ABSTRACT
Hodgkin lymphoma (HL) resistant to conventional therapies is increasing, making of interest the search for new schemes of treatment. Members of the “A Disintegrin And Metalloproteases” (ADAMs) family, mainly ADAM10 or ADAM17, have been proposed as therapeutic targets in solid tumors and some ADAMs inhibitors have been shown to exert antitumor effects. We have previously described an overexpression of ADAM10 in HL, together with increased release of NKG2D ligands (NKG2D-L) and reduced activation of effector T lymphocytes with anti-lymphoma capacity. Aim of the present work was to verify whether inhibition of ADAM10 in HL cells could restore the triggering of NKG2D-dependent anti-lymphoma T cell response. As no selective ADAM10 blockers have been reported so far, we synthesized the two hydroxamate compounds LT4 and MN8 with selectivity for ADAM10 over metalloproteases (MMPs), LT4 showing higher specificity for ADAM10 over ADAM17. We show that (i) HL lymph nodes (LN) and cultured HL cells express high levels of the mature active membrane form of ADAM10; (ii) ADAM10 is the major sheddase for the NKG2D-L in HL cells; (iii) the new LT4 and MN8 compounds strongly reduce the shedding of NKG2D-L by HL cell lines and enhance the binding of NKG2D receptor; (iv) of note, these new ADAM10 inhibitors increase the sensitivity of HL cell lines to NKG2D-dependent cell killing exerted by natural killer and γδ T cells. Overall, the biologic activity of LT4 and MN8 appears to be more potent than that of the commercial inhibitor GI254023X.
Introduction
The ADAMs family is composed of multidomain proteins, including ADAM10, involved in the so-called ectodomain shedding, a proteolytic process essential for cell development, migration and wound healing.Citation1,2 The active form of these enzymes is primarily located at the cell surface and is triggered by activation of protein kinase C, inhibition of phosphatases or increase in the intracellular calcium levels.Citation1-3 Among the best-known ADAM substrates, there are precursor forms of growth factors or cytokines, some of which, such as tumor necrosis factor (TNF)-α or epithelial growth factor (EGF), are involved in the pathogenesis and development of cancer.Citation4-6 As an example, ADAM10 and ADAM17 have been reported to promote epithelial tumor growth by releasing epidermal growth factor (HER)/EGF receptor ligands.Citation3,4 The evidence for ADAMs involvement in cancer is also supported by the finding that overexpression of these enzymes relates to parameters of tumor progression (tumor size, grade, metastasis and LN involvement).Citation2,3,5 ADAM10 and ADAM17 may also function as sheddases for the so-called “stress molecules,” including the MHC-class-I-related MIC-A and MIC-B, and the UL16-binding proteins (ULBPs).Citation7,8 These molecules are expressed at the cell surface during tumor transformation and can be upregulated by all-trans-retinoic acid (ATRA) or sodium valproate (VPA).Citation9-14 Once the natural killer group2 (NKG2D) is engaged by these antigens (called NKG2D ligands, NKG2D-L), NK and γδT lymphocytes initiate a rapid immune response against tumor cells preceding the expansion of specific αβT cells.Citation9-12 In particular, we described that this mechanism is active in chronic lymphocytic leukemia, acute myeloid leukemia, non-Hodgkin and HL.Citation13-16 In these cancers, NK and γδT cells proliferate in response to NKG2D-L bearing tumors and exert cytolytic activity against autologous cancer cells.Citation13-16 However, when NKG2D-L are shed by tumor cells, they interact with the NKG2D receptor on effector lymphocytes and hinder the recognition of neoplastic cells.Citation8,11,12,14-16 Soluble (s)NKG2D-L can also downregulate NKG2D expression on effector lymphocytes, contributing to tumor escape;Citation9 moreover, serum levels of sNKG2D-L have been shown to have prognostic significance in acute and chronic leukemias, multiple myeloma and lymphomas.Citation14-19 Along this line, we reported overexpression of ADAM10 in the LN microenvironment in HL, together with impaired T lymphocyte stimulation of antitumor activity and increased levels of sNKG2D-L.Citation16
Based on these data, ADAMs have been proposed as both biomarkers and therapeutic targets for cancer.Citation1-6 Some ADAM10 or ADAM17 inhibitors with antitumor effects have been described;Citation20-23 however, to our knowledge, no selective ADAM10 blockers and no data in HL have been reported so far. Thus, we developed inhibitors with progressively higher specificity for ADAM10 to enhance efficiency and selectivity of action.
In this paper, we show that (i) the mature active membrane form of ADAM10 is expressed in HL LN and on HL cells; (ii) ADAM10 is the major sheddase for the NKG2D-L in HL cells; (iii) the new hydroxamate compounds LT4 and MN8 reduce the shedding of NKG2D-L by HL cell lines and enhance the binding of NKG2D receptor; (iv) exposure to these new specific ADAM10 inhibitors increases the sensitivity of HL cell lines to NKG2D-dependent cell killing.
Results
The mature active membrane form of ADAM10 is expressed in HL LN and on HL cells and is relevant for NKG2D-L shedding
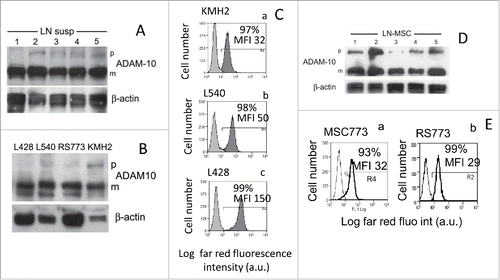
Cell suspensions from LN bioptic specimens were obtained from 10 HL (5 depicted in ) patients and subjected to Western blot analysis for ADAM10: in all samples, the mature form of ADAM10 was mainly detected (). The HL cell lines KMH2, L428, from pleural effusion, and L540, from bone marrow of HL patients, representative of the nodular sclerosis (L428, L540) and mixed cellularity (KMH2) HL histotypes, and the RS773 cell lines that we previously describedCitation16(mixed cellularity, obtained from LN5 of ) did express the mature ADAM10 form that is almost the only form detectable in L428 and RS773 cells (). All the HL cells analyzed showed surface expression of mature ADAM10 evidenced by immunofluorescence with the specific mAb and FACS analysis ( for KMH2, Cb for L540 and Cc for L428; for RS773,Citation16). This was also true for mesenchymal stromal cells (MSC) isolated from LN cell suspension ( for surface expression,Citation16). Thus, the mature membrane form of ADAM 10, that is described to display the enzymatic activity,Citation1-3 is prevalent in lymphoma cells, in keeping with our previous report of ADAM10 expression in situ in HL.Citation16 In the same report, we described a significant shedding of different NKG2D-L targets for ADAM10, including MIC-A and ULBPs, expressed by lymphoma cells.Citation16 The expression of MIC-A, MIC-B, ULBP2 and ULBP3 (with the exception of ULBP3 on L428) was low in all the HL cell lines tested (Fig. S1A); in particular, deglycosylation experiments showed that HL cell lines express mainly the non-truncated form of MIC-A (Fig. S1C). Likewise, the LN cell suspensions mostly displayed a dull expression of the NKG2D-L (Fig. S1B). Of note, soluble ULBP3 was found in the supernatant (SN) of LN cell suspensions (Fig. S1D).
Figure 1. The mature membrane form of ADAM10 is expressed on HL cells. Panels (A), (B), (D) Lysates obtained from HL LN cell suspensions (A) or HL cell lines (B) or LN MSCs obtained by culturing LN cell suspensions from HL patients (D) were subjected to Western blot as described in Materials and Methods; membranes were probed with the anti-ADAM10 or anti-β actin mAb followed by the relevant HRP-conjugated secondary antibodies and developed with the HRP substrate. In each blot, the precursor form (p) and the mature form (m) of ADAM10 molecule is indicated. Panels (C) and (E) Surface expression of ADAM10 on KMH2, L540, L428 (Ca, Cb, Cc, dark gray histograms) or MSC773 or RS773 (Ea, Eb) was evaluated with the specific mAb directed against the mature form of ADAM10 followed by APC-conjugated GAM and FACS analysis; results are expressed as Log far red fluorescence intensity, a.u., vs. number of cells.
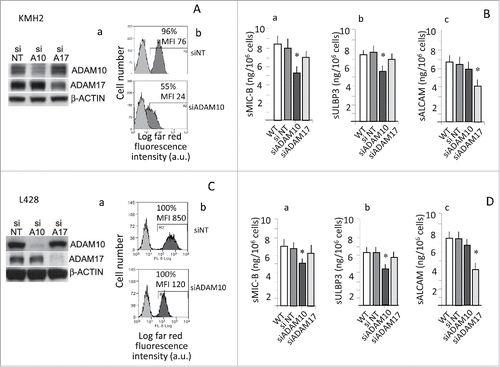
Since these ligands represent a substrate also for ADAM17,Citation7,8,24 we asked which sheddase was active in HL tumor cells. To address this question, either ADAM10 or ADAM17 were silenced in KMH2 () or L428 (). Silencing led to a significant reduction of ADAM10 mature form (panels Aa, Ca) (densitometric ratio: 0.59 for KMH2 and 0.1 for L428 vs siNT set to 1), and its expression at the cell surface (panels Ab, MFI: 20 vs. 100 a.u. and Cb, MFI: 100 vs. 800 a.u.). Likewise, ADAM17 silencing consistently reduced the protein (, 2Ca: densitometric ratio: 0.79 for KMH2 and 0.2 for L428). Of note, ADAM10 silencing resulted into decreased shedding of the NKG2D-L MIC-B () or ULBP3 (); although to a lesser extent, also MIC-A shedding was reduced (from 6 to 4 ng/106 cells in KMH2 cells and from 5 to 3 ng/106 in L428 cells, not shown). In turn, silencing of ADAM17 rather inhibited the release of sALCAM (), that is reported as preferential substrate for ADAM17.Citation25,26
Figure 2. ADAM10 silencing leads to decreased shedding of NKG2D-L. KMH2 and L428 cells were transfected with ADAM10 (siADAM10) or ADAM17 (siADAM17) siRNA or non-targeting siRNA (siNT) pool as negative control (KMH2: panels A, B; L428: panels C, D). Protein expression was analyzed by Western blot (panels Aa and Ca), and FACS analysis (Ab and Cb; in each histogram the percentage and MFI of positive cells is shown) with the specific anti-ADAM10 or anti-ADAM17 antibodies, 72 h after transfection. Soluble MIC-B (Ba, Da) or ULBP3 (Bb, Db) or sALCAM (Bc, Dc) were evaluated by ELISA in SN (collected upon further 24 h of culture 72 h after transfection). Results in (B) and (D) are expressed as pg/mL/105 cells and are the mean ± SD from three independent experiments. *p <0.001 vs siNT.
ADAM10 specific inhibitors reduce the shedding of NKG2D-L by HL cells and enhance the binding of NKG2D receptor
To counteract the enzymatic effect of ADAM10 in HL cells, the two hydroxamate compounds LT4 and MN8 were synthesized and tested in an in vitro enzymatic inhibition assay (Materials and Methods) to check the inhibitory effect of the two compounds on either ADAM10 or ADAM17. First, both inhibitors displayed high selectivity over MMPs, namely MMP-1 MMP-2, MMP-9 and MMP14, at variance with the commercial GI254023X (GIX, ). Moreover, LT4 showed high selectivity for ADAM10 over ADAM 17 (IC50 40 nM vs. 1500 nM on ADAM17, ). These inhibitors were compared to GIX and the reported compound 21 (JG26),Citation25 the latter showing a higher selectivity for ADAM17 over ADAM10 (IC501.9nM vs. 150nM, ). Soluble (s)MIC-A, sMIC-B, sULBP2, sULBP3 (as substrates for ADAM10 or ADAM17 sheddases) and sALCAM (as a preferential substrate for ADAM17) were measured by ELISA in the SN collected from HL cell lines untreated or exposed for 24 h to the ADAM10 inhibitors GIX, JG26, LT4, MN8, (from 10 μM to 1 μM) or to the solvent alone (DMSO). To maximize ADAMs activity, in some samples 100 μM sodium orthovanadate (Na3VO4) was added as pervanadate for 40 min before collecting SN. LT4 and MN8 could inhibit both constitutive () and pervanadate-induced () shedding of sMIC-A (), sMIC-B (), sULBP2 () and sULBP3 () by L428 cells with higher efficiency (inhibition detectable up to 5–2.5 μM) than JG26 or GIX (active at 5–10 μM concentration). In turn, JG26, GIX and MN8 were more efficient in reducing pervanadate induced release of sALCAM (). Thus, the ADAM10 inhibitor LT4 is the most efficient in preventing the shedding of NKG2D-L, with low activity on ALCAM release. Of note, LT4 pre-treatment of L428 cells could prevent the reduction of ULBP2 surface expression that follows pervanadate exposure (from 71% to 59% of positive cells in the presence of LT4, from 71% to 32% without LT4, ); moreover, LT4-treated L428 cells could efficiently bind the FcNKG2D chimeric receptor also after exposure to pervanadate (), suggesting that LT4 counteracted the sheddase activity of ADAM10, triggered by pervanadate, leading to stabilization of NKG2D-L expression and NKG2D receptor binding. Similar results were obtained with L540 (Fig. S1A and Ca) and KMH2 (Fig. S1B and Cb) cell lines: indeed, LT4 was the most efficient inhibitor of NKG2D-L shedding (Fig. S2Aa and Ba for sMIC-A, Ab and Bb for sMIC-B, Bc for sULBP2, Ac and Bd for ULBP3; note that L540 is ULBP2 negative, Fig. S1) enhancing the binding of FcNKG2D chimeric receptor as well (Fig. S2Ca for L540 and Cb for KMH2), compared to GIX, JG26 and MN8. We also tested the efficiency of LT4 on RS773 cells, derived in our laboratory from an LN of an HL patient and previously described.Citation16 As shown in Fig. S3, LT4 could inhibit (at 10 to 5 μM concentration) both constitutive (panels Aa, Ac) and pervanadate-induced (panels Ab, Ad) shedding of MIC-A (Aa, Ab) and MIC-B (Ac, Ad) by RS773 cells. Moreover, LT4 was able to increase the surface expression of MIC-A and MIC-B (Fig. S3B, second vs. first row) and prevent the reduction of surface MIC-A and MIC-B conceivably due to the activation of ADAM10 by pervanadate (Fig. S3B, fourth vs. third row). Accordingly, LT4 enhanced the binding of FcNKG2D chimeric receptor to RS773 cells (Fig. S3C, second vs. first histogram) and counteracted the effect of pervanadate (Fig. S3C, fourth vs. third histogram). MSC773 isolated from the same LN released detectable, but lower, amounts of sMIC-A and MIC-B (Fig. S3Da and Db); of note, LT4 was also able to inhibit pervanadate-induced increase of sMIC-A and MIC-B shedding by MSC (Fig. S3Da and Db).
Figure 3. ADAM10 inhibitors reduce the shedding of NKG2D-L by HL cell lines and maintain the binding of NKG2D receptor. L428 cells were exposed to culture medium alone, DMSO or GI254023X (GIX), JG26, MN8 or LT4 (at 10 to 2.5 μM concentration) for 24 h (panel A), followed by 100 μM Na3VO4 as pervanadate for 40 min at 37°C (panel B and C). Then, SN were harvested and sMIC-A (Aa, Ba), sMIC-B (Ab, Bb), sULBP2 (Ac, Bc), sULBP3 (Ad, Bd) or sALCAM (C) measured by specific ELISA. Results are expressed as ng/mL/106 cells and are representative of four independent experiments. *p <0.001 vs. DMSO. Panels (D) and (E) L428 cells exposed for 24 h to DMSO or 10 μM LT4 or 100 μM Na3VO4 as pervanadate, in the absence or presence of 10 μM LT4 as indicated, were harvested and evaluated for the expression of ULBP2 (D) with the specific mAb followed by APC-conjugated GAM or for the binding of the chimeric receptor (FcNKG2D, panel E) followed by APC-conjugated anti-human Fc antiserum, by FACS analysis; results are expressed as Log far red fluorescence intensity (arbitrary units, a.u.) vs. number of cells. In each subpanel: percentage and mean fluorescence intensity (MFI, a.u.) of positive cells. One representative experiment out of four.
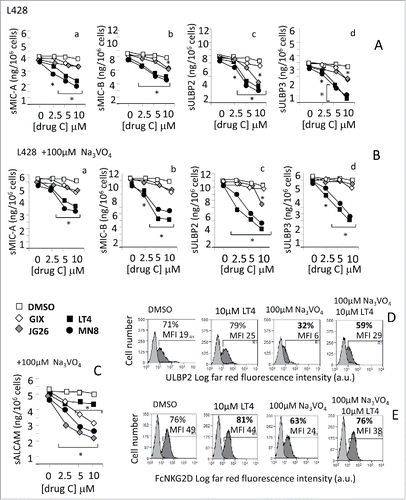
Table 1. In vitro enzymatic activity (IC50nM values)a of new compounds LT4 and MN8 and the reference compounds JG26 and GI254023X.
Exposure to ADAM10 inhibitors increases the sensitivity of HL cell lines to NKG2D-dependent cell killing
The above-reported data suggest that inhibition of ADAM10 leads to a decreased shedding of NKG2D-L with stabilization of their membrane expression and maintenance of NKG2D receptor binding to HL cells. Thus, we asked whether the various ADAM10 inhibitors improved the recognition of HL cell lines by effector lymphocytes and the NKG2D-mediated killing. To this aim, cytolytic activity of NK () and γδ T cells () was analyzed against L428, L540 or KMH2 cell lines at an E:T ratio of 10:1, before or after treatment for 24 h of the HL cell lines with the ADAM10 inhibitors LT4 or MN8 () compared to GIX and JG26 () at 10 μM concentration.
Figure 4. Exposure to ADAM10 inhibitors increases the sensitivity of HL cell lines to NKG2D-dependent cell killing. Cytolytic activity of NK cells (n = 6, panel A and B) or γδ T cells (n = 6, panel C) was analyzed against L428 (Aa, Ab, Ca) or L540 (Ba, Bb, Cb) cell lines at E:T ratio of 10:1 in a 4-h 51Cr-release assay. Some samples were set up after exposure of the target cell lines to either medium, or DMSO or LT4 or MN8 (Aa, Ba, Ca, Cb), GIX or JG26 (Ab, Bb) at 10 μM concentration for 24 h. In some samples, effector cells were exposed to saturating amounts (5 μg/mL) of the anti-NKG2D mAb at the onset of the cytotoxicity assay; an unrelated mAb, matched for the isotype, used as control, did not exert any effect (not shown). Results are expressed as % specific lysis calculated as described in Materials and Methods.
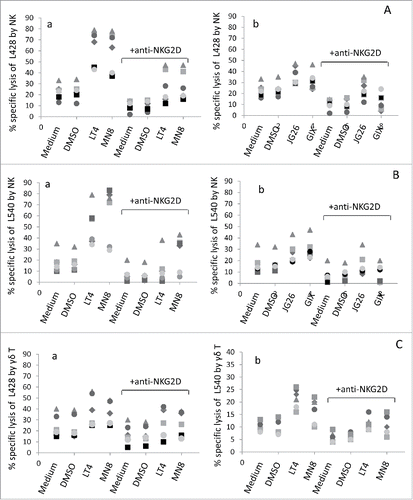
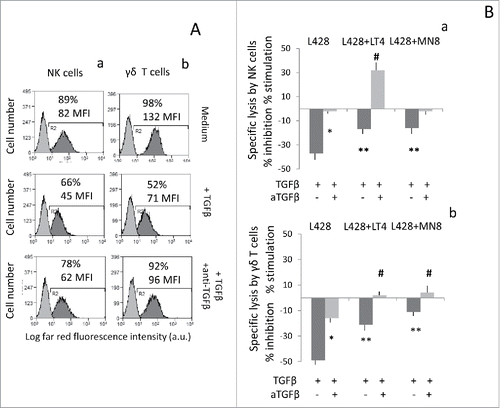
We found that exposure of HL cells to the new inhibitors LT4 and MN8 raised by 3-fold to 10-fold the NK cell killing of L428 (), L540 () and KMH2 (Fig. S4). On the other hand, the increase of NK cell killing of L428 and L540 treated with GIX or JG26 was 2–3-fold (). Of note, LT4 and MN8 could also enhance the cytotoxicity exerted by γδ T to L428 () and L540 (). Moreover, addition of the anti-NKG2D mAb at the onset of the cytotoxicity assay led to a reduction of the cytolytic activity of NK ( and Fig. S4) and γδ T cells (), both in the absence and in the presence of the inhibitors, suggesting that the cytotoxicity in this system is mainly NKG2D-mediated; an unrelated mAb, matched for the isotype, did not exert any effect (not shown for L428 or L540 and Fig. S4 A and B for KMH2). Thus, the new ADAM10 inhibitors LT4 and MN8 were more efficient than GIX and JG26 in sensitizing lymphoma cells to cytotoxic activity exerted by effector cells via NKG2D/NKG2D-L interaction. We previously reported that NKG2D-mediated killing of HL cells is impaired by TGFβ that downregulates the expression of the receptor at the surface of effector cells, and this effect can be prevented by neutralizing the cytokine.Citation16 Along this line, we asked whether the parallel treatment of cytotoxic effectors with an anti-TGFβ mAb and exposure of HL cells to ADAM10 inhibitors could achieve additional improvement of lymphoma cytolysis. As expected, TGFβ downregulated the expression of NKG2D at the surface of NK () or γδ T cells () and decreased the cytolytic activity of NK cells against L428 HL cell line ( for NK and for γδ T cells); both effects were neutralized by an anti-TGFβ mAb (). Of note, when assayed against L428 cells exposed to LT4, the cytotoxicity exerted by NK cells treated with TGFβ and neutralized with the anti-TGFβ mAb was potentiated, showing an additional significant improvement of cell lysis ().
Figure 5. Improvement of HL cell lysis by exposure to ADAM10 inhibitor LT4 and anti-TGFβ. Panel (A) NKG2D expression before (upper histograms) or after treatment with TGFβ (10 ng/mL), (middle histograms) or with TGFβ and anti-TGFβ mAb (1 µg/mL), on NK cells (Aa) or γδ T cells (Ab). In each subpanel: percentage of positive cells and MFI (a.u.). Panel (B) Cytolytic activity of NK cells (Ba) orγδ T cells (Bb) was analyzed against L428 cell line at E:T ratio of 5:1 in a 4-h 51Cr-release assay. Some samples were set up after exposure of the target cell lines to LT4 or MN8 at 10 μM concentration for 24 h. To some samples, we added effector cells exposed to TGFβ (10 ng/mL), with or without saturating amounts (1 μg/mL) of the anti-TGFβ mAb, as indicated. Results are expressed as % inhibition or stimulation of specific lysis calculated as described in Materials and Methods. *p <0.001 vs. TGFβ. **p <0.001 vs. TGFβ + anti-TGFβ. #p <0.001 vs. TGFβ + anti-TGFβ on untreated L428 cells.
Discussion
There is increasing evidence that sheddases of the ADAM family, in particular ADAM10 and 17 may become targets for anticancer therapies.Citation1-3,29 In particular, based on the literature and our previous data,Citation8,11,16 we hypothesized that modulation of NKG2D-L expression and/or release, by inhibiting the ADAM-sheddase enzymatic activity, could potentiate an anti-lymphoma stress-related immune response. Some ADAM10 or ADAM17 inhibitors have been shown to have antitumor effects;Citation20-23 however, no selective ADAM10 blockers have been reported and data in HL are not available so far. To this aim, we focused on the synthesis and use of specific inhibitors of either ADAM10 or ADAM17. In this paper, we show that (i) the mature active membrane form of ADAM10 is expressed in HL LN and at the surface of HL cells; (ii) ADAM10 is the major sheddase for NKG2D-L in HL cells; (iii) the new hydroxamate compounds LT4 and MN8 reduce the shedding of NKG2D-L by HL cell lines and enhance the binding of NKG2D receptor; (iv) exposure to these new specific ADAM10 inhibitors increases the sensitivity of HL cell lines to NKG2D-dependent cell killing.
In LN cell suspensions derived from HL patients, as well as in cultured HL cells, we found that the mature form of ADAM10 is highly expressed and localized at the cell membrane, supporting that the lymphoma microenvironment is enriched in the active form of ADAM10. These data are in keeping with our previous report on the overexpression of this enzyme at the tumor site in HL.Citation16 As reported, both ADAM10 and ADAM17 may function as sheddases for the so-called “stress molecules,” including MIC-A, MIC-B, and ULBPs.Citation7,8,28 Silencing of either ADAM10 or ADAM17 in HL tumor cells led to discrete results: indeed, while ADAM10 knock down resulted in reduced MIC-B and ULBP3 shedding, ADAM17 silencing downregulated ALCAM release. This suggests that ADAM10 is the main sheddase for NKG2D-L in HL; thus, two inhibitors with high (MN8 IC50 9.2 nM, LT4, IC50 40nM, ) and selective (LT4 IC50 40 nM vs. 1500 nM on ADAM17, ) activity on ADAM10 were developed to counteract the enzymatic effect of ADAM10 in HL cells. LT4 and MN8 could inhibit both constitutive and pervanadate-induced shedding of all the NKG2D-L tested, by the HL cell lines L428, L540, KMH2 and RS773, with higher efficiency than JG26 or the commercial GI254023X (GIX). In turn, JG26, GIX and MN8 were more efficient in reducing the release of sALCAM induced by ADAMs pervanadate stimulation. It has to be noted that the IC50 of GIX checked by our group is different to that previously described,Citation29 conceivably due to the more sensitive assay employed by us (fluorimetric assay, described in Materials and Methods, vs. scintillation proximity assay).Citation29
Overall, the selective ADAM10 inhibitor LT4 is the most efficient in preventing the shedding of NKG2D-L. Of interest, LT4 proved also to prevent the reduction of ULBP2 surface expression that follows pervanadate exposure in at least two HL cell lines, with the result of a preserved ability to bind the FcNKG2D chimeric receptor. This indicates that selective blocking of ADAM10 sheddase by LT4 leads to the stabilization of NKG2DL-NKG2D receptor interaction. Thus, we asked whether HL cell lines exposed to the various ADAM10 inhibitors become sensitive to NKG2D-mediated killing. Indeed, we found that LT4 and MN8 exposure increased NKG2D-dependent cell killing of HL cells with higher efficiency than less specific inhibitors such as GI254023X or JG26. Of note, LT4 and MN8 could sensitize lymphoma cells to the cytotoxicity exerted by NK and γδ T cells. This effect could also be improved by upregulation of NKG2D-L on target cells by ATRA or VPA;Citation13-15 neither drugs influenced NKG2D-L shedding, in acute myeloid leukemias or non-HLs or HL,Citation14-16 nevertheless, VPA pre-treated HL cells, besides upregulating NKG2D-L, appear to retain their expression with high efficiency than untreated cells when exposed to LT4 inhibitor. In addition to NKG2D/NKG2D-L, other receptor ligand interactions are involved in tumor cells killing by NK or T effector cells. In case of γδ T cells, they usually also kill different tumor cells via TCR-dependent recognition of tumor-derived pyrophosphates.Citation12,30,31 This can be exploited as an additional therapeutic tool as amino-bis-phosphonates, commonly administered as anti-osteopenic agents in multiple myeloma are known to activate γδ T cells and decrease regulatory signals that may occur in the microenvironment.Citation32,33
In conclusion, targeting selectively ADAM10 seems to stabilize the interaction between NKG2D receptor and their ligands expressed by HL cells and is sufficient to sensitize lymphoma cells to the NKG2D-mediated cytotoxicity exerted by effector lymphocytes. Possible major drawbacks of this therapeutic approach might be the accessibility of ADAM10+ cells at the tumor site, that we show to be abundant according to immunohistochemistry,Citation16 and the inhibition of cleavage of other ADAM10 substrates. Nevertheless, it is conceivable that effector lymphocytes resident in the LN can be activated and exert antitumor activity; furthermore, inhibition of cleavage of other ADAM10 substrates, such CD30, a target for antibody-based anti-lymphoma therapy,Citation34 or TNFα, a reported growth factor for lymphomas,Citation35 might also be useful in HL.
Thus, ADAM10 inibitors may be proposed to become part of anti-lymphoma therapeutic schemes and contribute to the enhancement of antitumor immune response.
Materials and methods
Cells and LN specimens. The HL cell lines KMH2, L428, obtained from pleural effusion, and L540, from bone marrow of HL patients, were purchased from DSMZ GmbH (Braunschweig, Germany). These cell lines are MIC-A/B+, ULBP2/3+and ALCAM+, except L540 that is ULBP2 negative and ALCAM low (Fig. S2), and do not express ULBP1 and 4 (not shown). The RS773 cell line was obtained from a HL LN and is MICA/B low and ULBP2/3 negative, as described.Citation16 LN MSCs were obtained by culturing LN cell suspensions from HL patients in six-well plates (5 × 106 cells/well) in MEM-α (GIBCO) complete medium: their phenotype and characterization has been reported.Citation16 After 3 d, non-adherent cells were washed away and adherent cells cultured for additional 7 d. LN biopsies of 10 HL patients, were obtained from the Unit of Pathology, IRCCS-AOU San Martino-IST, Genoa, under conventional diagnostic procedures, provided informed consent and the study was approved by the institutional ethical committee (IRB approval 0026910/07, renewal 03/2009). Samples taken as sentinel LN during surgical approaches and resulted free of neoplastic disease, showed little expression of ADAM10, not exceeding that of the housekeeping genes tested.Citation16 NK cell populations were separated from heparinized blood of healthy donors with the specific Rosettesep isolation kit (Stem Cell Technologies, Vancouver, Canada) according to manufacturer's instructions. γδT cells were purified with the specific Minimacs separation kit (MiltenyiBiotec, BergischGladbach, Germany) according to manufacturer's instruction. After this separation, NK or γδ T cells, always >96 percent of purity, were stimulated with 0.5 μg/mL of PHA in the presence of 10 ng/mL of recombinant interleukin-2 (rIL-2, PeproTechEC, London, UK) in 96 U-bottomed microwells; after 15 d of culture, NK or γδT cells were always 99–100 percent. Culture complete medium was composed of RPMI1640 (Biochrom, Berlin, Germany) with 10% fetal calf serum (FCS, Biochrom) supplemented with penicillin, streptomycin and L-glutamine (Biochrom) and rIL-2.
Inhibitors. GI254023X (GIX) was purchased from Sigma Chemicals Co. (St. Louis, MO). JG26 was synthesized as previously described,Citation25,26 and MN8 and LT4, showing a IC50 progressively more selective for ADAM10, were newly synthesized (). The various ADAM10 inhibitors (GIX, JG26, LT4 or MN8) used on the different HL cell lines, or on isolated MSC, at 2.5 up to 10 μM concentration for 48, 72 and 96 h (and the solvent DMSO at the same dilutions and time points) did not exert any toxic effect, as assessed by evaluating the mitochondrial potential upon staining with the dual emission fluorescent probe JC-1 (Molecular Probes, Life Technologies Italia, Monza) (not shown).
MMPs and ADAMs inhibition assays. Recombinant human MMP-14 catalytic domain was a kind gift of Prof. Gillian Murphy (Department of Oncology, University of Cambridge, UK). Pro-MMP-1, pro-MMP-2, pro-MMP-9, and recombinant human ADAM-17 (PF133) were purchased from Calbiochem (Calbiochem Italia, Milan). Recombinant human ADAM10 was from R&D Systems (Minneapolis, MN). Pro-enzymes were activated immediately prior to use with p-aminophenylmercuric acetate (APMA 2 mM for 1 h at 37°C for MMP-2, APMA 2 mM for 2 h at 37°C for MMP-1 and 1 mM for 1 h at 37°C for MMP-9). For assay measurements, the inhibitor stock solutions (DMSO, 10 mM) were further diluted in the fluorometric assay buffer (FAB: 50 mM Tris, pH = 7.5, 150 mM NaCl, 10 mM CaCl2, 0.05% Brij-35 and 1% DMSO). Activated enzyme (final concentration 0.56 nM for MMP-2, 1.3 nM for MMP-9, 1.0 nM for MMP-14cd, 2.0 nM for MMP-1, 5 nM for ADAM-17 and 20 nM for ADAM-10) and inhibitor solutions were incubated in the assay buffer for 3 h at 25°C. ADAM-17 was incubated for 30 min at 37°C and ADAM-10 for 1 h at 37°C in a different buffer at pH = 9 (25 mM Tris, 25 µM ZnCl2, 0.005% Brij-35). After the addition of 200 μM solution of the fluorogenic substrate Mca-Lys-Pro-Leu-Gly-Leu-Dap(Dnp)-Ala-Arg-NH2 (Bachem) for all the enzymes in DMSO (final concentration 2 μM for all enzymes, 10 μM for ADAM10), the hydrolysis was monitored every 15 sec for 20 min recording the increase in fluorescence (λex = 325 nm, λem = 400 nm) with a Molecular Devices SpectraMax Gemini XPS plate reader. The assays were performed in duplicate in a total volume of 200 μL per well in 96-well microtitre plates (Corning black, NBS). Control wells lack inhibitor. The MMP inhibition activity was expressed in relative fluorescent units (RFU). Percent of inhibition was calculated from control reactions without the inhibitor. IC50 was determined using the formula: vi/vo = 1/(1 + [I]/ IC50), where vi is the initial velocity of substrate cleavage in the presence of the inhibitor at concentration [I] and vo is the initial velocity in the absence of the inhibitor. Results were analyzed using SoftMax Pro software and GraFit software.
Small interfering RNA (siRNA) transfection. ON-TARGET plus SMART pool for human ADAM10 or ADAM17 (100 nM, Dharmacon CarloErba, Milan, Italy) were used to knockdown the expression of ADAM10 or ADAM17; siCONTROL non-targeting (NT) siRNA pool (Dharmacon) was used as negative control. L428 (2.5 × 105) were transfected by electroporation with Microporator MP-100 (Digital Bio) at the following electric conditions: Pulse voltage: 1400, pulse width: 10, pulse number: 3. Accel delivery system (Dharmacon) was used for KMH2 cell line, difficult to transfect. Briefly, 4 × 105cells were incubated with 1 μM Accell SMARTpool ADAM10 or ADAM17 siRNA or Accell non-targeting siRNA pool in Accell siRNA delivery medium for 72 h. Protein expression by western blot and immunofluorescence was analyzed 72 h after transfection. Soluble NKG2D-L were evaluated by ELISA in conditioned media collected from silenced cells upon further 24 h of culture after transfection.
Western blot. LN cell suspensions, prepared as described,Citation16or HL cell lines were harvested and lysed with ice-cold RIPA buffer containing protease and phosphatase inhibitors. Protein quantification in cell lysates was done with the DC Protein Assay Kit (Bio-Rad, Hercules, CA, USA). Equal amounts of protein (20 μg/lane) were loaded on precast 8–16% gradient gels (Thermo Fisher Scientific, Waltham, MA, USA) and then electro-transferred to PVDF membranes (GE Healthcare, Little Chalfont, UK). After blocking, membranes were probed overnight at 4°C with the rabbit polyclonal anti-ADAM17 (ab39163, Abcam, Cambridge, UK) or the anti-ADAM10 mAb (MAB1427 anti-ectodomain, R&D System) diluted according to the manufacturer's instructions. After washing, membranes were incubated for 1 h at room temperature with the relevant horseradish peroxidase (HRP)-conjugated secondary antibodies (Cell Signaling, EuroClone, Milan, Italy), and proteins were detected by Immobilon Western Chemiluminescent HRP Substrate (Millipore, Billerica, MA, USA). Anti-β-actin HRP-conjugated antibody (Cell Signaling) was used as a loading control. Densitometric analysis was performed using the ImageJ computer program (http://imagej.nih.gov/ij/download.html) and it is reported as fold change relative to controls set to 1.
Some samples were deglycosylated with N-glycosidase F (PNGase F, New England Biolabs) according to the manufacturer's instructions. Briefly, 35 µg of HL cell lysates were digested overnight with PNGase F before western immunoblot with the specific anti-MICA/B antibody (BAMO1, Axxora GmbH, Grafelfing Germany). HeLa cells, homozygotes for the allelic variant MICA*008, and MDA-MB-231 were used as references.Citation28
Immunofluorescence and cytofluorimetric analysis. Immunofluorescence on HL cells or on HL LN suspensions was performed with the anti-ADAM10 mAb (MAB1427) or the anti- CD30 mAb or the Ig-NKG2D chimera (FcNKG2D), purchased from R&D System, the anti-MIC-A mAb M2032B5Citation28 or the anti-MIC-B mAb (clone 12, Sino Biologicals Inc., Beijing, China) and the anti-ULPBs mAbs (anti-huULBP2 M311 and anti-huULBP3 M551) kindly provided by Amgen (Seattle, WA, M.T.A. no. 200309766-001) or the anti-ALCAM antibody (MAB6561, R&D System). In some experiments, TGFβ (recombinant human TGFβ1, R&D) was added to either NK or γδ T cells at 10 ng/mL for 12 h, in the presence or absence of saturating amounts of the anti TGFβ mAb (1 µg/mL, clone 1D11, R&D), before staining with the anti-NKG2D mAb (MAB139, R&D System). Some experiments were performed after cell exposure to the ADAM10 inhibitors GIX, JG26, LT4, MN8, at 10 μM or 5 μM concentration; in some samples 100 μM sodium orthovanadate (Na3VO4, Sigma Chemicals Co., St Louis, MO) was added as pervanadate (obtained with 100 μM H2O2) for 40 min at 37°C, to maximize the ADAMs enzymatic activity.Citation25,26 This dose and time of treatment were not toxic for the HL cell lines tested, as evaluated by JC1 staining (not shown). Samples were stained with the indicated mAbs for 30 min at 4°C, followed by anti-isotype goat anti-mouse (GAM) or anti-rabbit (GAR) antiserum conjugated with the appropriate fluorochromes and analyzed by CyAn ADP flow cytometer (Beckman Coulter, Inc.). Control aliquots were stained with isotype-matched irrelevant mAbs. Results are expressed as log of mean fluorescence intensity (MFI) or percentage of positive cells, or as the ratio between the MFI of each sample and the negative control, as indicated in the figure legend.
Cytotoxicity assay. Cytolytic activity of NK and γδ T cells was analyzed against L540, L428 or KMH2 cell lines at an E:T ratio of 10:1, in V-bottomed microwells, in a 4-h 51Cr-release assay as described.Citation15,16 Some samples were set up after exposure of the target cell lines to each ADAM10 inhibitor (GIX, JG26, LT4 or MN8) at 10 μM concentration for 24 h. In some samples, the effector cells were exposed to saturating amounts (5 μg/mL) of the anti-NKG2D mAb at the onset of the cytotoxicity assay; an unrelated mAb, matched for the isotype (BD PharMingen, BD Italia, Milan, Italy), was used as control. Other experiments were performed by adding to the effector cells TGFβ (at 10 ng/mL for 12 h, in the presence or absence of saturating amounts of the anti TGFβ mAb (1 µg/mL), before starting the cytotoxicity assay. One hundred microliters of SN were measured in a gamma counter and the percentage of 51Cr-specific release was calculated as described previously. The percentage of specific release was calculated with the following formula: experimental release (counts)-spontaneous release (counts)/maximum release (counts)-spontaneous release (counts). Maximum and spontaneous release was calculated as described.
ELISA for sMIC-A/B, sULBP2/3, and sALCAM. Soluble (s)MIC-A, sMIC-B, sULBP2, sULBP3 and sALCAM were measured in SN by ELISA as described.Citation15,16 SN were collected from cell cultures (HL cell lines), before or after 24 h exposure to the various ADAM10 inhibitors (GIX, JG26, LT4, MN8, from 10 μM to 1 μM). In some samples, 100 μM sodium orthovanadate was added as pervanadate for 40 min before collecting SN. The anti-MIC-A mAbs AMO1 and BAMO3 were from Immatics Biotechnologies (Tubingen, Germany) and the anti-ULPBs mAbs (anti-ULBP2 M311, IgG1; anti-ULBP3 M551, IgG1) were provided by Amgen (Seattle, WA). The anti-ULBP2 and anti-ULBP3 detection mAbs (MAB1298, IgG2a; MAB15171, IgG2a) and the ELISA detection kit for sMIC-B and sALCAM (DuoSet) were from R&D System. Anti-mouse IgG2a HRP was from Southern Biotechnology Associates (Birmingham, AL). Plates were developed with 2,2-azinobis (2-ethylbenzothiazoline-6-sulfonic acid) (Sigma) and read at OD450nm. Results are expressed as ng/mL and referred to a standard curve obtained with the MIC-A/B/Fc, ULPB2/Fc or ULBP3/Fc chimeras (R&D System). sMIC-A and sULBP3 content was measured by ELISA also in SN from LN cell suspensions cultured overnight at 106/mL.
Statistical analysis. Data are presented as mean ± SD. Statistical analysis was performed using two-tailed student's t test. The cut-off value of significance is indicated in each legend to figure.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KONI_A_1123367_supplemental_material.zip
Download Zip (852.3 KB)Funding
This study was supported by research funding from the Italian Association for Cancer Research (AIRC IG-12759) to MRZ. CC is recipient of a fellowship based on the same AIRC grant.
References
- Rocks N, Paulissen G, El Hour M, Quesada F, Crahay C, Gueders M, Foidart JM, Noel A, Cataldo D. Emerging roles of ADAM and ADAMTS metalloproteinases in cancer. Biochimie 2008; 90:369-79; PMID:17920749; http://dx.doi.org/10.1016/j.biochi.2007.08.008
- Saftig P, Reiss K. The “A Disintegrin And Metalloproteases” ADAM10 and ADAM17: novel drug targets with therapeutic potential? Eur J Cell Biol 2011; 90:527-35; PMID:21194787; http://dx.doi.org/10.1016/j.ejcb.2010.11.005
- Duffy MJ, Mullooly M, O'Donovan N, Sukor S, Crown J, Pierce A, McGowan PM. The ADAMs family of proteases: new biomarkers and therapeutic targets for cancer? Clin Proteomics 2011; 8:9-13; PMID:21906355; http://dx.doi.org/10.1186/1559-0275-8-9
- Blobel CP. ADAMs: key components in EGFR signalling and development. Nature Rev Cancer 2005; 6:32-43; PMID:15688065; http://dx.doi.org/10.1038/nrm1548
- Duffy MJ, McKiernan E, O'Donovan N, McGowan P. Role of ADAMs in cancer formation and progression. Clin Cancer Res 2007; 13:2335-43; PMID:17438092; http://dx.doi.org/10.1158/1078-0432.CCR-06-2092
- Murphy G. The ADAMs: Signalling scissors in the tumor microenvironment. Nature Rev Cancer 2008; 8:929-41; PMID:19005493; http://dx.doi.org/10.1038/nrc2459.
- Waldhauer I, Steinle A. Proteolytic release of soluble UL16-binding protein 2 from tumor cells. Cancer Res 2006; 66(5):2520-26; PMID:16510567; http://dx.doi.org/10.1158/0008-5472.CAN-05-2520
- Waldhauer I, Goehlsdorf D, Gieseke F, Weinschenk T, Wittenbrink M, Ludwig A, Stevanovic S, Rammensee HG, Steinle A. Tumor-associated MICA is shed by ADAM proteases. Cancer Res 2008; 68(15):6368-76; PMID:18676862; http://dx.doi.org/10.1158/0008-5472.CAN-07-6768
- Nausch N, Cerwenka A. NKG2D ligands in tumor immunity. Oncogene 2008; 27:5944-58; PMID:18836475; http://dx.doi.org/10.1038/onc.2008.272
- Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunol Rev 2010; 235:267-85; PMID:20536569; http://dx.doi.org/10.1111/j.0105-2896.2010.00893.x
- Salih HR, Antropius H, Gieseke F, Lutz SZ, Kanz L, Rammensee HG, Steinle A. Functional expression and release of ligands for the activating immunoreceptor NKG2D in leukemia. Blood 2003; 102:1389-96; PMID:12714493; http://dx.doi.org/10.1182/blood-2003-01-0019
- Hayday AC. γδT cells and the lymphoid stress-surveillance response. Immunity 2009; 31:184-96; PMID:19699170; http://dx.doi.org/10.1016/j.immuni.2009.08.006
- Poggi A, Venturino C, Catellani S, Clavio M, Miglino M, Gobbi M, Steinle A, Ghia P, Stella S, Caligaris-Cappio F et al. Vδ1 T lymphocytes from B-CLL patients recognize ULBP3 expressed on leukemic B cells and up-regulated by trans-retinoic acid. Cancer Res 2004; 64:9172-9; PMID:15604289; http://dx.doi.org/10.1158/0008-5472.CAN-04-2417
- Poggi A, Catellani S, Garuti A, Pierri I, Gobbi M, Zocchi MR. Effective in vivo induction of NKG2D ligands in acute myeloid leukaemias by ATRA acid or VPA. Leukemia 2009; 23:641-8; PMID:19151770; http://dx.doi.org/10.1038/leu.2008.354
- Catellani S, Poggi A, Bruzzone A, Dadati P, Ravetti JL, Gobbi M, Zocchi MR. Expansion of Vdelta1 T lymphocytes producing IL-4 in low-grade non-Hodgkin lymphomas ex pressing UL-16-binding proteins. Blood 2007; 109:2078-85; PMID:16973957; http://dx.doi.org/10.1182/blood-2006-06-028985
- Zocchi MR, Catellani S, Canevali P, Tavella S, Garuti A, Villaggio B, Zunino A, Gobbi M, Fraternali-Orcioni G, Kunkl A et al. High ERp5/ADAM10 expression in lymph node microenvironment and impaired NKG2D-ligands recognition in Hodgkin lymphomas. Blood 2012; 119:1479-89; PMID:22167753; http://dx.doi.org/10.1182/blood-2011-07-370841
- Groh V, Wu J, Yee C, Spies T. Tumor-derived soluble MIC ligands impair expression of NKG2D and T cell activation. Nature 2002; 419:734-8; PMID:12384702; http://dx.doi.org/10.1038/nature01112
- Jinushi M, Vanneman M, Munshi NC, Tai YT, Prabhala RH, Ritz J, Neuberg D, Anderson KC, Carrasco DR, Dranoff G. MIC-A antibodies and shedding are associated with the progression of multiple myeloma. Proc Natl Acad Sci USA 2008; 105:1285-90; PMID:18202175; http://dx.doi.org/10.1073/pnas.0711293105
- Nückel H, Switala M, Sellmann L, Horn PA, Dürig J, Dührsen U, Küppers R, Grosse-Wilde H, Rebmann V. The prognostic significance of soluble NKG2D ligands in B-cell chronic lymphocytic leukemia. Leukemia 2010; 24:1152-59; PMID:20428196; http://dx.doi.org/10.1038/leu.2010.74
- Zhou BB, Peyton M, He B, Liu C, Girard L, Caudler E, Lo Y, Baribaud F, Mikami I, Reguart N et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell 2006; 10:39-50; PMID:16843264; http://dx.doi.org/10.1016/j.ccr.2006.05.024
- Witters L, Scherle P, Friedman S, Fridman J, Caulder E, Newton R, Lipton A. Synergistic inhibition with a dual epidermal growth factor receptor/HER-2/neu tyrosine kinase inhibitor and a disintegrin and metallo proteiase inhibitor. Cancer Res 2008; 68:7082-89; PMID:18757423; http://dx.doi.org/10.1158/0008-5472.CAN-08-0739
- Fridman JS, Scherle PA, Liu X, Caulder E, Hansbury M, Yang G, Wang Q, Lo Y, Zhou J, Yao W et al. Preclinical characterization of INCB7839, a potent and selective inhibitor of Erb ligand and HER2 receptor shedding: inhibition of ADAM10 and ADAM17 for the treatment of breast cancer. Breast Cancer Res Treat 2007; 106(Suppl1):S82.
- Moss ML, Stoeck A, Yan W, Dempsey PJ. ADAM10 as a target for anti-cancer therapy. Curr Pharm Biotechnol 2008; 9:2-8; PMID:18289051; http://dx.doi.org/10.2174/138920108783497613
- Theocharis AD, Gialeli C, Bouris P, Giannopoulou E, Skandalis SS, Aletras AJ, Iozzo RV, Karamanos NK. Cell-matrix interactions: focus on proteoglycan-proteinase interplay and pharmacological targeting in cancer. FEBS J 2014; 281:5023-42; PMID:25333340; http://dx.doi.org/10.1111/febs.12927
- Nuti E, Casalini F, Santamaria S, Fabbi M, Carbotti G, Ferrini S, Marinelli L, La Pietra V, Novellino E, Camodeca C et al. Selective Arylsulfonamide Inhibitors of ADAM-17:Hit Optimization and Activity in Ovarian Cancer Cell. Models J Med Chem 2013; 56:8089-103; PMID:24044434; http://dx.doi.org/10.1021/jm4011753
- Nuti E, Casalini F, Avramova SI, Santamaria S, Fabbi M, Ferrini S, Marinelli L, La Pietra V, Limongelli V, Novellino E et al. Potent arylsulfonamide inhibitors of tumor necrosis factor-α converting enzyme able to reduce activated leukocyte cell adhesion molecule shedding in cancer cell models. J Med Chem 2010; 53:2622-35; PMID:20180536; http://dx.doi.org/10.1021/jm901868z
- Poggi A, Zancolli M, Boero S, Catellani S, Musso A, Zocchi MR. Differential survival of γδT cells, αβT cells and NK cells upon engagement of NKG2D by NKG2DL-expressing leukemic cells. Int J Cancer 2011; 129:387-96; PMID:20853320; http://dx.doi.org/10.1002/ijc.25682
- Chitadze G, Lettau M, Bhat J, Wesch D, Steinle A, Fürst D, Mytilineos J, Kalthoff H, Janssen O, Oberg HH et al. Shedding of endogenous MHC class I-related chain molecules A and B from different human tumor entities: heterogeneous involvement of the “a disintegrin and metalloproteases” 10 and 17. Int J Cancer 2013; 133:1557-67; PMID:23526433; http://dx.doi.org/10.1002/ijc.28174
- Hundhausen C, Misztela D, Berkhout TA, Broadway N, Saftig P, Reiss K, Hartmann D, Fahrenholz F, Postina R, Matthews V et al. The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1-mediated cell-cell adhesion. Blood 2003; 102:1186-95; PMID:12714508; http://dx.doi.org/10.1182/blood-2002-12-3775
- Gober Hj, Kistowska M, Angman L, Jenö P, Mori L, De Libero G. Human T cell receptor gammadelta cells recognize endogenous mevalonate metabolites in tumor cells. J Exp Med 2003; 197:163-68; PMID:12538656; http://dx.doi.org/10.1084/jem.20021500
- Bonneville M, O'Brien RL, Born, WK. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nat Rev Immunol 2010; 10:467-78; PMID:20539306; http://dx.doi.org/10.1038/nri2781
- Kunzman V, Bauer E, Feurle J, Weissinger F, Tony HP, Wilhelm M. Stimulation of T cells by aminobiphosphonates and induction of anti-plasma cell activity in multiple myeloma. Blood 2000; 96:384-92; PMID:10887096.
- Musso A, Catellani S, Canevali P, Tavella S, Venè R, Boero S, Pierri I, Gobbi M, Kunkl A, Ravetti JL et al. Aminobisphosphonates prevent the inhibitory effects exerted by lymph node stromal cells on anti-tumor Vδ2 T lymphocytes in non-Hodgkin lymphomas. Haematologica 2014; 99:131-9; PMID:24162786; http://dx.doi.org/10.3324/haematol.2013.097311
- Nakayama S, Yokote T, Tsuji M, Akioka T, Miyoshi T, Hirata Y, Hiraoka N, Iwaki N, Takayama A, Nishiwaki U et al. Expression of tumour necrosis factor-α and its receptors in Hodgkin lymphoma. Br J Haematol 2014; 167:574-7; PMID:25039986; http://dx.doi.org/10.1111/bjh.13015
- Eichenauer DA, Simhadri VL, von Strandmann EP, Ludwig A, Matthews V, Reiners KS, von Tresckow B, Saftig P, Rose-John S, Engert A et al. ADAM10 inhibition of human CD30 shedding increases specificity of targeted immunotherapy in vitro. Cancer Res. 2007; 67:332-8; PMID:17210715; http://dx.doi.org/10.1158/0008-5472.CAN-06-2470
