ABSTRACT
Chemotaxis, which is chemoattractant-guided directional cell migration, plays major roles in recruitment of neutrophils, the metastasis of cancer cells, and the development of the model organism Dictyostelium discoideum. These cells share remarkable similarities in the signaling pathways by which they control chemotaxis. They all use a G protein-coupled receptor (GPCR)-mediated signal transduction pathway to sense the chemotactic gradient to guide cell migration. Diverse chemokines activate Rac through conserved GPCR signaling pathways. ELMO proteins are an evolutionarily conserved, essential component of the ELMO/Dock complex, which functions as a guanine nucleotide exchange factor (GEF) for small G protein Rac activation. The linkages between the GPCR-initiated gradient sensing compass and the Rac-mediated migrating machinery have long been missing. Here, we summarize recent findings on ELMO proteins that directly interact with G protein and transduce GPCR signaling to control the reorganization of actin-based cytoskeleton through regulating Rac activation during chemotaxis, first in D. discoideum and then in mammalian cancer cells. This represents an evolutionarily conserved signaling shortcut from GPCR to the actin cytoskeleton.
Introduction
Chemotaxis, which is directional cell migration guided by chemoattractant gradients, plays major roles in the recruitment of neutrophils, the metastasis of cancer cells, and the development of the model organism Dictyostelium discoideum. Citation03,Citation1 The molecular mechanisms of chemotaxis in mammalian cells and in D. discoideum are evolutionarily conserved, and D. discoideum provides a powerful model system in which to identify new components and to reveal their functions in chemotaxis. Neutrophils, cancer cells, and D. discoideum all utilize G protein-coupled receptor (GPCR)-regulated signal transduction pathways to sense chemotactic gradients and control directional cell migration.Citation06,Citation4 Binding of chemoattractants to their receptors induces the activation of heterotrimeric G-proteins.Citation09,Citation7 Active Gα and Gβγ activate their downstream effectors, and this leads to the activation of small GTPase Rac, a subclass of small GTPase Rho that regulates many aspects of intracellular actin dynamics.Citation10 Active Rac then activates Wiskott-Aldrich syndrome proteins, such as WASP, N-WASP, WAVE, and SCAR.Citation11 Activated WASP proteins interact with the Arp2/3 complex, a 7-subunit protein complex that includes actin-related proteins Arp2 and Arp3 and controls nucleation of actin polymerization and branching of filaments, and increase its nucleating activity.Citation12 Consequently, activation of GPCRs promotes actin polymerization. However, the linkages between the GPCR/G protein-initiated gradient sensing apparatus and the Rac-controlled migration machinery is just beginning to be revealed.
Activation of Rac proteins promotes the growth of actin filaments that drive cell migration. Rac proteins, like any other small GTPases including Ras and Rho, cycle between GDP- and GTP-bound states. They activate their effectors when they are in their GTP-bound (active) state. Two large classes of regulatory proteins control the activation state of small GTPases. The GTPase-activating proteins (GAPs) bind to GTP-bound Rac proteins to enhance their GTPase activity, thereby converting them to the inactive GDP-bound state. Conversely, guanine exchange factors (GEFs) promote nucleotide exchange from GDP to GTP on small GTPases. The GEFs for Rho/Rac GTPases are divided into the Dbl and Dock (Dictator of cytokinesis) families. The Dbl family of GEFs is characterized by the presence of the Dbl domain, which is critical for GDP/GTP exchange activity.Citation13 The Dock GEFs were discovered later and are characterized by the presence of 2 evolutionarily conserved domains: the lipid-binding Dock homology region-1 (DHR-1) and the GEF DHR-2 modules.Citation16,Citation14 Because they lack a Dbl domain, Dock GEFs are often referred to as “atypical GEFs.” Based on sequence similarity and domain organization, the 11 Dock members are classified into 4 subgroups: DockA (Dock1/2/5; Dock1 is also called Dock180); DockB (Dock3/4; Dock3 is also called presenilin binding partner, PBP or Modifier of Cell Adhesion, MOCA); DockC (Dock6/7/8); and DockD (Dock9/10/11, also called Zizimin (Ziz)-related proteins).Citation16 A key feature of the Dock GEFs is their specificity toward Rac and/or Cdc42, in contrast to Dbl family members, which can also act as GEFs for other members of the Rho family including RhoA.Citation15 Active GTP-bound Rac proteins activate the activators of Arp2/3 complexes, such as WASP, N-WASP, and WAVE, and promote actin polymerization for cell migration.
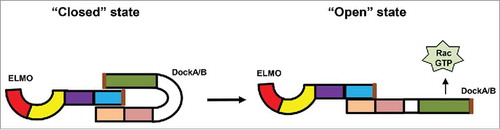
ELMO proteins, identified as the binding partner of Dock proteins, form an evolutionarily conserved and ancient family that includes members in D. discoideum, C. elegans, fungi, Drosophila, and higher vertebrates.Citation17,Citation18 The first member identified, Ced-12, was discovered in a genetic screen to identify components required for engulfment of dead cells during development of C. elegans.Citation19 The Ced-12 gene encodes a protein that is essential for engulfment and cell motility, and thus it was named ELMO. Other members of the ELMO family were later found in many organisms.Citation18,Citation14 Extensive studies in C. elegans and in mammalian cells showed that ELMO and Dock proteins form a complex that serves as a GEF for Rac proteins.Citation14,Citation19 Although ELMO proteins do not have Rac GEF activity, formation of the complex of ELMO and DockA/B proteins is essential for the GEF activity of Dock proteins and for Rac-induced cell migration. It appears that ELMO and Dock often form a complex, which exists in 2 different states: a “closed” state that has little GEF activity for Rac proteins, and an “open” state that enables the complex to function as a Rac GEF [20] (). Activation of cell surface receptors, such as GPCRs, stimulates the GEF activity of ELMO/Dock complexes, which in turn activate Rac proteins for cell migration. However, the mechanism by which the GPCR/G-protein machinery regulates the ELMO/Dock complex has not been fully understood.
In this review, we summarize the function of ELMO proteins as a linkage that transduces GPCR signaling to Rac activation, as discovered first in D. discoideum and later in the mammalian system.Citation23,Citation21 These findings present an evolutionarily conserved signaling pathway that connects the GPCR-mediated gradient-sensing compass and the moving machinery of eukaryotic cells for directional cell migration.
GPCR-mediated functions of ELMO/Dock complexes in the model organism D. discoideum
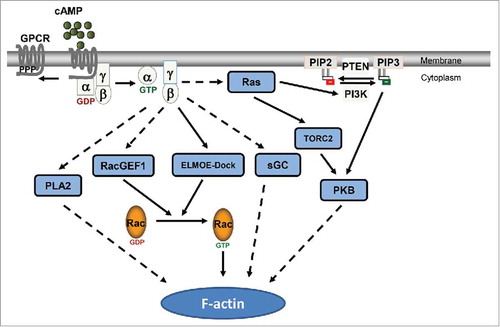
The signaling mechanisms linking the activation of GPCR to the reorganization of actin cytoskeleton are well-characterized in D. discoideum. Activation of cAR1, the receptor for chemoattractant cAMP in D. discoideum, leads to the dissociation of the G-proteins into Gα2 and Gβγ subunits,Citation8 which in turn control several pathways that regulate the actin-based movement apparatus for chemotaxis. For example, the cAR1/G-protein machinery activates Ras proteins that regulate 4 pivotal effectors: PI3K, PLA2, TORC2 and sGC ().Citation24,Citation25,Citation26,Citation27,Citation28 Four of the effectors have been shown to contribute to chemotaxis, while it is still not clear how they regulate Rac activity. Active Rac stimulates Arp2/3 complex, which initiates the branching of actin filaments from existing ones; this results in the growth of the dendritic actin-network pushing the membrane forward in the leading front of the chemotaxing cells.Citation10,Citation29 Thus, it is important to establish the linkage between chemoattractant GPCRs and the ELMO/Dock complex, which guides the reorganization of actin-based migration machinery during chemotaxis.
ELMO and Dock proteins are evolutionarily conserved and present in a wide variety of eukaryotes, including D. discoideum, C. elegans, fungi, Drosophila, and higher vertebrates.Citation14,Citation18 D. discoideum encodes 8 genes for Dock-like proteins.Citation30 Among them, DdDockA–D appear to belong to the subfamily of Dock1-related proteins (DockA/B), and DdZizA–D belong to the subfamily of DockC, zizimin-related proteins (). As noted, DdDock proteins do not possess the evolutionarily conserved DHR-1 domain, which binds PIP3 and plays a critical role in PIP3 membrane recruitment.Citation16 D. discoideum has 6 genes (ELMOA–F) with the ELMO domain and distinct featuresCitation17,Citation31 (). ELMOA and ELMOB are similar to mammalian ELMOD1 and ELMOD2.Citation17 A <70 aa C-terminal domain in ELMOA is conserved in human ELMO1-3. ELMOC has a coiled coil domain in its N terminus. In ELMOD, 4 ankyrin repeat regions are located toward its C-terminus. Both ELMOE and ELMOF have internal repeats (IR), a feature typically found in many D. discoideum proteins, and their ELMO domains are most closely related to human ELMO1-3.
Figure 3. ELMO and Dock proteins in Dictyostelium discoideum. (A) Six ELMO domain-containing proteins with distinct domain composition in D. discoideum. (B) Eight Dock domain-containing proteins in D. discoideum.

ELMOE interacts directly with the Gβ subunit and transduces GPCR signal to the actin cytoskeleton by regulating RacB activation. ELMOE is the most closely related to human ELMO1-3. Not surprisingly, we found that ELMOE consistently associates with DdDockC and DdZizA to form an ELMO/Dock complex that serves as a GEF for RacB in D. discoideum.Citation23 Activation of cAR1 results in ELMOE-mediated activation of RacB, which in turn leads to actin-polymerization during chemotaxis. In resting D. discoideum cells, ELMOE/DdDock complexes reside in the cytoplasm. To define how they signal, it is key to understand how they reach the membrane. DdDock proteins do not have a PIP3-binding DHR-1 domain or a Ras binding domain (RBD).Citation16,Citation32 Consistent with the above, cAMP-induced membrane translocation of ELMOE is also observed in rasC−:rasG− cells, in which both rasC and rasG genes are disrupted and little PIP3 is generated in response to cAMP stimulation.Citation33 This result indicates that Ras and PIP3 play no role in membrane recruitment of the ELMOE/Dock complex. Importantly, the membrane recruitment of ELMOE requires Gβ.Citation23 GPCR activation also promotes the association between Gβ and ELMOE, providing the first direct link between the ELMO/Dock complexes and GPCR signaling. Lacking the N-terminal part, ELMOE fails to associate with Gβ or be recruited to the membrane, but it still forms a complex with Dock1. This result indicates that Gβ might play an essential role in membrane recruitment and subsequent activation of ELMOE/DdDock GEF activity. It is possible that, as with human ELMO1, the binding of Gb to ELMOE through its N-terminal facilitates recruitment and relieves autoinhibition of the ELMOE/DdDock complex and properly positions the complex for optimal Rac activation,Citation34,Citation20 although ELMOE lacks a clearly definable ELMO inhibitory domain (EIM) in its N-terminal part.Citation23 Future work is required to understand the molecular mechanism of Gβ and ELMOE/DdDock interaction. In mammalian cells, it was speculated and has been recently verified that the G protein/ELMO/Dock complex is also conserved and plays a crucial role in chemokine GPCR-controlled directional cell migration and metastasis of breast cancer.Citation21,Citation22
Of note, ELMOA, the other studied ELMO protein in D. discoideum, functions unexpectedly as a negative regulator of actin polymerization during phagocytosis and cell migration.Citation31 Loss of elmoA (elmoA− ) results in an overall increase in phagocytosis, which is dissimilar to the defects in engulfment of dead cells exhibited by the C. elegans ced-12 (ELMO) mutant. In addition, elmoA− cells have increased pseudopod formation and elevated F-actin localization within the pseudopods. The mutant is defective in maintaining cell polarity and in suppressing the formation of lateral pseudopods, which are essential for effective chemotaxis. ELMOA associates with cortical actin and myosin II heavy chain, which are known to enhance cell cortical rigidity, thereby ensuring cell polarity.Citation31 ELMOA forms a strong clade with human ELMOD1–D2,Citation17 which are different from ELMOs functioning as Rac GEF and which display GAP activity for the small GTPases Arls and Arfs.Citation37,Citation35 While GAP activity has not been analyzed, ELMOA negatively regulates actin polymerization, a downstream target of Rac. We speculate that ELMOA may function to inhibit localized Dock-mediated GEF activity and/or may act as a GAP, similar to the function of human ELMODs and consistent with its evolutionary proximity to these human proteins.
GPCR-mediated functions of ELMO/Dock complexes in mammalian systems
The mammalian family of ELMO domain-containing proteins is defined by the presence of the ELMO domain and consists of 6 members. These 6 proteins, based on protein size and domain architecture, have been divided into 2 subgroups, ELMOs and ELMODs.Citation35 ELMOs (ELMO1-3) contain multiple domains (). The N-terminal Ras binding domain (RBD) and ERM binding domain (ERM) bind to RhoG, IpgB1, and ERM proteins.Citation40,Citation38 The middle ELMO domain has no assigned function, but is conserved in the ELMO family. The C-terminal PH domain mediates the association of ELMO and Dock to form an ELMO/Dock complex. The pro-rich C-terminus interacts with the SH3 domain of Dock proteins and maintains autoinhibition of Dock GEF activity.Citation41 The RacGEF function of ELMO proteins was mostly obtained from ELMO1 and ELMO2, and little is known about ELMO3. The ELMODs (ELMOD1-3) consist of little more than the ELMO domain. Not surprisingly, then, the sequence homology between ELMOs and ELMODs lies only within the ELMO domain. Identified as the binding partner of Dock proteins, ELMOs are required for ELMO/Dock complex formation and Dock RhoGEF activity.Citation16 Excellent reviews have summarized the domain composition and function of Dock proteins.Citation16,Citation14 Of the 4 subgroups, DockA/B have conserved domain composition: an N-terminal SH3 domain, a middle phospholipid-binding DHR-1 domain and RacGEF DHR-2 domain, and a C-terminal proline-reach motif (). DockA/B associate with ELMOs to activate Rac and are involved in cell migration, adhesion, and invasion and metastasis of cancer.Citation22,Citation42,Citation43,Citation44,Citation45 The N-terminal SH3 domain of DockA/B functions as an intramolecular inhibitor of the exchange factor; this inhibition can be relieved by various mechanisms, including the binding of Dock and ELMO proteins.Citation15,Citation41 In contrast to the ELMOs, which form complexes with Dock proteins and function as Rho GEFs, ELMODs have GAP activity toward Arls and Arfs.Citation37,Citation35
Figure 4. ELMO proteins in mammals. (A) Six ELMO domain-containing proteins in mammals are divided into 2 subgroups: ELMOs (ELMO1-3) and ELMODs (ELMOD1-3). Conserved domain structures are shared among ELMOs, while ELMODs have little more than an ELMO domain. (B) The conserved domain structure of DockA/DockB: a N-terminal SH3 domain, a middle PIP3-binding DHR-1 domain and RacGEF catalytic domain of DHR-2, and a C-terminal proline-rich motif (PxxP). (C) Various mechanisms of GPCR-mediated membrane recruitment and activation of the ELMO/Dock complex.
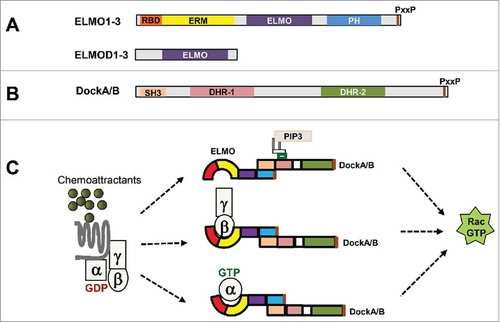
ELMO proteins regulate the activity and spatiotemporal localization of Dock GEFs in GPCR-mediated chemotaxis.Citation14,Citation21,Citation22,Citation46 As was well-summarized in excellent reviews,Citation14,Citation16 the activity of Dock GEFs is regulated through a variety of mechanisms, including phosphorylation and association with membrane proteins and/or lipids. GPCR activation also recruits and activates Dock GEFs to the membrane through their lipid binding. Dock2 is a major Rac GEF that controls motility and polarity during chemotaxis in neutrophils.Citation47 Chemoattractants activate PI3K and generate PIP3 on the inner leaflet of the membrane; the PIP3 binds with the DHR1 domain of Dock2 and consequently recruits Dock2 from the cytoplasm to the membrane. However, the later accumulation of Dock2 in the leading edge and the subsequent local actin polymerization is through binding to phosphatidic acid (PA).Citation46 Dock1 has also been shown to be recruited and to function as a Rac GEF in a PA-dependent fashion.Citation48 It is still not clear whether ELMO proteins play any role in lipid-mediated membrane recruitment of the Dock/ELMO complex. We and others have shown that ELMO1 plays a critical role in recruiting the ELMO1/Dock1 complex to the membrane and subsequently activating GEF activity though its interaction with G protein.Citation21,Citation22,Citation23,Citation49 In HL60, Hela, HEK293 and breast cancer cells, ELMO1 consistently associates with Dock1 and localizes predominantly in the cytoplasm.Citation21,Citation22 Various chemoattractants induce the membrane translocation of ELMO1. Activation of GPCRs also promotes the association of ELMO1 and Gαi and/or Gβ subunits in these cells. The association of ELMO1 and G protein subunits is required for chemotaxis and invasion during metastasis of breast cancer cells in vivo. By determining the domain requirement of ELMO1 for the association capability with Gβ, we found that the N-terminal region of ELMO1, including a Ras binding domain (RBD) and an ELMO inhibitory domain (EIM), is required for ELMO1/Gβγ association.Citation22 Although the domain of ELMO1 required for its interaction with Gαi remains unclear, the association between ELMO1 and G subunits is required for the GEF activity of the ELMO1/Dock1 complex for Rac activation. This is consistent with the findings that activated GTPases of the Rho and Arf families, RhoG and Arl4a, use ELMO1 as effectors by binding to the Ras-binding domain (RBD), and this contributes to both relieving the autoinhibition of Dock1 and also positioning the ELMO/Dock1 complex at the membrane for optimal Rac activation.Citation32,Citation34 New questions are emerging from the above studies, such as, how does an ELMO protein prioritize its interaction with G subunits? If they coexist in the same cells, which interaction is more important in the defined microenvironment? Future work is required to reveal the molecular mechanism of the association of Gαi and ELMO proteins and of the preference or conditions of the association of ELMO protein with G protein subunits.
Recent studies indicate that ELMO2 also has a Rac GEF function and is involved in polarity, adhesion, and cell migration, while little is known about ELMO3. In Madin-Darby canine kidney cells, ELMO2 recruits Dock1 to initial cell-cell contact.Citation50 In cell-cell contact, both ELMO2 and Dock1 are essential for the rapid recruitment and spreading of E-cadherin, actin reorganization, localized Rac and Rho GTPase activities, and the development of strong cell-cell adhesion. Sun et al. identified ELMO2 as the interacting protein of ClipR-59 using a yeast 2-hybrid screen, and found that interaction of ClipR-59 with ELMO2 enhanced Rac1 activation.Citation51 Recently, it has been shown that ELMO2 is required for insulin-induced Rac1 GTP loading.Citation52 It has been reported that there is differential distribution of ELMO1 and ELMO2 in the developing mouse brain.Citation53 Little is known about GPCR-mediated ELMO2 function in these processes.
ELMODs, in contrast to the ELMOs involved in unconventional Rac GEF function, have GAP activity for small GTPases Arls and Arfs. The Arf family consists of at least 30 members in mammals, including 6 Arfs, 22 Arls (Arf-like proteins), and 2 Sar proteins.Citation54 Arf and Sar regulate membrane trafficking,Citation55 while Arls are involved in diverse cellular functions, including energy metabolism, cytoskeleton dynamics, cytokinesis, lipid droplet formation, cilia function, and recruitment of Golgins to the Golgi.Citation58,Citation56 ELMOD2 was first purified based on its GAP activity for Arl2 and is the first GAP identified that shows activity toward 22 mammalian Arl proteins.Citation35 Consistent with the above, ELMOD1 also shows GAP activity toward Arl2. Surprisingly, ELMOD2 also exhibits GAP activity toward Arf1 and Arf6, although it lacks the unique canonical cysteine-rich zinc finger Arf GAP signature found in every other known Arf GAP protein.Citation59,Citation60 Recently, it was revealed that the Arf GAP activity of the ELMOD family lies within the ELMO domain and that a highly conserved arginine residue is critical for both the biochemical and cellular GAP activity of ELMODs.Citation36 It has also been shown that ELMOD1-3 display wide variations in GAP activity toward 6 different Arf members.Citation35,Citation37 The specific activities of ELMODs toward multiple members of the Arf family might contribute to their different roles in cellular functions. Neutrophils express Arf1 and Arf3.Citation61 Chemoattractant fMLP and leukotriene B4 trigger the membrane recruitment of Arf1 and the activation of phospholipase D (PLD) in neutrophils.Citation62,Citation63 Activated PLD hydrolyzes to produce phosphatidic acid (PA), a key molecule that recruits the ELMO/Dock2 complex.Citation46 These results indicate potential crosstalk between ELMOs and ELMODs in GPCR signaling. Further work is required to uncover the functions of ELMODs in GPCR-mediated Arf signaling and the functions of ELMOs in GPCR-mediated chemotaxis.
From the model organism D. discoideum to mammalian cells, cells utilize an evolutionarily conserved ELMO protein as the mediator to transduce GPCR signal through direct interaction with heterotrimeric G protein. Because very little is known about how Gα and Gβγ subunits interact and activate their effectors, future investigation of the molecular mechanisms of ELMO/G protein interactions, the preference of ELMO proteins for the defined G protein subunits, and the unconventional GAP activity of ELMO proteins will shed new light on the mechanism by which ELMO protein transduces GPCR/G protein signal.
Closing remarks
Eukaryotic cells often use GPCR-mediated signal transduction pathways to sense chemotactic gradients and guide directed cell migration. ELMO proteins are an evolutionarily conserved, essential component of the ELMO/Dock complex, which functions as a guanine nucleotide exchange factor (GEF) for small G protein Rac activation. The linkages between the GPCR/G protein-initiated gradient sensing machinery and the Rac-mediated actin polymerization machinery are not fully understood. Here, we summarize that eukaryotic cells use the direct association of ELMO and G proteins to transduce GPCR signal to effect dynamic remodeling of actin cytoskeleton through regulating Rac activation during chemotaxis as a general strategy in eukaryotic cells.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Dr. Xu is supported by the Intramural Fund of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
References
- Nourshargh S, Alon R. Leukocyte migration into inflamed tissues. Immunity 2014; 41:694-707.
- Jin T. GPCR-controlled chemotaxis in Dictyostelium discoideum. Wiley Interdiscip Rev Syst Biol Med 2011; 3:717-27; PMID:21381217; https://doi.org/10.1002/wsbm.143
- Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer 2011; 11:573-87; https://doi.org/10.1038/nrc3078
- Devreotes PN. G protein-linked signaling pathways control the developmental program of Dictyostelium. Neuron 1994; 12:235-41; PMID:8110455; https://doi.org/10.1016/0896-6273(94)90267-4
- Murphy PM. The molecular biology of leukocyte chemoattractant receptors. Annu Rev Immunol 1994; 12:593-633.
- Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001; 410:50-6; PMID:11242036; https://doi.org/10.1038/35065016
- Jin T, Xu X, Hereld D. Chemotaxis, chemokine receptors and human disease. Cytokine 2008; 44:1-8; PMID:18722135; https://doi.org/10.1016/j.cyto.2008.06.017
- Xu X, Meckel T, Brzostowski JA, Yan J, Meier-Schellersheim M, Jin T. Coupling mechanism of a GPCR and a heterotrimeric G protein during chemoattractant gradient sensing in Dictyostelium. Sci Signal 2010; 3:ra71; PMID:20876874
- Xu X, Meier-Schellersheim M, Jiao X, Nelson LE, Jin T. Quantitative imaging of single live cells reveals spatiotemporal dynamics of multistep signaling events of chemoattractant gradient sensing in Dictyostelium. Mol Biol Cell 2005; 16:676-88; PMID:15563608; https://doi.org/10.1091/mbc.E04-07-0544
- Sun CX, Magalhaes MA, Glogauer M. Rac1 and Rac2 differentially regulate actin free barbed end formation downstream of the fMLP receptor. J Cell Biol 2007; 179:239-45; PMID:17954607; https://doi.org/10.1083/jcb.200705122
- Pollitt AY, Insall RH. WASP and SCAR/WAVE proteins: the drivers of actin assembly. J Cell Sci 2009; 122:2575-8; PMID:19625501; https://doi.org/10.1242/jcs.023879
- Burianek LE, Soderling SH. Under lock and key: spatiotemporal regulation of WASP family proteins coordinates separate dynamic cellular processes. Semin Cell Dev Biol 2013; 24:258-66; PMID:23291261; https://doi.org/10.1016/j.semcdb.2012.12.005
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol 2005; 6:167-80; PMID:15688002; https://doi.org/10.1038/nrm1587
- Cote JF, Vuori K. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol 2007; 17:383-93; PMID:17765544; https://doi.org/10.1016/j.tcb.2007.05.001
- Gadea G, Blangy A. Dock-family exchange factors in cell migration and disease. Eur J Cell Biol 2014; 93:466-77; PMID:25022758; https://doi.org/10.1016/j.ejcb.2014.06.003
- Laurin M, Cote JF. Insights into the biological functions of Dock family guanine nucleotide exchange factors. Genes Dev 2014; 28:533-47; PMID:24637113; https://doi.org/10.1101/gad.236349.113
- Brzostowski JA, Fey P, Yan J, Isik N, Jin T. The Elmo family forms an ancient group of actin-regulating proteins. Communicative Integrative Biol 2009; 2:337-40; https://doi.org/10.4161/cib.2.4.8549
- Meller N, Merlot S, Guda C. CZH proteins: a new family of Rho-GEFs. J Cell Sci 2005; 118:4937-46; PMID:16254241; https://doi.org/10.1242/jcs.02671
- Reddien PW, Horvitz HR. The engulfment process of programmed cell death in caenorhabditis elegans. Annu Rev Cell Dev Biol 2004; 20:193-221; PMID:15473839; https://doi.org/10.1146/annurev.cellbio.20.022003.114619
- Patel M, Pelletier A, Cote JF. Opening up on ELMO regulation: New insights into the control of Rac signaling by the DOCK180/ELMO complex. Small GTPases 2011; 2:268-275; PMID:22292130; https://doi.org/10.4161/sgtp.2.5.17716
- Li H, Yang L, Fu H, Yan J, Wang Y, Guo H, Hao X, Xu X, Jin T, Zhang N. Association between Galphai2 and ELMO1/Dock180 connects chemokine signalling with Rac activation and metastasis. Nat Commun 2013; 4:1706; PMID:23591873; https://doi.org/10.1038/ncomms2680
- Wang Y, Xu X, Pan M, Jin T. ELMO1 directly interacts with Gbetagamma subunit to Transduce GPCR signaling to Rac1 activation in Chemotaxis. J Cancer 2016; 7:973-83; PMID:27313788; https://doi.org/10.7150/jca.15118
- Yan J, Mihaylov V, Xu X, Brzostowski JA, Li H, Liu L, Veenstra TD, Parent CA, Jin T. A Gbetagamma effector, ElmoE, transduces GPCR signaling to the actin network during chemotaxis. Dev Cell 2012; 22:92-103; PMID:22264729; https://doi.org/10.1016/j.devcel.2011.11.007
- Cai H, Das S, Kamimura Y, Long Y, Parent CA, Devreotes PN. Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis. J Cell Biol 2010; 190:233-45; PMID:20660630; https://doi.org/10.1083/jcb.201001129
- Charest PG, Shen Z, Lakoduk A, Sasaki AT, Briggs SP, Firtel RA. A Ras signaling complex controls the RasC-TORC2 pathway and directed cell migration. Dev Cell 2010; 18:737-49; PMID:20493808; https://doi.org/10.1016/j.devcel.2010.03.017
- Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell 2002; 109:611-23; PMID:12062104; https://doi.org/10.1016/S0092-8674(02)00755-9
- Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell 2002; 109:599-610; PMID:12062103; https://doi.org/10.1016/S0092-8674(02)00745-6
- Veltman DM, van Haastert PJ. The role of cGMP and the rear of the cell in Dictyostelium chemotaxis and cell streaming. J Cell Sci 2008; 121:120-7; PMID:18073238; https://doi.org/10.1242/jcs.015602
- Insall RH, Machesky LM. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev Cell 2009; 17:310-22; PMID:19758556; https://doi.org/10.1016/j.devcel.2009.08.012
- Meller N, Westbrook MJ, Shannon JD, Guda C, Schwartz MA. Function of the N-terminus of zizimin1: autoinhibition and membrane targeting. Biochem J 2008; 409:525-33; PMID:17935486; https://doi.org/10.1042/BJ20071263
- Isik N, Brzostowski JA, Jin T. An Elmo-like protein associated with myosin II restricts spurious F-actin events to coordinate phagocytosis and chemotaxis. Dev Cell 2008; 15:590-602; PMID:18854143; https://doi.org/10.1016/j.devcel.2008.08.006
- Patel M, Chiang TC, Tran V, Lee FJ, Cote JF. The Arf family GTPase Arl4A complexes with ELMO proteins to promote actin cytoskeleton remodeling and reveals a versatile Ras-binding domain in the ELMO proteins family. J Biol Chem 2011; 286:38969-79; PMID:21930703; https://doi.org/10.1074/jbc.M111.274191
- Kae H, Lim CJ, Spiegelman GB, Weeks G. Chemoattractant-induced Ras activation during Dictyostelium aggregation. EMBO Rep 2004; 5:602-6; PMID:15143344; https://doi.org/10.1038/sj.embor.7400151
- Patel M, Margaron Y, Fradet N, Yang Q, Wilkes B, Bouvier M, Hofmann K, Cote JF. An evolutionarily conserved autoinhibitory molecular switch in ELMO proteins regulates Rac signaling. Curr Biol 2010; 20:2021-7; PMID:21035343; https://doi.org/10.1016/j.cub.2010.10.028
- Bowzard JB, Cheng D, Peng J, Kahn RA. ELMOD2 is an Arl2 GTPase-activating protein that also acts on Arfs. J Biol Chem 2007; 282:17568-80; PMID:17452337; https://doi.org/10.1074/jbc.M701347200
- East MP, Bowzard JB, Dacks JB, Kahn RA. ELMO domains, evolutionary and functional characterization of a novel GTPase-activating protein (GAP) domain for Arf protein family GTPases. J Biol Chem 2012; 287:39538-53; PMID:23014990; https://doi.org/10.1074/jbc.M112.417477
- Ivanova AA, East MP, Yi SL, Kahn RA. Characterization of recombinant ELMOD (cell engulfment and motility domain) proteins as GTPase-activating proteins (GAPs) for ARF family GTPases. J Biol Chem 2014; 289:11111-21; PMID:24616099; https://doi.org/10.1074/jbc.M114.548529
- Grimsley CM, Lu M, Haney LB, Kinchen JM, Ravichandran KS. Characterization of a novel interaction between ELMO1 and ERM proteins. J Biol Chem 2006; 281:5928-37; PMID:16377631; https://doi.org/10.1074/jbc.M510647200
- Handa Y, Suzuki M, Ohya K, Iwai H, Ishijima N, Koleske AJ, Fukui Y, Sasakawa C. Shigella IpgB1 promotes bacterial entry through the ELMO-Dock180 machinery. Nat Cell Biol 2007; 9:121-8; PMID:17173036; https://doi.org/10.1038/ncb1526
- Katoh H, Negishi M. RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature 2003; 424:461-4; PMID:12879077; https://doi.org/10.1038/nature01817
- Komander D, Patel M, Laurin M, Fradet N, Pelletier A, Barford D, Cote JF. An alpha-helical extension of the ELMO1 pleckstrin homology domain mediates direct interaction to DOCK180 and is critical in Rac signaling. Mol Biol Cell 2008; 19:4837-51; PMID:18768751; https://doi.org/10.1091/mbc.E08-04-0345
- Hiramoto-Yamaki N, Takeuchi S, Ueda S, Harada K, Fujimoto S, Negishi M, Katoh H. Ephexin4 and EphA2 mediate cell migration through a RhoG-dependent mechanism. J Cell Biol 2010; 190:461-77; PMID:20679435; https://doi.org/10.1083/jcb.201005141
- Vives V, Laurin M, Cres G, Larrousse P, Morichaud Z, Noel D, Cote JF, Blangy A. The Rac1 exchange factor Dock5 is essential for bone resorption by osteoclasts. J Bone Miner Res 2011; 26:1099-110; PMID:21542010; https://doi.org/10.1002/jbmr.282
- Xiao Y, Peng Y, Wan J, Tang G, Chen Y, Tang J, Ye WC, Ip NY, Shi L. The atypical guanine nucleotide exchange factor Dock4 regulates neurite differentiation through modulation of Rac1 GTPase and actin dynamics. J Biol Chem 2013; 288:20034-45; PMID:23720743; https://doi.org/10.1074/jbc.M113.458612
- Li Z, Jiang H, Xie W, Zhang Z, Smrcka AV, Wu D. Roles of PLC-beta2 and -beta3 and PI3Kgamma in chemoattractant-mediated signal transduction. Science 2000; 287:1046-9; PMID:10669417; https://doi.org/10.1126/science.287.5455.1046
- Nishikimi A, Fukuhara H, Su W, Hongu T, Takasuga S, Mihara H, Cao Q, Sanematsu F, Kanai M, Hasegawa H, et al. Sequential regulation of DOCK2 dynamics by two phospholipids during neutrophil chemotaxis. Science 2009; 324:384-7; PMID:19325080; https://doi.org/10.1126/science.1170179
- Kunisaki Y, Nishikimi A, Tanaka Y, Takii R, Noda M, Inayoshi A, Watanabe K, Sanematsu F, Sasazuki T, Sasaki T, et al. DOCK2 is a Rac activator that regulates motility and polarity during neutrophil chemotaxis. J Cell Biol 2006; 174:647-52; PMID:16943182; https://doi.org/10.1083/jcb.200602142
- Sanematsu F, Nishikimi A, Watanabe M, Hongu T, Tanaka Y, Kanaho Y, Cote JF, Fukui Y. Phosphatidic acid-dependent recruitment and function of the Rac activator DOCK1 during dorsal ruffle formation. J Biol Chem 2013; 288:8092-100; PMID:23362269; https://doi.org/10.1074/jbc.M112.410423
- Margaron Y, Fradet N, Cote JF. ELMO recruits actin cross-linking family 7 (ACF7) at the cell membrane for microtubule capture and stabilization of cellular protrusions. J Biol Chem 2013; 288:1184-99; PMID:23184944; https://doi.org/10.1074/jbc.M112.431825
- Toret CP, Collins C, Nelson WJ. An Elmo-Dock complex locally controls Rho GTPases and actin remodeling during cadherin-mediated adhesion. J Cell Biol 2014; 207:577-87; PMID:25452388; https://doi.org/10.1083/jcb.201406135
- Sun Y, Ren W, Cote JF, Hinds PW, Hu X, Du K. ClipR-59 interacts with Elmo2 and modulates myoblast fusion. J Biol Chem 2015; 290:6130-40; PMID:25572395; https://doi.org/10.1074/jbc.M114.616680
- Sun Y, Cote JF, Du K. Elmo2 is a regulator of Insulin-dependent Glut4 membrane translocation. J Biol Chem 2016; 291:16150-61; PMID:27226625; https://doi.org/10.1074/jbc.M116.731521
- Katoh H, Fujimoto S, Ishida C, Ishikawa Y, Negishi M. Differential distribution of ELMO1 and ELMO2 mRNAs in the developing mouse brain. Brain Res 2006; 1073-4:103-8; PMID:16443196; https://doi.org/10.1016/j.brainres.2005.12.085
- Kahn RA, Cherfils J, Elias M, Lovering RC, Munro S, Schurmann A. Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J Cell Biol 2006; 172:645-50; PMID:16505163; https://doi.org/10.1083/jcb.200512057
- Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol 2011; 12:362-75; PMID:21587297; https://doi.org/10.1038/nrm3117
- Zhou C, Cunningham L, Marcus AI, Li Y, Kahn RA. Arl2 and Arl3 regulate different microtubule-dependent processes. Mol Biol Cell 2006; 17:2476-87; PMID:16525022; https://doi.org/10.1091/mbc.E05-10-0929
- Li Y, Wei Q, Zhang Y, Ling K, Hu J. The small GTPases ARL-13 and ARL-3 coordinate intraflagellar transport and ciliogenesis. J Cell Biol 2010; 189:1039-51; PMID:20530210; https://doi.org/10.1083/jcb.200912001
- Larkins CE, Aviles GD, East MP, Kahn RA, Caspary T. Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol Biol Cell 2011; 22:4694-703; PMID:21976698; https://doi.org/10.1091/mbc.E10-12-0994
- Cukierman E, Huber I, Rotman M, Cassel D. The ARF1 GTPase-activating protein: zinc finger motif and Golgi complex localization. Science 1995; 270:1999-2002; PMID:8533093; https://doi.org/10.1126/science.270.5244.1999
- Kahn RA, Bruford E, Inoue H, Logsdon JM, Jr, Nie Z, Premont RT, Randazzo PA, Satake M, Theibert AB, Zapp ML, et al. Consensus nomenclature for the human ArfGAP domain-containing proteins. J Cell Biol 2008; 182:1039-44; PMID:18809720; https://doi.org/10.1083/jcb.200806041
- Gamara J, Chouinard F, Davis L, Aoudjit F, Bourgoin SG. Regulators and Effectors of Arf GTPases in Neutrophils. J Immunol Res 2015; 2015:235170; PMID:26609537; https://doi.org/10.1155/2015/235170
- Grenier S, Flamand N, Pelletier J, Naccache PH, Borgeat P, Bourgoin SG. Arachidonic acid activates phospholipase D in human neutrophils; essential role of endogenous leukotriene B4 and inhibition by adenosine A2A receptor engagement. J Leukoc Biol 2003; 73:530-9; PMID:12660228; https://doi.org/10.1189/jlb.0702371
- Thibault N, Harbour D, Borgeat P, Naccache PH, Bourgoin SG. Adenosine receptor occupancy suppresses chemoattractant-induced phospholipase D activity by diminishing membrane recruitment of small GTPases. Blood 2000; 95:519-27; PMID:10627457