Abstract
Effective antigen-specific cancer immunotherapy requires exact knowledge of tumor-associated epitopes that can act as rejection antigens. Although the current paradigm views mutation-derived neoantigens as the most promising targets, we have recently demonstrated that leukemia-specific T-cell responses associated with survival benefit in CLL patients target a panel of non-mutated tumor-associated antigens.
With decades of research into non-mutated tumor-associated antigens (TAAs) translating into only limited clinical success, more recently the focus of T cell-based immunotherapy has shifted to actively personalized approaches targeting tumor-specific mutation-derived neoantigens.Citation1 Recent data demonstrate that actively targeting neoepitopes in human cancers is feasible and clinically effectiveCitation2 and implicate their involvement in tumor control in patients after immune checkpoint inhibitor therapy.Citation3,4 However, the current neoepitope-centric paradigm of T cell-based immunotherapy might restrict the spectrum of tumor entities deemed eligible for immunotherapeutic intervention to malignancies with high mutational loads and, furthermore, faces challenges of timing, target identification, and validation. Off-the-shelf immunotherapeutic approaches targeting broadly presented non-mutated TAAs might offer immediate bridging therapy closing the time gap between tumor debulking and patient-individualized immunotherapy, and furthermore lend themselves as targets for the immunotherapy of malignancies with low numbers of mutations. On the other hand, the underwhelming clinical effectiveness of non-mutated targets to date might, at least in part, be attributable to suboptimal antigen selection strategies lacking direct evidence of tumor-associated epitope presentation. As previously reported by our group, the highly distorted correlation of gene expression and HLA-restricted antigen presentation calls for the direct analysis of naturally presented HLA ligands by mass spectrometry.Citation5 Recent technological advances now enable comprehensive mapping of the HLA ligandome landscape of primary patient material in unprecedented depth, which in turn allows for the implementation of novel strategies of antigen identification based solely on HLA ligandome data.Citation6
In our most recent study we analyzed and characterized the non-mutant HLA-presented antigenome of chronic lymphocytic leukemia (CLL) as an archetypical example of malignancy with a low mutational burden.Citation7
Based on cohorts of 30 CLL patients and 30 healthy individuals we identified a panel of 49 CLL-associated antigens that were represented exclusively on malignant cells in more than 20% of analyzed patients. To ensure broad clinical applicability, these novel antigens were validated to be broadly and frequently presented across different stages and mutational subtypes of CLL and were found to be robustly represented in HLA ligandomes of patients undergoing standard chemoimmunotherapy. Notably, functional annotation clustering and gene expression analysis of these novel antigens did not identify any comprehensive unifying characteristics that might have enabled the development of computational approaches for the prediction of such TAAs. This indicates the unique character of the HLA ligandome and underscores the importance of defining T-cell antigens by direct HLA ligandome profiling.
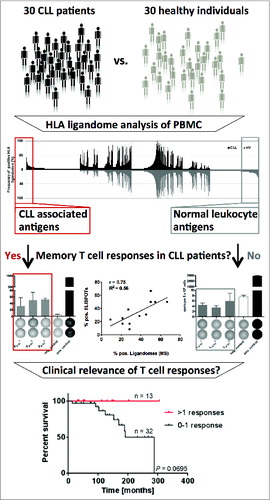
Surprisingly, for 14/15 of these novel antigens functional characterization revealed spontaneous, pre-existing immune responses exclusively in CLL patients. These immune responses were further verified to be strictly leukemia-directed and mediated by functional CLL patient-derived T cells, thus indicating tumor-dependent priming of T cells specific for non-mutated tumor epitopes in vivo in CLL patients. A direct correlation between the frequency of presentation of these epitopes in CLL patient ligandomes and immune recognition by CLL patient T cells was observed. Strikingly, retrospective survival analysis in a cohort of 45 CLL patients who were dichotomized according to their number of CLL antigen-specific T-cell responses revealed significantly improved overall survival for the patient group recognizing more than one antigen (Fig. 1). Together, this suggests that disease control in CLL might be mediated, at least in part, by spontaneous T-cell responses targeting non-mutated self-antigens.
These findings were surprising in multiple aspects and have the following important implications for the immunotherapy of CLL: (1) Non-mutated TAAs are frequently targeted by spontaneous T-cell responses in CLL patients in spite of the absence of mutation-induced “foreignness.” Although the notion of “autoreactive” T cells targeting tumor-associated self-antigens has long been established, the high frequency of immunogenicity of antigens identified in our study is encouraging and suggests that the direct HLA ligandome-centric approach might be highly effective and indispensable for the identification of immunologically relevant tumor antigens. Importantly, our findings also indicate that CLL and other hematologic malignancies might be effectively treated by immune checkpoint modulation in spite of their reportedly low number of somatic mutations. (2) The direct correlation between the frequencies of epitope detection in CLL ligandomes and frequencies of peptide-specific immune recognition might be useful for predicting the immunogenicity of novel epitopes based on HLA ligandomics data, but it also indicates that antigen presentation on cancer cells might be the limiting factor for the priming of specific immune responses. We are currently conducting further experiments investigating the relative roles of the naïve T cell repertoire and antigen presentation on cancer cells in the manifestation of antigen-specific T-cell responses in CLL patients. (3) The association of CLL antigen-specific T-cell responses with improved patient survival suggests that the characterized epitopes are underlying targets of pathophysiologically relevant antileukemia immune responses mediating disease control in CLL patients. Knowledge of such relevant antigens may directly be harnessed for the induction and guidance of specific antileukemia immune responses by off-the-shelf peptide-specific immunotherapy.
While these findings are encouraging, ultimately prospective clinical trials are needed to investigate the clinical potential and limitations of these novel targets and identify optimal protocols with respect to patient stratification, timing, combination, and sequence of therapeutic intervention.
Figure 1. HLA ligandome-centric workflow used for the identification of non-mutated targets of antileukemia T-cell responses in patients with chronic lymphocytic leukemia. Comparative analysis of non-mutated HLA ligands presented on peripheral blood mononuclear cells of CLL patients and healthy individuals by mass spectrometry identified the most frequently represented CLL-exclusive antigens. Functional characterization by ELSIPOT assay revealed pre-existing T-cell responses strictly directed against CLL-associated antigens exclusively in CLL patients. Retrospective survival analysis in 45 CLL patients revealed an association of these T-cell responses with improved overall survival.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, Lin JC, Teer JK, Cliften P, Tycksen E, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med 2013; 19:747-52; PMID:23644516; http://dx.doi.org/10.1038/nm.3161.
- Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, Wunderlich JR, Somerville RP, Hogan K, Hinrichs CS, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 2014; 344:641-5; PMID:24812403; http://dx.doi.org/10.1126/science.1251102.
- van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, van Dijk LJ, Behjati S, Hilkmann H, El Atmioui D, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol 2013; 31:e439-42; PMID:24043743; http://dx.doi.org/10.1200/JCO.2012.47.7521.
- Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014; 371:2189-99; PMID:25409260; http://dx.doi.org/10.1056/NEJMoa1406498.
- Weinzierl AO, Lemmel C, Schoor O, Muller M, Kruger T, Wernet D, Hennenlotter J, Stenzl A, Klingel K, Rammensee HG, et al. Distorted relation between mRNA copy number and corresponding major histocompatibility complex ligand density on the cell surface. Mol Cell Proteomics 2007; 6:102-13; PMID:17074750; http://dx.doi.org/10.1074/mcp.M600310-MCP200.
- Berlin C, Kowalewski DJ, Schuster H, Mirza N, Walz S, Handel M, Schmid-Horch B, Salih HR, Kanz L, Rammensee HG, et al. Mapping the HLA ligandome landscape of acute myeloid leukemia: a targeted approach toward peptide-based immunotherapy. Leukemia. 2015 Mar;29(3):647-59. doi: 10.1038/leu.2014.233. PMID:25092142; http://dx.doi.org/10.1038/leu.2014.233
- Kowalewski DJ, Schuster H, Backert L, Berlin C, Kahn S, Kanz L, Salih HR, Rammensee HG, Stevanovic S, Stickel JS. HLA ligandome analysis identifies the underlying specificities of spontaneous antileukemia immune responses in chronic lymphocytic leukemia (CLL). Proc Natl Acad Sci U S A 2014; 112(2):E166-75; PMID:25548167.
