ABSTRACT
The RAF-MAPK signaling pathway regulates several very diverse cellular processes such as proliferation, differentiation, apoptosis, and transformation. While the canonical function of RAF kinases within the MAPK pathway is the activation of MEK, our group could demonstrate an important crosstalk between RAF signaling and the pro-apoptotic mammalian sterile 20-like kinase (MST2) tumor suppressor pathway in several cancer entities, including head and neck, colon, and breast. Here, the RAF kinases CRAF and ARAF sequester and inhibit the pro-apoptotic kinase MST2 independently of their own kinase activity. In our recent study, we showed that the ARAF-MST2 complex is regulated by subcellular compartmentalization during epithelial differentiation. Proliferating cells of the basal cell layer in squamous epithelia and tumor cells express ARAF at the mitochondria thus allowing for efficient sequestration of MST2. In contrast, non-malignant squamous epithelia have ARAF localized at the plasma membrane, where the control of MST2-mediated apoptosis is compromised. This re-distribution is regulated by the scaffold protein kinase suppressor of Ras 2 (KSR2). Here, we summarize how spatial and temporal regulation of RAF signaling complexes affect cellular signaling and functions.
A brief introduction to RAF signaling
ARAF is member of the RAF family of serine/threonine protein kinases which also comprises BRAF and CRAF. RAF kinases are at the apex of the 3-tiered RAF/MEK/ERK pathway, also referred to as the classic mitogen-activated protein kinase (MAPK) cascade. Upstream of this pathway, RAS GTPases activate RAF, which in turn phosphorylates and activates MEK, and MEK subsequently phosphorylates and activates ERK.Citation1-Citation3 This well-studied signaling pathway links receptor activation at the plasma membrane to >150 substrates in the cytosol and nucleus, which in turn regulate fundamental cellular functions such proliferation, transformation, metabolism, and apoptosis.
With this functional repertoire and important role in cellular regulation, it comes as no surprise that de-regulation of the MAPK pathway has major implications for the cell. BRAF is by far the most frequently mutated RAF isoform in human cancers.Citation4 The most prevalent oncogenic mutation at position V600 is located in its kinase domain and results in increased kinase activity.Citation5 The discovery of this and other oncogenic BRAF mutations in a variety of human malignancies,Citation6 including a high prevalence of mutations in melanomaCitation7 and papillary thyroid carcinoma,Citation8 has put RAF kinases in the limelight for therapeutic intervention.
RAF kinases are comprised of 3 conserved regions (CR), where the N-terminal CR1 contains the RAS-binding domain (RBD) and the cysteine-rich motif (CRD). CR2 features a short cluster of serine and threonine residues, and the C-terminal CR3 contains the kinase domain. In contrast to CRAF and ARAF, BRAF possesses higher basal kinase activity due to a motif called the N-region (negative charge regulatory region).Citation9 MEK1 and MEK2 are considered the main bona fide physiologic RAF substrates.Citation1
The RAF activation and deactivation cycle and spatial regulation is a highly complex sequence of dynamic protein modifications and interactions, but despite years of research is still not completely understood in al details (recently reviewed in refs. Citation5, Citation10).
In quiescent cells, RAF resides in the cytoplasm in an inactive state. Here, auto-inhibited RAF exists in a closed conformation with the N-terminal regulatory domain folding over the catalytic C-terminus, which is stabilized by the adaptor 14–3–3. Upon stimulation with mitogens, activated RAS binds RAF via the RBD thereby allowing for the dephosphorylation of Ser259 (CRFA) by phosphatases including PP2A, PP1, and PP5, subsequent release of 14–3–3 and membrane anchoring. Here, membrane bound RAF is phosphorylated by SRC family kinases (SFKs) and casein kinase 2 (CK2) in the N-region, which, in combination with RAS nanoclustering, results in RAF dimerization, activation segment phosphorylation, and allosteric RAF transactivation. Active RAF is able to bind and activate MEK to facilitate signaling down the 3-tiered MAPK module. As part of a negative feedback loop, activated ERK in turn phosphorylates RAF inhibitory sites, resulting in the release from activated RAS and the disruption of active RAF dimers.
Over the last decade, it has become clear that the initial view of the ERK pathway as a linear pathway is not accurate, but that its components are rather embedded in a web of other signaling and metabolic networks. MAPK components mediate crosstalk with other pathways and regulate many different proteins outside the linear pathway thereby adding positive and negative feedback mechanisms to the pathwayCitation11 A number of scaffold proteins contribute to this signaling crosstalk, the temporal and spatial specificity of the pathway, and fine-tuning of the signaling flux .12 Among those, kinase suppressor of RAS 1 (KSR1) and KSR2 are among the best characterized scaffolds of the MAPK pathway. While both KSRs have overlapping functions by binding to RAF, MEK, and ERK thereby facilitating their phosphorylation and activation, KSR2 was shown to have functions outside the canonical MAPK pathway. KSR2 controls AMP-activated protein kinase (AMPK),Citation13 which acts as a fuel sensor and master regulator of energy homeostasis. KSR2 knockout mice are characterized by impaired glucose tolerance and high insulin levels, leading to an obese phenotype, suggesting a clear role and crosstalk to energy metabolism.
ARAF signaling
Among the 3 mammalian RAF kinases, ARAF is clearly the “ugly duckling,” with most of the research focusing on CRAF and BRAF over the last decades. For their canonical functions, RAF kinases are being regulated in a similar fashion, however, important differences have emerged over the years. While binding to active RAS is sufficient for BRAF activation, CRAF and ARAF require the presence of both activated RAS and SRC family tyrosine kinases.Citation14,Citation15 These are thought to phosphorylate tyrosine residues 301/302 (ARAF) in the regulatory N-region upstream of the kinase domain. In BRAF, these tyrosines are replaced by aspartates, whose negative charge substitute for N-region phosphorylation activated by RAS binding. ARAF possesses by far the lowest kinase activity toward MEK which is due to (i) a substitution of a critical residue (arginine 22 for lysine) in its RBD, causing a weaker affinity to RAS, and (ii), a non-conserved tyrosine 296, whose mutation to glycine induced constitutive kinase activity.Citation16 Recent reports suggest that, despite its low kinase activity, ARAF still plays a crucial role in MAPK signaling. Here, ARAF acts as a scaffold by stabilizing CRAF-BRAF complexes in RAF-inhibited cells to ensure efficient signaling.Citation17 In addition, ARAF depletion prevents MEK activation and cell migration in a cell-type dependent manner, and importantly, dimerization seems to be instrumental for ARAF kinase activity.Citation18
ARAF regulates the MST2/Hippo pathway
With its low kinase activity toward MEK, physiologic functions outside the canonical MAPK pathway have been discussed for ARAF for a long time. Our group demonstrated that ARAF is able to bind and inhibit the pro-apoptotic kinase mammalian sterile 20-like kinase (MST2) independently of its own kinase activity.Citation19 While sequestration by CRAF is induced by stress and relieved by mitogens,Citation20,Citation21 ARAF binds constitutively to MST2, thereby inhibiting MST2 dimerization and activation.
Upstream, the tumor suppressor RASSF1A is able to inhibit the RAF-MST2 complex and promote pro-apoptotic signaling through activation of downstream LATS1 and YAP.Citation22 In this context, YAP in turn forms a complex with p73, leading to transcription and expression of the pro-apoptotic BH3 gene PUMA and induction of apoptosis.Citation22
With this anti-apoptotic role, ARAF is able to promote the survival of cancer cells and elevated levels were detected in several human malignancies including head and neck cancers and late stage colon cancers.Citation19,Citation23 The alternative splice factors HNRNPHCitation23 and HNRNP A1/A224 were identified to regulate ARAF pre-mRNA alternative splicing, resulting in 2 functional isoforms, which we termed ARAFwildtype and ARAFshort. Enhanced expression of HNRNPH in several malignancies including colon, head and neck, and hepatocellular carcinoma result primarily in the expression of ARAFwildtype, while low levels of these splice factors are detected in non-malignant tissues, thereby favoring the expression of the alternative splice form ARAFshort.Citation23 In contrast to wildtype ARAF, this splice form retains intronic sequences, and generates a shortened protein lacking the kinase domain. While ARAFshort is not able to control and bind MST2, our group was able to demonstrate a function as a dominant-negative antagonist by binding and blocking activated RAS, inhibiting ERK signaling, and cellular transformation. With decreased levels detected in several human malignancies, ARAFshort seems to act as a tumor suppressor. A number of recent studies report activating as well as inactivating mutations for the ARAF gene in several human cancer types, but how these mutations affect the signaling events described here is currently not known.
These differential roles of ARAF in MAPK and MST2 signaling lead to an interesting hypothesis for RAF evolution: While BRAF as the oldest family member has the strongest MEK activity, but very little affinity for MST2, the youngest member ARAF possesses poor MEK kinase activity but a strong capacity for MST2/Hippo control. This inverse control of MST2 signaling might suggest that during evolution the role of RAF has shifted from exclusively activating the ERK pathway to regulating additional cellular processes such as MST2-mediated apoptosis.
While RAF proteins are generally considered cytoplasmatic proteins which can bind to the inner surface of the plasma membrane, the ARAF-MST2 complex also localizes to the surface of the mitochondria in tumor cell lines as well as primary tumors.Citation19 Interaction studies suggest that this localization is mediated by hTOM and hTIM, 2 proteins involved in the mitochondrial transport system, however, the exact role is not clear so far.Citation25
Spatial control of ARAF during differentiation
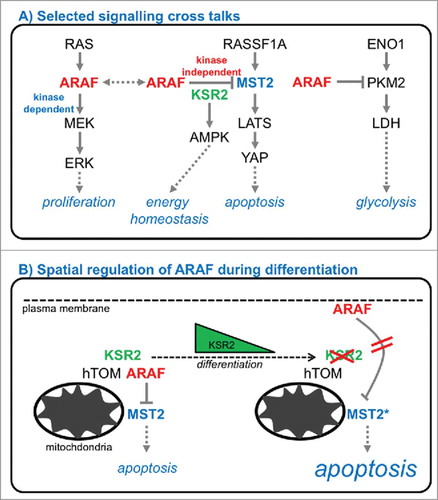
In our recent study we demonstrated, that the ARAF-MST2 complex is regulated by subcellular compartmentalization ().Citation26 In highly proliferating cells of the basal cell layer of non-malignant stratified, non-keratinized squamous epithelia, ARAF controls MST2 in the cytoplasm and at the mitochondria. In contrast, differentiated cells of these squamous epithelia have ARAF localized at the plasma membrane. Of importance, here, ARAF is no longer able to keep MST2 function in check thus rendering cells susceptible to apoptosis. ARAF expression levels and anti-apoptotic function correlate with previous reports from our group: While the splice factor HNRNPH controlling ARAF alternative splicing is highly expressed in basal cells, hardly any expression is detectable during epithelial differentiation.Citation19
To study the re-localization of ARAF observed in head and neck epithelia, we established a constitutive and an inducible cell system to mimic the re-localization system of ARAF to the plasma-membrane. In both cases, re-localized ARAF loses the ability to efficiently sequester and inactivate MST2, which does not follow this re-localization. As mentioned above, several phosphorylations regulate ARAF activity.Citation27 While phosphorylation of serine 432 was described to be important for binding to MEK, phosphorylation of serines 257, 262, and 264 in the so-called Isoform-specific Hinge (IH) segment are crucial for activation, but, importantly, also localization. From a structural point of view, phosphorylation of this IH segment leads to an accumulation of negative surface charges, resulting in electrostatic destabilization of ARAF localization at the plasma membrane and release into the cytoplasm. Whether the full phosphorylation of the IH segment is involved in localization changes during epithelial differentiation is currently not understood. Also, membrane-localized ARAF seems to have no impact on MAPK signaling, as no effect on bulk ERK signaling or dynamics could be observed.
In addition to the observed ARAF re-localization to the plasma membrane during epithelial differentiation in head and neck tissues and growth factor induced differentiation of MCF7 cancer cell lines, ARAF overexpression also correlates with the increased differentiation in MCF7 cells. This is in line with a previous report,Citation28 where ARAF overexpression inhibits MST2 signaling thereby promoting HGF-induced Epithelial-Mesenchymal Transition (EMT). Other groups have reported also that ARAF expression correlates with adipocyteCitation29 and myogenicCitation30 differentiation, suggesting a clear role of ARAF during differentiation.
In contrast to spatial regulation of ARAF during differentiation, MST2 expression and localization in the cytoplasm or around the mitochondria seems not to undergo changes. The MST2 tumor suppressor pathway regulates fundamental cellular processes including apoptosis, proliferation, and differentiation thereby controlling organ size and tissue development.Citation31-Citation33 MST1 and MST2 are directly involved in several differentiation processes and downstream signaling (reviewed in ref. Citation34) including embryonic stem cell differentiation,35 trophoblast differentiation,Citation36 pancreatic acinar differentiation,Citation37 myoblast differentiation,Citation38 and monocytic differentiation of myeloid leukemia HL60 cells.Citation39 In addition, both MST2 and MST1 regulate junction formation in epithelial cells through interaction with members of the angiomotin (AMOT) family.Citation40,Citation41
For the involvement of ARAF and MST2 during epithelial differentiation, we suggest the following model: During differentiation of stratified, non-keratinized squamous epithelia, basal cells withdraw from cell cycle and differentiate toward the surface of the epithelium. Within this process, the differentiated cells flatten and lose their nucleus, before they are finally shed from the surface. For these processes and terminal differentiation, apoptotic processes are instrumental controlling caspase cleavage, enculeation, and internucleosomal DNA cleavage.Citation42-Citation44 During differentiation, spatial regulation of ARAF activates MST2, leading to activation of the core apoptotic machinery including Caspase 3, PARP, and transcriptional activation of the apoptotic effector PUMA.
Kinase suppressor of Ras 2
Instrumental for the functional regulation of the ARAF-MST2 complex seems to be the action of Kinase Suppressor of Ras 2 (KSR2), one of the scaffold proteins that organize RAF and MAPK functions (). These scaffolds serve as binding platforms and control the spatial and temporal aspects of the signaling flux (reviewed in refs. Citation1, Citation45, Citation46). In our recent work, we demonstrated a loss of KSR2 expression during epithelial differentiation, thereby releasing ARAF to the plasma membrane leading to activation of MST2 and subsequent apoptosis. KSR2 was described recently to interact with ARAF, however only in response to TNFα47 and not in quiescent cells or upon stimulation with EGF. Interestingly, KSR2 also provides a link to energy homeostasis by binding to AMPK and mediating its stimulatory effects on glucose uptake and fatty acid oxidationCitation13 How KSR2 expression levels are regulated during differentiation and how loss of KSR2 induces ARAF re-localization is currently not understood. Of note, ARAF also binds and inhibits the glycolytic enzyme pyruvate kinase M2 (PKM2)Citation48 suggesting further links to both energy and glucose metabolism.
Taken together, our data and other reports suggest that ARAF is a central signaling hub regulating very diverse processes including MAPK signaling (MEK, RAF, KSR2), apoptosis (MST2, KSR2), energy homeostasis (AMPK), glycolysis (PKM2), and mitochondrial transport (hTIM, hTOM) (). How this interesting signaling crosstalk and these dynamic signaling complexes are fine-tuned is not understood and warrants further investigation.
Conclusions
In summary, the spatial and temporal regulation of signaling complexes seems instrumental for fine-tuning signaling events and achieving robust cellular functions and phenotypes. This is orchestrated by post-translational modifications such as phosphorylations, adjusted expression levels in normal cells and disease, alternative splice form selection, and crosstalk with other molecules of the cell.
The signaling crosstalk described here is a good example for the bewildering complexity of cellular signaling. Despite intensive research on RAF-MAPK signaling for decades, we still lack a systems-level understanding and, in particular, should focus on deciphering the composition of parallel, diverse signaling complexes, and their dynamic and spatial regulation to regulate specific cell fate decisions.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Science Foundation Ireland under Grant No. 14/IA/2395.
References
- Matallanas D, Birtwistle M, Romano D, Zebisch A, Rauch J, von Kriegsheim A, Kolch W. Raf family kinases: old dogs have learned new tricks. Genes Cancer 2011; 2:232-60; PMID:21779496; http://dx.doi.org/10.1177/1947601911407323
- Yoon S, Seger R. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors 2006; 24:21-44; PMID:16393692; http://dx.doi.org/10.1080/02699050500284218
- Marais R, Light Y, Paterson HF, Mason CS, Marshall CJ. Differential regulation of Raf-1, A-Raf, and B-Raf by oncogenic ras and tyrosine kinases. J Biol Chem 1997; 272:4378-83; PMID:9020159; http://dx.doi.org/10.1074/jbc.272.7.4378
- Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic acids Res 2015; 43:D805-11; PMID:25355519; http://dx.doi.org/10.1093/nar/gku1075
- Lavoie H, Therrien M. Regulation of RAF protein kinases in ERK signalling. Nat Rev Mol Cell Biol 2015; 16:281-98; PMID:25907612; http://dx.doi.org/10.1038/nrm3979
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature 2002; 417:949-54; PMID:12068308; http://dx.doi.org/10.1038/nature00766
- Cancer Genome Atlas N. Genomic Classification of Cutaneous Melanoma. Cell 2015; 161:1681-96; PMID:26091043; http://dx.doi.org/10.1016/j.cell.2015.05.044
- Cancer Genome Atlas Research N. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014; 159:676-90; PMID:25417114; http://dx.doi.org/10.1016/j.cell.2014.09.050
- Hu J, Stites EC, Yu H, Germino EA, Meharena HS, Stork PJS, Kornev AP, Taylor SS, Shaw AS. Allosteric Activation of Functionally Asymmetric RAF Kinase Dimers. Cell 2013; 154:1036-46; PMID:23993095; http://dx.doi.org/10.1016/j.cell.2013.07.046
- Baljuls A, Kholodenko BN, Kolch W. It takes two to tango–signalling by dimeric Raf kinases. Mol Biosyst 2013; 9:551-8; PMID:23212737; http://dx.doi.org/10.1039/C2MB25393C
- Kholodenko B, Hancock J, Kolch W. Signalling ballet in space and time. Nat Rev Mol Cell Biol 2010; 11:414-26; PMID:20495582; http://dx.doi.org/10.1038/nrm2901
- Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol 2005; 6:827-37; PMID:16227978; http://dx.doi.org/10.1038/nrm1743
- Costanzo-Garvey DL, Pfluger PT, Dougherty MK, Stock JL, Boehm M, Chaika O, Fernandez MR, Fisher K, Kortum RL, Hong EG, et al. KSR2 is an essential regulator of AMP kinase, energy expenditure, and insulin sensitivity. Cell Metab 2009; 10:366-78; PMID:19883615; http://dx.doi.org/10.1016/j.cmet.2009.09.010
- Fabian JR, Daar IO, Morrison DK. Critical tyrosine residues regulate the enzymatic and biological activity of Raf-1 kinase. Mol Cell Biol 1993; 13:7170-9; PMID:7692235; http://dx.doi.org/10.1128/MCB.13.11.7170
- Marais R, Light Y, Paterson HF, Marshall CJ. Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. Embo J 1995; 14:3136-45. PMID:7542586.
- Baljuls A, Mueller T, Drexler HC, Hekman M, Rapp UR. Unique N-region determines low basal activity and limited inducibility of A-RAF kinase: the role of N-region in the evolutionary divergence of RAF kinase function in vertebrates. J Biol Chem 2007; 282:26575-90; PMID:17613527; http://dx.doi.org/10.1074/jbc.M702429200
- Rebocho AP, Marais R. ARAF acts as a scaffold to stabilize BRAF:CRAF heterodimers. Oncogene 2013; 32:3207-12; PMID:22926515; http://dx.doi.org/10.1038/onc.2012.330
- Mooz J, Oberoi-Khanuja TK, Harms GS, Wang W, Jaiswal BS, Seshagiri S, Tikkanen R, Rajalingam K. Dimerization of the kinase ARAF promotes MAPK pathway activation and cell migration. Sci Signal 2014; 7:ra73; PMID:25097033; http://dx.doi.org/10.1126/scisignal.2005484
- Rauch J, O'Neill E, Mack B, Matthias C, Munz M, Kolch W, Gires O. Heterogeneous nuclear ribonucleoprotein H blocks MST2-mediated apoptosis in cancer cells by regulating A-Raf transcription. Cancer Res 2010; 70:1679-88; PMID:20145135; http://dx.doi.org/10.1158/0008-5472.CAN-09-2740
- O'Neill E, Rushworth L, Baccarini M, Kolch W. Role of the kinase MST2 in suppression of apoptosis by the proto-oncogene product Raf-1. Science 2004; 306:2267-70; PMID:15618521; http://dx.doi.org/10.1126/science.1103233
- Romano D, Nguyen LK, Matallanas D, Halasz M, Doherty C, Kholodenko BN, Kolch W. Protein interaction switches coordinate Raf-1 and MST2/Hippo signalling. Nat Cell Biol 2014; 16:673-84; PMID:24929361; http://dx.doi.org/10.1038/ncb2986
- Matallanas D, Romano D, Yee K, Meissl K, Kucerova L, Piazzolla D, Baccarini M, Vass JK, Kolch W, O'neill E. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol Cell 2007; 27:962-75; PMID:17889669; http://dx.doi.org/10.1016/j.molcel.2007.08.008
- Rauch J, Moran-Jones K, Albrecht V, Schwarzl T, Hunter K, Gires O, Kolch W. c-Myc regulates RNA splicing of the A-Raf kinase and its activation of the ERK pathway. Cancer Res 2011; 71:4664-74; PMID:21512137; http://dx.doi.org/10.1158/0008-5472.CAN-10-4447
- Shilo A, Ben Hur V, Denichenko P, Stein I, Pikarsky E, Rauch J, Kolch W, Zender L, Karni R. Splicing factor hnRNP A2 activates the Ras-MAPK-ERK pathway by controlling A-Raf splicing in hepatocellular carcinoma development. RNA 2014; 20:505-15; PMID:24572810; http://dx.doi.org/10.1261/rna.042259.113
- Yuryev A, Ono M, Goff SA, Macaluso F, Wennogle LP. Isoform-specific localization of A-RAF in mitochondria. Mol Cell Biol 2000; 20:4870-8; PMID:10848612; http://dx.doi.org/10.1128/MCB.20.13.4870-4878.2000
- Rauch J, Vandamme D, Mack B, McCann B, Volinsky N, Blanco A, Gires O, Kolch W. Differential localization of A-Raf regulates MST2-mediated apoptosis during epithelial differentiation. Cell Death Differ 2016; 23:1283-95; PMID:26891695; http://dx.doi.org/10.1038/cdd.2016.2
- Baljuls A, Schmitz W, Mueller T, Zahedi RP, Sickmann A, Hekman M, Rapp UR. Positive regulation of A-RAF by phosphorylation of isoform-specific hinge segment and identification of novel phosphorylation sites. J Biol Chem 2008; 283:27239-54; PMID:18662992; http://dx.doi.org/10.1074/jbc.M801782200
- Farrell J, Kelly C, Rauch J, Kida K, Garcia-Munoz A, Monsefi N, Turriziani B, Doherty C, Mehta JP, Matallanas D, et al. HGF induces epithelial-to-mesenchymal transition by modulating the mammalian hippo/MST2 and ISG15 pathways. J Proteome Res 2014; 13:2874-86; PMID:24766643; http://dx.doi.org/10.1021/pr5000285
- Zmuidzinas A, Gould GW, Yager JD. Expression of c-raf-1 and A-raf-1 during differentiation of 3T3-L1 preadipocyte fibroblasts into adipocytes. Biochem Biophys Res Commun 1989; 162:1180-7; PMID:2669746; http://dx.doi.org/10.1016/0006-291X(89)90798-5
- Yokoyama T, Takano K, Yoshida A, Katada F, Sun P, Takenawa T, Andoh T, Endo T. DA-Raf1, a competent intrinsic dominant-negative antagonist of the Ras-ERK pathway, is required for myogenic differentiation. J Cell Biol 2007; 177:781-93; PMID:17535970; http://dx.doi.org/10.1083/jcb.200703195
- Gomez M, Gomez V, Hergovich A. The Hippo pathway in disease and therapy: cancer and beyond. Clin Transl Med 2014; 3:22; PMID:25097725; http://dx.doi.org/10.1186/2001-1326-3-22
- Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev 2013; 27:355-71; PMID:23431053; http://dx.doi.org/10.1101/gad.210773.112
- Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer 2013; 13:246-57; PMID:23467301; http://dx.doi.org/10.1038/nrc3458
- Bernascone I, Martin-Belmonte F. Crossroads of Wnt and Hippo in epithelial tissues. Trends in cell biology 2013; 23:380-9; PMID:23607968; http://dx.doi.org/10.1016/j.tcb.2013.03.007
- Li P, Chen Y, Mak KK, Wong CK, Wang CC, Yuan P. Functional role of Mst1/Mst2 in embryonic stem cell differentiation. PloS one 2013; 8:e79867; PMID:24224013; http://dx.doi.org/10.1371/journal.pone.0079867
- Du X, Dong Y, Shi H, Li J, Kong S, Shi D, Sun LV, Xu T, Deng K, Tao W. Mst1 and mst2 are essential regulators of trophoblast differentiation and placenta morphogenesis. PloS one 2014; 9:e90701; PMID:24595170; http://dx.doi.org/10.1371/journal.pone.0090701
- Gao T, Zhou D, Yang C, Singh T, Penzo-Mendez A, Maddipati R, Tzatsos A, Bardeesy N, Avruch J, Stanger BZ. Hippo signaling regulates differentiation and maintenance in the exocrine pancreas. Gastroenterology 2013; 144:1543-53, 53 e1.
- Graves JD, Gotoh Y, Draves KE, Ambrose D, Han DK, Wright M, Chernoff J, Clark EA, Krebs EG. Caspase-mediated activation and induction of apoptosis by the mammalian Ste20-like kinase Mst1. EMBO J 1998; 17:2224-34; PMID:9545236; http://dx.doi.org/10.1093/emboj/17.8.2224
- Wang X, Wang T-T, White JH, Studzinski GP. Expression of human kinase suppressor of Ras 2 (hKSR-2) gene in HL60 leukemia cells is directly upregulated by 1,25-dihydroxyvitamin D(3) and is required for optimal cell differentiation. Exp Cell Res 2007; 313:3034-45; PMID:17599832; http://dx.doi.org/10.1016/j.yexcr.2007.05.021
- Wang Y, Li Z, Xu P, Huang L, Tong J, Huang H, Meng A. Angiomotin-like2 gene (amotl2) is required for migration and proliferation of endothelial cells during angiogenesis. J Biol Chem 2011; 286:41095-104; PMID:21937427; http://dx.doi.org/10.1074/jbc.M111.296806
- Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev 2011; 25:51-63; PMID:21205866; http://dx.doi.org/10.1101/gad.2000111
- McCall CA, Cohen JJ. Programmed cell death in terminally differentiating keratinocytes: role of endogenous endonuclease. J Investigative Dermatol 1991; 97:111-4; PMID:1647418; http://dx.doi.org/10.1111/1523-1747.ep12478519
- Weil M, Raff MC, Braga VM. Caspase activation in the terminal differentiation of human epidermal keratinocytes. Curr Biol 1999; 9:361-4; PMID:10209121; http://dx.doi.org/10.1016/S0960-9822(99)80162-6
- Lu QL, Abel P, Foster CS, Lalani EN. bcl-2: role in epithelial differentiation and oncogenesis. Human pathology 1996; 27:102-10; PMID:8617450; http://dx.doi.org/10.1016/S0046-8177(96)90362-7
- Kholodenko BN, Birtwistle MR. Four-dimensional dynamics of MAPK information processing systems. Wiley Interdiscip Rev Syst Biol Med 2009; 1:28-44; PMID:20182652; http://dx.doi.org/10.1002/wsbm.16
- Witzel F, Maddison L, Bluthgen N. How scaffolds shape MAPK signaling: what we know and opportunities for systems approaches. Front Physiol 2012; 3:475; PMID:23267331; http://dx.doi.org/10.3389/fphys.2012.00475
- Liu L, Channavajhala PL, Rao VR, Moutsatsos I, Wu L, Zhang Y, Lin LL, Qiu Y. Proteomic characterization of the dynamic KSR-2 interactome, a signaling scaffold complex in MAPK pathway. Biochim Biophys Acta 2009; 1794:1485-95; PMID:19563921; http://dx.doi.org/10.1016/j.bbapap.2009.06.016
- Le Mellay V, Houben R, Troppmair J, Hagemann C, Mazurek S, Frey U, Beigel J, Weber C, Benz R, Eigenbrodt E, et al. Regulation of glycolysis by Raf protein serine/threonine kinases. Adv Enzyme Regul 2002; 42:317-32; PMID:12123723; http://dx.doi.org/10.1016/S0065-2571(01)00036-X