ABSTRACT
The small GTPase Rab7 is the main regulator of membrane trafficking at late endosomes. This small GTPase regulates endosome-to-trans Golgi Network trafficking of sorting receptors, membrane fusion of late endosomes to lysosomes, and autophagosomes to lysosomes during autophagy. Rab7, like all Rab GTPases, binds downstream effectors coordinating several divergent pathways. How cells regulate these interactions and downstream functions is not well understood. Recent evidence suggests that Rab7 function can be modulated by the combination of several post-translational modifications that facilitate interactions with one effector while preventing binding to another one. In this review, we discuss recent data on how phosphorylation, palmitoylation and ubiquitination modulate the ability of this small GTPase to orchestrate membrane trafficking at the late endosomes.
Introduction
The small GTPases of the Rab family play crucial roles in determining the identity and destination of vesicles. As such, they are key regulators of the formation, trafficking, and fusion of transport vesicles at the endoplasmic reticulum (ER), Golgi apparatus and early and late endosomes.Citation1 Cells tightly regulate the function of Rab GTPases through various mechanisms. Indeed increasing evidence suggests that alterations in the physiological functions of Rab GTPases can lead to the development of a wide range of diseases, spanning from cancerCitation2 to neuropathyCitation3 and neurodegenerative disorders.Citation4
After their cytosolic synthesis, Rab GTPases bind Rab Escort Protein (REP)Citation5 and are presented to the geranylgeranyltransferase (RabGGTase)Citation6 to be prenylated (geranylgeranylated) on their C-terminal cysteines.Citation7 Prenylation is a non-reversible post-translational modification consisting in the attachment of a lipidic moiety to a cysteine residue. It was the first post-translational modification identified on Rab GTPases and is required for Rabs to bind membranes as mutations affecting C-terminal cysteine prenylation preclude membrane localization of the protein, which appears almost completely localized in the cytosol.Citation8 Rab GTPases cycle between a GTP-bound active state and a GDP-bound inactive state. In their inactive state, Rab GTPases are bound by a guanine-nucleotide dissociation inhibitor (GDI) that masks the prenylated tail and keeps Rabs soluble in the cytosol.Citation9
The exact mechanisms governing membrane translocation of Rabs, as well as how specific membrane targeting is achieved is not completely understood. Evidence indicates that guanine exchange factors (GEFs) are able to induce GDI release and Rab targeting onto specific membranes,Citation10,11 suggesting that each GEF activates and recruits a specific Rab at a defined subcellular compartment.Citation12 This GDP to GTP switch also enables Rab GTPases to interact with specific downstream effectors.Citation13 GTPase activating proteins (GAPs) terminate the activity of Rab GTPases by hydrolyzing GTP to GDP.Citation14 In a GDP bound form, Rabs bind to GDI, are returned to the cytosol, and the cycle can begin anew.
The small GTPase Rab7 is a regulator of membrane trafficking at the late endosome and also plays other key roles by interacting with a variety of downstream effectors. Rab7 regulates the spatiotemporal recruitment of retromer, an evolutionary conserved multimeric complex composed of a trimer of vacuolar protein sorting (Vps)-26, Vps35, and Vps29, and of a dimer composed of a varying combination of sorting nexins (SNX).Citation15,16 In mammalian cells, retromer is responsible for the retrieval of Vps10-domain family receptors (Sortilin, SorLA, SorCS)Citation17,18 and cation-independent mannose phosphate receptor (CI-MPR) from the late endosome to the trans-Golgi Network (TGN).Citation19,20 In what appears to be a Rab7-independent pathway, retromer is also responsible for recycling of integral membrane proteins from the endosome to the plasma membrane.Citation21 Rab7 is also required for the degradation of cell surface receptors such as the Epidermal Growth Factor Receptor (EGFR).Citation22,23 Finally, Rab7 also participates in autophagosome-lysosome fusion through the homotypic fusion and vacuole protein sorting (HOPS) complex,Citation24 thereby regulating autophagy. For more detailed information on Rab7 functions, we refer the reader to a recent review by Guerra and Bucci.Citation25 How Rab7 is able to specifically interact with several effectors to regulate these various pathways is still not completely understood. In the last few years, the role of post-translational modifications including phosphorylation, ubiquitination and palmitoylation, in modulating Rab7 function has been increasingly explored. In this review, we will highlight recent advances on how post-translational modifications regulate Rab7 function.
Phosphorylation
Phosphoproteomic analysis identified at least two phosphorylatable sites on Rab7, serine 72 (S72) and tyrosine 183 (Y183).Citation26,27 Although the phosphorylation of these two residues has now been demonstrated, it is still not fully understood how this post-translational modification at these sites modulate Rab7 function(s) and which kinase(s) and phosphatase(s) are involved. However recent work has begun to unravel the role of phosphorylation on Rab7 function.
Serine 72
Phosphorylation on Serine 72 appears to negatively regulate Rab7 activity in the cell. Indeed, the phosphomimetic mutant Rab7S72E, which mimics a constitutively phosphorylated protein at this site, does not localize to endosomes and is almost completely cytosolic.Citation28 On the contrary, the phosphonull Rab7 mutant Rab7S72A, that cannot be phosphorylated, shows a similar localization to wild-type Rab7.Citation28 Functionally, HeLa cells transfected with Rab7S72E demonstrate delayed EGFR degradation, compared to non-transfected cells or cells expressing wild-type Rab7.28 In turn, this resulted in increased EGFR signaling.Citation28 Further supporting a role of phosphorylation at S72 functioning as a negative regulator of Rab7 function, Rab7S72E does not interact with its effector Rab interacting lysosomal protein (RILP) as efficiently as wild-type Rab7 or Rab7S72A. Taken together, this data suggests a possible role of S72 phosphorylation as a molecular switch that, together with GAP-mediated GTP to GDP hydrolysis, contributes to termination of Rab7 activity in the cell.
Tyrosine 183
A recent publication showed that the phosphorylation of Y183 prevents the protein from localizing to endosomes. Indeed, phosphomimetic Rab7Y183D was found in the cytosol compared to wild-type Rab7 or phosphonull Rab7Y183F which showed punctate staining.Citation28 However, a subsequent study found phosphomimetic Rab7Y183E displaying a similar pattern of localization to phosphonull Rab7Y183F, suggesting that constitutive phosphorylation of Y183 did not interfere with the ability of Rab7 to bind endosomal membranes.Citation29 The discrepancy in these results could possibly be explained by the use of two different phosphomimetic mutations, Y183D versus Y183E. However, in both cases, quantification experiments were not performed. A robust statistical analysis of the distribution of these mutants or membrane separation assays could be used to determine the localization of these mutants, in side-by-side comparison to determine whether of not phosphorylation at Y183 affects membrane binding.
Functionally, three studies have investigated the impact of Y183 phosphorylation on the degradation kinetics of EGFR. In two cases, it was found that overexpressing phosphomimetic Rab7Y183D or Rab7Y183E in HeLa cells impaired EGFR degradation.Citation28, 29 However, in contrast to these finding, another group reported that phosphorylation on Y183 is required to efficiently degrade EGFR.Citation30 Indeed, these authors demonstrated that subsequent to EGF stimulation, EGFR promotes Rab7Y183 phosphorylation enabling its degradation in lysosome and preventing its recycling to the plasma membrane.Citation30 In that same study, expression of the phosphonull mutant, Rab7Y183F, blocked EGFR degradation and sustained extracellular signal–regulated kinases (ERK) signaling.Citation30 It is important to point out that all three groups performed experiments in cell expressing endogenous Rab7 that could affect the data obtained, especially in the overexpression studies.
Studies to determine the role of Y183 phosphorylation in mediating downstream effector interactions found that Y183 phosphorylation inhibits Rab7 binding with RILP. Indeed, the phosphomutant Rab7Y183F was able to co-immunoprecipitate more RILP than wild-type Rab7, while the phosphomimetic mutants Rab7Y183D and Rab7Y183E do not co-immunoprecipitate this effector.Citation28,29 If Y183 mediates membrane localization as was shown for Rab7Y183D, and membrane localization is a prerequisite for activation, this would explain why the interaction with RILP was lost and this would also suggest that all effector binding would be lost. If phosphorylation at Y183 does not affect membrane binding as was shown with Rab7Y183E, this would suggest that phosphorylation at this site could be used to prevent the Rab7/RILP interaction and redirect the activity of Rab7 toward a different effector(s), while not affecting its GTP loading. It will be important to further characterize the functional role of Y183 phosphorylation by testing more effector interactions and functional pathways where Rab7 plays a crucial function.
Kinases and Phosphatases
To date, little is known about the kinases and phosphatases regulating the Rab7 phosphorylation cycle. For example, no kinase(s) have been identified that would phosphorylate S72. However, Src kinase has been identified as responsible for the phosphorylation on Y183. Indeed, a significant decrease in the phosphorylation level of Rab7 was observed in cells treated with an inhibitor of the Src-kinase family or shRNA depletion of Src kinase.Citation29
The phosphatase and tensin homologous 10 (PTEN) seems to have a role in de-phosphorylating Rab7 on both serine 72 and tyrosine 183Citation28 as shRNA-mediated knock-down of PTEN was associated with increased Rab7 phosphorylation on both S72 and Y183. Moreover, depletion of PTEN induced a re-localization of the small GTPase from late-endosomes to the cytosol, recapitulating the subcellular localization of the Rab7 phosphomimetic mutants.Citation28 More work will be required to identify and characterize other enzymes in the Rab7 phosphorylation cycle and identify which functions of Rab7 these proteins regulate.
Ubiquitination
Ubiquitination is the attachment of a small protein, ubiquitin, to a lysine residue of a target protein substrate.Citation31 This reversible post-translational modification is mediated by the coordinated action of three enzymes, E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes and E3 ubiquitin-ligases, while a family of deubiquitinating (DUBs) enzymes is responsible for ubiquitin deconjugation.Citation32,33 Ubiquitination regulates several aspects of cellular function, such as endocytosis,Citation34 intracellular signaling pathwaysCitation35 and protein degradation in lysosomes.Citation36,37 A target protein can be mono or poly-ubiquitinated at one or multiple sites and linear or branched ubiquitin chains can be attached, finely modulating the substrate function.Citation38 Global analysis of the ubiquitinated proteome identified Rab7 lysine K38, K191 and K194 as possible target for ubiquitination.Citation39–Citation41 Interestingly, Rab7 ubiquitination is mediated by Parkin, which belongs to the family of E3 ubiquitin ligase. Loss-of-function mutations on parkin have been linked to the development of familial, early onset Parkinson's disease (PD), demonstrating a possible role of Rab7 in this disease. Parkin mediates the ubiquitination of all three ubiquitinatable Rab7 lysine residues, with preference for K38. In particular this modification appears to be important to stabilize Rab7, favoring the interaction of the small GTPase with its effector RILP. Indeed, mutation of K38 on Rab7 or mutations inactivating the ubiquitin ligase-activity of Parkin are associated with reduced binding between Rab7 and RILP.Citation40 Furthermore, Rab7K38R mutant is displaced to the cytosol, suggesting that K38 ubiquitination is required for efficient translocation of Rab7 to membranes.Citation40 Functionally, the expression of Rab7K38R partially recapitulates the phenotype associated with Parkin loss of function, highlighting the importance of a strictly regulated control of Rab7 activity in the context of PD.Citation25,40 To date, no DUBs have been identified as Rab7 deubiquitinating enzymes.
Palmitoylation
Palmitoylation (also known as S-Acylation) is the reversible attachment of a palmitate group to a cysteine residue of a protein via a reversible thioester bond.Citation42 This post-translational modification can modulate the function of a protein in different ways including stability, protein-protein interactions and membrane binding.Citation43 Palmitoyltransferases (PATs) are enzymes that mediate palmitoylation and contain a conserved catalytic domain, characterized by the amino acid sequence aspartic acid-histidine-histidine-cysteine.Citation44 In humans, 24 PATs (also referred as DHHCs) have been identified,Citation44–Citation46 and proteomic analysis of S-acylated proteins in mammals revealed that several hundred proteins are palmitoylated.Citation44 We do not yet know the substrate specificity of each DHHC, but it is clear that some proteins can be palmitoylated by different DHHCs, while others require the action of a defined enzyme.Citation44 Much less is known about the mechanisms regulating protein depalmitoylation. Acyl-Protein thioesterases (APT) 1 has been shown to have a role in G protein alpha subunit, SNAP-23 and H-Ras depalmitoylation,Citation47–Citation50 while APT2 has been shown to be responsible for deacylation of GAP-43.Citation48 Recently, members of the ABHD family have been identified as thioesterases for N-Ras and PSD-95,Citation51,52 but little is known on the cellular localization and substrate specificity of these enzymes.
We have recently shown that Rab7 is palmitoylated on two cysteine residues, C83 and C84. This modification is not required for membrane binding as non-palmitoylatable Rab7, Rab7C83,83S, was still membrane bound and localized to endosomes.Citation8 Palmitoylation regulates the ability of Rab7 to interact with and recruit retromer to endosomes.Citation8 Rab7 palmitoylation is essential for efficient retromer function in cells as the expression of Rab7C83,84S in a Rab7-KO HEK293 cell line was not able to properly recruit retromer to late endosomes. This lead to impaired endosome-to-TGN cargo receptors recycling that ultimately caused the miss-localization of lysosomal enzymes, which are secreted into the extracellular media.Citation8
Interestingly, palmitoylation is required for the Rab7/retromer interaction and recruitment, but is not required for the Rab7/RILP interaction. Indeed, not only did the non-palmitoylatable mutant Rab7C83,84S interact with RILP as efficiently as wild-type Rab7, but its expression in Rab7-KO cell restored the degradation of EGFR as efficiently as wild-type Rab7, suggesting that palmitoylation is involved specifically in regulating the Rab7/retromer interaction and function.Citation8
There is no data yet on the enzymes regulating the Rab7 palmitoylation/depalmitoylation cycle. DHHC5 was found in a screen to identify proteins involved in endosome-to-TGN traffickingCitation53 and it would be interesting to investigate if Rab7 is one of the targets of this enzyme.
Conclusion and Perspectives
In the last few years, increasing evidence has pointed to how the functions of Rab7 are modulated by different post-translational modifications. Indeed the combination of palmitoylation, phosphorylation and ubiquitination enables the protein to interact specifically with one effector and not others, making Rab7 extremely versatile and able to coordinate several different pathways at the late endosome. We are only at the beginning of our understanding of the mechanisms behind Rab7 post-translational modifications. The enzymes involved in these processes have not been fully identified or characterized and the function of these modifications requires further work. Although one kinase (Src),Citation29 one phosphatase (PTEN)Citation28 and an E3 ubiquitin-ligase (Parkin)Citation40 have been identified (), the enzymes in the palmitoylation cycle have not and we predict that other kinases, phosphatases and ubiquitin ligases may yet be identified and characterized. Once we have a greater understanding of the players involved in controlling these modifications, this machinery could be used as pharmaceutical target to differentially modulate the function of Rab7.
Rab7 and its downstream effectors such as retromer have been implicated in neurological diseases such as Charcot-Marie-Tooth diseaseCitation54 and in the development of neurodegenerative diseases such as Alzheimer's (AD),Citation55 Parkinson's (PD)Citation56 and Batten disease (BD).Citation57 Indeed, even small reductions in retromer function can progressively impair the lysosome degradative system leading in the long term to neurodegeneration. Post-mortem analysis of brain sections of AD patients revealed a decrease in the expression level of retromer subunits.Citation58,59 Interestingly, Rab7 expression was upregulated in AD patients.Citation60 Moreover, mutations affecting subunits of the retromer heterotrimer have been linked to familial PDCitation61–Citation63 and atypical Parkinsonism.Citation64 Due to its crucial role in the pathogenesis of neurodegenerative disorders, modulating the function of retromer via Rab7 post-translational modification could offer a novel therapeutic approach to treat these diseases. In vitro, the use of pharmaceutical chaperones stabilizing retromer has been shown to improve retromer functions ameliorating ADCitation65 and PD phenotypesCitation66 highlighting the feasibility of using this approach.
Acknowledgments
We would like to thank Dr. Peter McCormick and members from our research team for helpful discussions. We apologize to our colleagues whose work was not cited in this review due to space constraints. Work from our group included in this review was supported by a grant from the Canadian Institutes of Health Research (MOP-102754) to SL. SL is a recipient of salary support from Fonds de Recherche du Québec – Santé. GM is the recipient of a scholarship from the Fondation Universitaire Armand-Frappier.
Disclosure Statement
The authors declare no conflicts
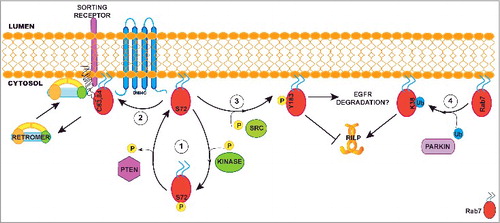
Figure 1. Role of post-translational modifications in modulating Rab7 functions. (1) Phosphorylation on Serine 72 by a non-identified kinase displaces Rab7 from the membrane to the cytosol. Here, the phosphatase PTEN can remove the phosphorylation on Serine 72 enabling Rab7 to bind the membrane again. (2) On the membrane, Rab7 can be palmitoylated by a yet unknown palmitoyltransferase. Palmitoylation on cysteines 83 and 84 is required to recruit retromer at the late endosome to mediate sorting receptor retrieval to the trans-Golgi Network. (3) Src mediated phosphorylation of Y183 blocks Rab7 interaction with its effector RILP. The role of this modification on EGFR degradation is controversial as some groups found that phosphorylation is required for degradation while other found it must not be phosphorylated. (4) Ubiquitination of K38 mediated by the E3-ligase parkin stabilizes Rab7 on the membrane and is required for the interaction with the Rab7 effector RILP. In this schematic view, Rab7 is represented with one post-translational modification at time, but we cannot exclude the possibility that palmitoylation, phosphorylation and ubiquitination are mutually exclusive. In fact, it is feasible that various combinations of post-translational modifications determine the specific function of Rab7 on late-endosomal membranes.
Additional information
Funding
References
- Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–49. doi:10.1152/physrev.00059.2009. PMID:21248164.
- Tzeng H-T, Wang Y-C. Rab-mediated vesicle trafficking in cancer. J Biomed Sci. 2016;23:70. doi:10.1186/s12929-016-0287-7. PMID:27716280.
- Cogli L, Piro F, Bucci C. Rab7 and the CMT2B disease. Biochem Soc Trans. 2009;37:1027–31. doi:10.1042/BST0371027. PMID:19754445.
- Tang BL. Rabs, Membrane Dynamics, and Parkinson's Disease. J Cell Physiol. 2017;232:1626–33. doi:10.1002/jcp.25713. PMID:27925204.
- Alexandrov K, Horiuchi H, Steele-Mortimer O, Seabra MC, Zerial M. Rab escort protein-1 is a multifunctional protein that accompanies newly prenylated rab proteins to their target membranes. EMBO J 1994;13:5262–73. PMID:7957092.
- Andres DA, Seabra MC, Brown MS, Armstrong SA, Smeland TE, Cremers FP, Goldstein JL. cDNA cloning of component A of Rab geranylgeranyl transferase and demonstration of its role as a Rab escort protein. Cell 1993;73:1091–9. doi:10.1016/0092-8674(93)90639-8. PMID:8513495.
- Joberty G, Tavitian A, Zahraoui A. Isoprenylation of Rab proteins possessing a C-terminal CaaX motif. FEBS Lett 1993;330:323–8. doi:10.1016/0014-5793(93)80897-4. PMID:8375503.
- Modica G, Skorobogata O, Sauvageau E, Vissa A, Yip CM, Kim PK, Wurtele H, Lefrancois S. Rab7 palmitoylation is required for efficient endosome-to-TGN trafficking. J Cell Sci. 2017;130:2579–90. doi:10.1242/jcs.199729. PMID:28600323.
- Muller MP, Goody RS. Molecular control of Rab activity by GEFs, GAPs and GDI. Small GTPases. 2017:1–17.
- Blumer J, Rey J, Dehmelt L, Mazel T, Wu YW, Bastiaens P, Goody RS, Itzen A. RabGEFs are a major determinant for specific Rab membrane targeting. J Cell Biol. 2013;200:287–300. doi:10.1083/jcb.201209113. PMID:23382462.
- Wu Y-W, Oesterlin LK, Tan K-T, Waldmann H, Alexandrov K, Goody RS. Membrane targeting mechanism of Rab GTPases elucidated by semisynthetic protein probes. Nat Chem Biol. 2010;6:534–40. doi:10.1038/nchembio.386. PMID:20512138.
- Pfeffer S, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol. 2004;5:886–96. doi:10.1038/nrm1500. PMID:15520808.
- Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci U S A. 2006;103:11821–7. doi:10.1073/pnas.0601617103. PMID:16882731.
- Barr F, Lambright DG. Rab GEFs and GAPs. Curr Opin Cell Biol. 2010;22:461–70. doi:10.1016/j.ceb.2010.04.007. PMID:20466531.
- Rojas R, van Vlijmen T, Mardones GA, Prabhu Y, Rojas AL, Mohammed S, Heck AJ, Raposo G, van der Sluijs P, Bonifacino JS. Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J Cell Biol. 2008;183:513–26. doi:10.1083/jcb.200804048. PMID:18981234.
- Seaman MN, Harbour ME, Tattersall D, Read E, Bright N. Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J Cell Sci. 2009;122:2371–82. doi:10.1242/jcs.048686. PMID:19531583.
- Canuel M, Lefrancois S, Zeng J, Morales CR. AP-1 and retromer play opposite roles in the trafficking of sortilin between the Golgi apparatus and the lysosomes. Biochem Biophys Res Commun. 2008;366:724–30. doi:10.1016/j.bbrc.2007.12.015. PMID:18078806.
- Fjorback AW, Seaman M, Gustafsen C, Mehmedbasic A, Gokool S, Wu C, Militz D, Schmidt V, Madsen P, Nyengaard JR, et al. Retromer binds the FANSHY sorting motif in SorLA to regulate amyloid precursor protein sorting and processing. J Neurosci. 2012;32:1467–80. doi:10.1523/JNEUROSCI.2272-11.2012. PMID:22279231.
- Seaman MN. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol. 2004;165:111–22. doi:10.1083/jcb.200312034. PMID:15078902.
- Arighi CN, Hartnell LM, Aguilar RC, Haft CR, Bonifacino JS. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J Cell Biol. 2004;165:123–33. doi:10.1083/jcb.200312055. PMID:15078903.
- Temkin P, Lauffer B, Jager S, Cimermancic P, Krogan NJ, von Zastrow M. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat Cell Biol. 2011;13:715–21. doi:10.1038/ncb2252. PMID:21602791.
- Ceresa BP, Bahr SJ. rab7 activity affects epidermal growth factor:epidermal growth factor receptor degradation by regulating endocytic trafficking from the late endosome. J Biol Chem. 2006;281:1099–106. doi:10.1074/jbc.M504175200. PMID:16282324.
- Vanlandingham PA, Ceresa BP. Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J Biol Chem. 2009;284:12110–24. doi:10.1074/jbc.M809277200. PMID:19265192.
- McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, Coxon FP, Miranda de Stegmann D, Bhogaraju S, Maddi K, et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell. 2015;57:39–54. doi:10.1016/j.molcel.2014.11.006. PMID:25498145.
- Guerra F, Bucci C. Multiple Roles of the Small GTPase Rab7. Cells. 2016;5. doi:10.3390/cells5030034. PMID:27548222.
- Zhou H, Di Palma S, Preisinger C, Peng M, Polat AN, Heck AJ, Mohammed S. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J Proteome Res. 2013;12:260–71. doi:10.1021/pr300630k. PMID:23186163.
- Sharma K, D'Souza Rochelle CJ, Tyanova S, Schaab C, Wiśniewski Jacek R, Cox J, Mann M. Ultradeep Human Phosphoproteome Reveals a Distinct Regulatory Nature of Tyr and Ser/Thr-Based Signaling. Cell Reports. 2014;8:1583–94. doi:10.1016/j.celrep.2014.07.036. PMID:25159151.
- Shinde SR, Maddika S. PTEN modulates EGFR late endocytic trafficking and degradation by dephosphorylating Rab7. Nat Commun. 2016;7:10689. doi:10.1038/ncomms10689. PMID:26869029.
- Lin X, Zhang J, Chen L, Chen Y, Xu X, Hong W, Wang T. Tyrosine phosphorylation of Rab7 by Src kinase. Cell Signal. 2017;35:84–94. doi:10.1016/j.cellsig.2017.03.006. PMID:28336235.
- Francavilla C, Papetti M, Rigbolt KT, Pedersen AK, Sigurdsson JO, Cazzamali G, Karemore G, Blagoev B, Olsen JV. Multilayered proteomics reveals molecular switches dictating ligand-dependent EGFR trafficking. Nat Struct Mol Biol. 2016;23:608–18. doi:10.1038/nsmb.3218. PMID:27136326.
- Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi:10.1038/nrm1700. PMID:16064136.
- Yamano K, Matsuda N, Tanaka K. The ubiquitin signal and autophagy: an orchestrated dance leading to mitochondrial degradation. EMBO Rep. 2016;17:300–16. doi:10.15252/embr.201541486. PMID:26882551.
- Hanpude P, Bhattacharya S, Dey AK, Maiti TK. Deubiquitinating enzymes in cellular signaling and disease regulation. IUBMB Life. 2015;67:544–55. doi:10.1002/iub.1402. PMID:26178252.
- Urbe S. Ubiquitin and endocytic protein sorting. Essays Biochem. 2005;41:81–98. doi:10.1042/bse0410081. PMID:16250899.
- Devoy A, Soane T, Welchman R, Mayer RJ. The ubiquitin-proteasome system and cancer. Essays Biochem. 2005;41:187–203. doi:10.1042/bse0410187. PMID:16250906.
- d'Azzo A, Bongiovanni A, Nastasi T. E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic. 2005;6:429–41. doi:10.1111/j.1600-0854.2005.00294.x. PMID:15882441.
- Dumaresq-Doiron K, Jules F, Lefrancois S. Sortilin turnover is mediated by ubiquitination. Biochem Biophys Res Commun. 2013;433:90–5. doi:10.1016/j.bbrc.2013.02.059. PMID:23485461.
- Ohtake F, Tsuchiya H. The emerging complexity of ubiquitin architecture. J Biochem. 2017;161:125–33. PMID:28011818.
- Mertins P, Qiao JW, Patel J, Udeshi ND, Clauser KR, Mani DR, Burgess MW, Gillette MA, Jaffe JD, Carr SA. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods. 2013;10:634–7. doi:10.1038/nmeth.2518. PMID:23749302.
- Song P, Trajkovic K, Tsunemi T, Krainc D. Parkin Modulates Endosomal Organization and Function of the Endo-Lysosomal Pathway. J Neurosci. 2016;36:2425–37. doi:10.1523/JNEUROSCI.2569-15.2016. PMID:26911690.
- Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10:M111. 013284. doi:10.1074/mcp.M111.013284..
- Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. Protein palmitoylation by a family of DHHC protein S-acyltransferases. Journal of lipid research. 2006;47:1118–27. doi:10.1194/jlr.R600007-JLR200. PMID:16582420.
- Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi:10.1038/nrm2084. PMID:17183362.
- Chamberlain LH, Shipston MJ. The physiology of protein S-acylation. Physiol Rev. 2015;95:341–76. doi:10.1152/physrev.00032.2014. PMID:25834228.
- Fukata Y, Iwanaga T, Fukata M. Systematic screening for palmitoyl transferase activity of the DHHC protein family in mammalian cells. Methods. 2006;40:177–82. doi:10.1016/j.ymeth.2006.05.015. PMID:17012030.
- Korycka J, Lach A, Heger E, Boguslawska DM, Wolny M, Toporkiewicz M, Augoff K, Korzeniewski J, Sikorski AF. Human DHHC proteins: a spotlight on the hidden player of palmitoylation. Eur J Cell Biol. 2012;91:107–17. doi:10.1016/j.ejcb.2011.09.013. PMID:22178113.
- Dekker FJ, Rocks O, Vartak N, Menninger S, Hedberg C, Balamurugan R, Wetzel S, Renner S, Gerauer M, Schölermann B, et al. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat Chem Biol. 2010;6:449–56. doi:10.1038/nchembio.362. PMID:20418879.
- Tomatis VM, Trenchi A, Gomez GA, Daniotti JL. Acyl-protein thioesterase 2 catalyzes the deacylation of peripheral membrane-associated GAP-43. PLoS One. 2010;5:e15045. doi:10.1371/journal.pone.0015045. PMID:21152083.
- Duncan JA, Gilman AG. Characterization of Saccharomyces cerevisiae acyl-protein thioesterase 1, the enzyme responsible for G protein alpha subunit deacylation in vivo. J Biol Chem. 2002;277:31740–52. doi:10.1074/jbc.M202505200. PMID:12080046.
- Duncan JA, Gilman AG. A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS). J Biol Chem 1998;273:15830–7. doi:10.1074/jbc.273.25.15830. PMID:9624183.
- Lin DT, Conibear E. ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. eLife. 2015;4:e11306. doi:10.7554/eLife.11306. PMID:26701913.
- Yokoi N, Fukata Y, Sekiya A, Murakami T, Kobayashi K, Fukata M. Identification of PSD-95 Depalmitoylating Enzymes. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2016;36:6431–44. doi:10.1523/JNEUROSCI.0419-16.2016. PMID:27307232.
- Breusegem SY, Seaman MN. Genome-wide RNAi screen reveals a role for multipass membrane proteins in endosome-to-golgi retrieval. Cell Rep. 2014;9:1931–45. doi:10.1016/j.celrep.2014.10.053. PMID:25464851.
- Spinosa MR, Progida C, De Luca A, Colucci AM, Alifano P, Bucci C. Functional characterization of Rab7 mutant proteins associated with Charcot-Marie-Tooth type 2B disease. J Neurosci. 2008;28:1640–8. doi:10.1523/JNEUROSCI.3677-07.2008. PMID:18272684.
- Li C, Shah SZ, Zhao D, Yang L. Role of the Retromer Complex in Neurodegenerative Diseases. Front Aging Neurosci. 2016;8:42. doi:10.3389/fnagi.2016.00042. PMID:26973516.
- McMillan KJ, Korswagen HC, Cullen PJ. The emerging role of retromer in neuroprotection. Curr Opin Cell Biol. 2017;47:72–82. doi:10.1016/j.ceb.2017.02.004. PMID:28399507.
- Mamo A, Jules F, Dumaresq-Doiron K, Costantino S, Lefrancois S. The role of ceroid lipofuscinosis neuronal protein 5 (CLN5) in endosomal sorting. Mol Cell Biol. 2012;32:1855–66. doi:10.1128/MCB.06726-11. PMID:22431521.
- Small SA, Kent K, Pierce A, Leung C, Kang MS, Okada H, Honig L, Vonsattel JP, Kim TW. Model-guided microarray implicates the retromer complex in Alzheimer's disease. Ann Neurol. 2005;58:909–19. doi:10.1002/ana.20667. PMID:16315276.
- Small SA, Petsko GA. Retromer in Alzheimer disease, Parkinson disease and other neurological disorders. Nat Rev Neurosci. 2015;16:126–32. doi:10.1038/nrn3896. PMID:25669742.
- Ginsberg SD, Mufson EJ, Alldred MJ, Counts SE, Wuu J, Nixon RA, Che S. Upregulation of select rab GTPases in cholinergic basal forebrain neurons in mild cognitive impairment and Alzheimer's disease. J Chem Neuroanat. 2011;42:102–10. doi:10.1016/j.jchemneu.2011.05.012. PMID:21669283.
- Vilariño-Güell C, Wider C, Ross Owen A, Dachsel Justus C, Kachergus Jennifer M, Lincoln Sarah J, et al. VPS35 Mutations in Parkinson Disease. Am J Hum Genet. 2011;89:162–7. doi:10.1016/j.ajhg.2011.06.001. PMID:21763482.
- Zimprich A, Benet-Pagès A, Struhal W, Graf E, Eck Sebastian H, Offman Marc N, Haubenberger D, Spielberger S, Schulte EC, Lichtner P, et al. A Mutation in VPS35, Encoding a Subunit of the Retromer Complex, Causes Late-Onset Parkinson Disease. Am J Hum Genet. 2011;89:168–75. doi:10.1016/j.ajhg.2011.06.008. PMID:21763483.
- Follett J, Norwood SJ, Hamilton NA, Mohan M, Kovtun O, Tay S, Zhe Y, Wood SA, Mellick GD, Silburn PA, et al. The Vps35 D620N mutation linked to Parkinson's disease disrupts the cargo sorting function of retromer. Traffic. 2014;15:230–44. doi:10.1111/tra.12136. PMID:24152121.
- McMillan KJ, Gallon M, Jellett AP, Clairfeuille T, Tilley FC, McGough I, Danson CM, Heesom KJ, Wilkinson KA, Collins BM, et al. Atypical parkinsonism-associated retromer mutant alters endosomal sorting of specific cargo proteins. J Cell Biol. 2016;214:389–99. doi:10.1083/jcb.201604057. PMID:27528657.
- Mecozzi VJ, Berman DE, Simoes S, Vetanovetz C, Awal MR, Patel VM, Schneider RT, Petsko GA, Ringe D, Small SA. Pharmacological chaperones stabilize retromer to limit APP processing. Nat Chem Biol. 2014;10:443–9. doi:10.1038/nchembio.1508. PMID:24747528.
- Follett J, Bugarcic A, Yang Z, Ariotti N, Norwood SJ, Collins BM, Parton RG, Teasdale RD. Parkinson Disease-linked Vps35 R524W Mutation Impairs the Endosomal Association of Retromer and Induces alpha-Synuclein Aggregation. J Biol Chem. 2016;291:18283–98. doi:10.1074/jbc.M115.703157. PMID:27385586.
