Posttranslational modifications of Rab proteins cause effective displacement of GDP dissociation inhibitor. Proc Natl Acad Sci U S A 2012; 109(15):5621-6; PMID:22411835; http://dx.doi.org/10.1073/pnas.1121161109
- Seixas E, Barros M, Seabra MC, Barral DC. Rab and Arf proteins in genetic diseases. Traffic 2013; 14(8):871-85; PMID:23565987; http://dx.doi.org/10.1111/tra.12072
- Aligianis IA, Johnson CA, Gissen P, Chen D, Hampshire D, Hoffmann K, Maina EN, Morgan NV, Tee L, Morton J, et al. Mutations of the catalytic subunit of RAB3GAP cause Warburg Micro syndrome. Nat Genet 2005; 37(3):221-3; PMID:15696165; http://dx.doi.org/10.1038/ng1517
- Liegel RP, Handley MT, Ronchetti A, Brown S, Langemeyer L, Linford A, Chang B, Morris-Rosendahl DJ, Carpanini S, Posmyk R, et al. Loss-of-function mutations in TBC1D20 cause cataracts and male infertility in blind sterile mice and Warburg micro syndrome in humans. Am J Hum Genet 2013; 93(6):1001-14; PMID:24239381; http://dx.doi.org/10.1016/j.ajhg.2013.10.011
- Aligianis IA, Morgan NV, Mione M, Johnson CA, Rosser E, Hennekam RC, Adams G, Trembath RC, Pilz DT, Stoodley N, et al. Mutation in Rab3 GTPase-activating protein (RAB3GAP) noncatalytic subunit in a kindred with Martsolf syndrome. Am J Hum Genet 2006; 78(4):702-7; PMID:16532399; http://dx.doi.org/10.1086/502681
- Corbier C, Sellier C. C9ORF72 is a GDP/GTP exchange factor for Rab8 and Rab39 and regulates autophagy. Small GTPases 2016; 5:1-6. PMID:27494456; http://dx.doi.org/10.1080/21541248.2016.1212688
- Ciura S, Sellier C, Campanari ML, Charlet-Berguerand N, Kabashi E. The most prevalent genetic cause of ALS-FTD, C9orf72 synergizes the toxicity of ATXN2 intermediate polyglutamine repeats through the autophagy pathway. Autophagy 2016; 12(8):1406-8; PMID:27245636; http://dx.doi.org/10.1080/15548627.2016.1189070
- Coxon FP, Taylor A, Stewart CA, Baron R, Seabra MC, Ebetino FH, Rogers MJ. The gunmetal mouse reveals Rab geranylgeranyl transferase to be the major molecular target of phosphonocarboxylate analogues of bisphosphonates. Bone 2011; 49(1):111-21; PMID:21419243; http://dx.doi.org/10.1016/j.bone.2011.03.686
- Seabra MC. New insights into the pathogenesis of choroideremia: A tale of two REPs. Ophthalmic Genetics 1996; 17(2):43-46; PMID:8832719; http://dx.doi.org/10.3109/13816819609057869
- van den Hurk JA, Schwartz M, van Bokhoven H, van de Pol TJ, Bogerd L, Pinckers AJ, Bleeker-Wagemakers EM, Pawlowitzki IH, Rüther K, Ropers HH, et al. Molecular basis of choroideremia (CHM): mutations involving the Rab escort protein-1 (REP-1) gene. Hum Mutat 1997; 9(2):110-7; PMID:9067750; http://dx.doi.org/10.1002/(SICI)1098-1004(1997)9:2%3c110::AID-HUMU2%3e3.0.CO;2-D
- D'Adamo P, Menegon A, Lo Nigro C, Grasso M, Gulisano M, Tamanini F, Bienvenu T, Gedeon AK, Oostra B, Wu SK, et al. Mutations in GDI1 are responsible for X-linked non-specific mental retardation. Nat Genet 1998; 19(2):134-9; PMID:9620768; http://dx.doi.org/10.1038/487
- Klopper TH, Kienle N, Fasshauer D, Munro S. Untangling the evolution of Rab G proteins: implications of a comprehensive genomic analysis. BMC Biol 2012; 10:71; PMID:22873208; http://dx.doi.org/10.1186/1741-7007-10-71
- Yoshimura S, Gerondopoulos A, Linford A, Rigden DJ, Barr FA. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J Cell Biol 2010; 191(2):367-81; PMID:20937701; http://dx.doi.org/10.1083/jcb.201008051
- Ishida M, Oguchi ME, Fukuda M. Multiple types of guanine nucleotide exchange factors (GEFs) for Rab small GTPases. Cell Struct Funct 2016; 41:61-79; PMID:27246931
- Clabecq A, Henry JP, Darchen F. Biochemical characterization of Rab3-GTPase-activating protein reveals a mechanism similar to that of Ras-GAP. J Biol Chem 2000; 275(41):31786-91; PMID:10859313; http://dx.doi.org/10.1074/jbc.M003705200
- Delprato A, Merithew E, Lambright DG. Structure, exchange determinants, and family-wide rab specificity of the tandem helical bundle and Vps9 domains of Rabex-5. Cell 2004; 118(5):607-17; PMID:15339665; http://dx.doi.org/10.1016/j.cell.2004.08.009
- Park SY, Jin W, Woo JR, Shoelson SE. Crystal structures of human TBC1D1 and TBC1D4 (AS160) RabGTPase-activating protein (RabGAP) domains reveal critical elements for GLUT4 translocation. J Biol Chem 2011; 286(20):18130-8; PMID:21454505; http://dx.doi.org/10.1074/jbc.M110.217323
- Moya M, Roberts D, Novick P. DSS4-1 is a dominant suppressor of sec 4-8 that encodes a nucleotide exchange protein that aids Sec 4p function. Nature 1993; 361(6411):460-3; PMID:8429886; http://dx.doi.org/10.1038/361460a0
- Chin HF, Cai Y, Menon S, Ferro-Novick S, Reinisch KM, De La Cruz EM. Kinetic analysis of the guanine nucleotide exchange activity of TRAPP, a multimeric Ypt1p exchange factor. J Mol Biol 2009; 389(2):275-88; PMID:19361519; http://dx.doi.org/10.1016/j.jmb.2009.03.068
- Esters H, Alexandrov K, Iakovenko A, Ivanova T, Thomä N, Rybin V, Zerial M, Scheidig AJ, Goody RS. Vps9, Rabex-5 and DSS4: proteins with weak but distinct nucleotide-exchange activities for Rab proteins. J Mol Biol 2001; 310(1):141-56; PMID:11419942; http://dx.doi.org/10.1006/jmbi.2001.4735
- Du LL, Novick P. Yeast rab GTPase-activating protein Gyp1p localizes to the Golgi apparatus and is a negative regulator of Ypt1p. Mol Biol Cell 2001; 12(5):1215-26; PMID:11359917; http://dx.doi.org/10.1091/mbc.12.5.1215
- Albert S, Gallwitz D. Two new members of a family of Ypt/Rab GTPase activating proteins. Promiscuity of substrate recognition. J Biol Chem 1999; 274(47):33186-9; PMID:10559187; http://dx.doi.org/10.1074/jbc.274.47.33186
- Will E, Gallwitz D. Biochemical characterization of Gyp6p, a Ypt/Rab-specific GTPase-activating protein from yeast. J Biol Chem 2001; 276(15):12135-9; PMID:11278907; http://dx.doi.org/10.1074/jbc.M011451200
- Vollmer P, Will E, Scheglmann D, Strom M, Gallwitz D. Primary structure and biochemical characterization of yeast GTPase-activating proteins with substrate preference for the transport GTPase Ypt7p. Eur J Biochem 1999; 260(1):284-90; PMID:10091609; http://dx.doi.org/10.1046/j.1432-1327.1999.00192.x
- Will E, Albert S, Gallwitz D. Expression, purification, and biochemical properties of Ypt/Rab GTPase-activating proteins of Gyp family. Methods Enzymol 2001; 329:50-8; PMID:11210571
- Itzen A, Rak A, Goody RS. Sec 2 is a highly efficient exchange factor for the Rab protein Sec 4. J Mol Biol 2007; 365(5):1359-67; PMID:17134721; http://dx.doi.org/10.1016/j.jmb.2006.10.096
- Albert S, Will E, Gallwitz D. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. Embo J 1999; 18(19):5216-5225; PMID:10508155; http://dx.doi.org/10.1093/emboj/18.19.5216
- Nickerson DP, Russell MR, Lo SY, Chapin HC, Milnes JM, Merz AJ. Termination of isoform-selective Vps21/Rab5 signaling at endolysosomal organelles by Msb3/Gyp3. Traffic 2012; 13(10):1411-28; PMID:22748138; http://dx.doi.org/10.1111/j.1600-0854.2012.01390.x
