ABSTRACT
Immature myeloid cells such as myeloid-derived suppressor cells (MDSCs) and M2 macrophages play a vital role in the tumor immune escape and tumor progression. Cytotoxic T lymphocyte-associated antigen 4 (CTLA4), as a negative immune checkpoint, is highly expressed in numerous solid tumors. However, precise functions of CTLA4 in head and neck squamous cell carcinoma (HNSCC) have not yet been elucidated. In this study, we demonstrated that the ratio of CD8+/CTLA4 can be used as a potential index with a clinical prognostic value for HNSCC. Using immunocompetent transgenic mouse model with spontaneous HNSCC, we directly observed that targeting CTLA4 decreases MDSCs and M2 macrophages and promotes T cell activation in both tumor microenvironment and macro-environment. In all, our study provides direct evidence in vivo and proposes a rationale for CTLA4 inhibition as a future therapeutic strategy in patients with HNSCC.
Abbreviations
| aCTLA4 | = | anti-CTLA4 |
| CTLA4 | = | Cytotoxic T lymphocyte associated antigen 4 |
| Dys | = | dysplasia |
| HNSCC | = | Head and neck squamous cell carcinoma |
| HPV | = | human papillomavirus |
| LN | = | lymph node |
| MDSCs | = | Myeloid-derived suppressor cells |
| TAMs | = | Tumor-associated macrophages |
| TIL | = | Tumor infiltrate lymphocytes. |
Introduction
HNSCC has been identified as a significant cause of morbidity and mortality.Citation1 Smoking, alcohol use, and human papillomavirus (HPV) infections have been cited as major risk factors for HNSCC.Citation1 The 5-year survival rate of advanced patients has been improved only marginally over the past decades despite recent advances in surgery and target therapy.Citation2 HNSCC has recently been recognized as an immunosuppressive disease,Citation3 suggesting the potential use of immunotherapy to improve patient survival.
The host immune system, which functions as an important defense against cancer, can identify and destroy tumor cells. However, this process is controlled by inhibitory receptors and ligands, including CTLA4.Citation4 The phenotype of Ctla-4 knockout mice can lead to lethal systemic immune hyperactivation, demonstrating the central role of CTLA4 in downregulating T cell activation and maintaining immunologic homeostasis.Citation5 CTLA4 largely downregulates the amplitude of T cells responses by inhibiting co-stimulation molecular CD28, with which it shares the ligands B7.1 (CD80) and B7.2 (CD86).Citation6 Moreover, compared with CD28, CTLA4 exhibits a markedly higher affinity for both B7.1 and B7.2.Citation7 This fundamental research led to the clinical development and approval anti-CTLA4 (aCTLA4) as a treatment for patients with advanced melanoma.Citation8
The tumor microenvironment comprises a complex network, in which myeloid cells play key roles in tumor development.Citation9 MDSCs, a heterogeneous immature population of myeloid lineage, contribute to tumor-mediated immune escape.Citation10 In mice, MDSCs are identified as CD11b positive and Gr-1 positive; in humans, MDSCs are identified as co-expression of CD11b and CD33.Citation11 Recent research has shown that blood and intratumor MDSCs were significantly increased in patients with HNSCC.Citation12 M2 macrophages (tumor-associated macrophages, TAMs) are among the widely known component in tumor microenvironment.Citation13 Compared with M1 macrophages, M2 macrophages express high levels of immunosuppressive cytokines such as TGF-β and IL-10. In the tumor microenvironment, the M1–M2 switch accelerates tumor progression and aggressiveness by inducing T cell recruitment and activation.Citation14 Recent clinical data show that increased accumulation of M2 macrophages in the tumor indicates a poor prognosis for cancer patients.Citation15
In this study, we demonstrated that a significant increase in CTLA4 expression is an important immunosuppressive mechanism in humans and mice HNSCC. Oncogene activation by the conditional knockout of Pten and Tgfbr1 may contribute to the tumor immunosuppressive microenvironment with concomitantly significant increase in MDSCs and M2 macrophages. Moreover, we found that the blockade of CTLA4 significantly reduces MDSCs and M2 macrophages, promoting antigen presentation in mouse models. These findings indicated that targeting CTLA4 is a potentially effective therapeutic approach patients with HNSCC.
Results
High expression of CTLA4 in HNSCC and high CD8+/CTLA4 ratio in tumor predict improved overall survival in patients with HNSCC
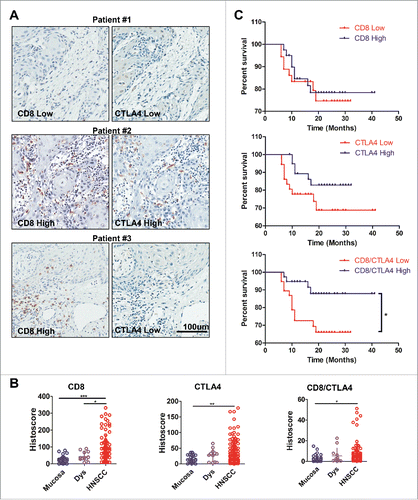
To identify CTLA4 expression in human HNSCC, we searched the publicly available cancer dataset by using the Oncomine database.Citation16 In a meta-analysis consisting of five data sets of head and neck cancer gene expression profiling, the mRNA level of CTLA4 was significantly increased in HNSCC compared with the control counterpart (p < 0.05, Fig. S1). Further, we constructed a tissue microarray from clinical specimens including 34 oral mucosa, 12 Dysplasia (Dys) and 86 HNSCC specimens (Table S1). Given the negative regulatory role of CTLA4 for T cells, we analyzed CTLA4 and CD8+ expressions in serial section tissue array (). We observed three types of CTLA4 and CD8+ expression in tumor. The first type, both CD8+ and CTLA4 were lowly expressed in the tumor-infiltrate lymphocytes (TIL) of HNSCC ( patient #1). In the second type, CD8+ and CTLA4 were highly expressed in TIL of HNSCC ( patient #2). In the third type, CD8+ was highly expressed, unlike CTLA4, in TIL of HNSCC ( patient #3). The last type may suggest a preferable antitumor immune reactive in intratumor. Quantification of the results of immunohistochemical staining indicated that the expression of CD8+ was increased in HNSCC compared with Dys (p < 0.001; ) and normal oral mucosa (p < 0.05; ). CTLA4 expression was significantly increased in HNSCC than in oral mucosa (p < 0.01; ). Of interest, the CD8+/CTLA4 ratio was remarkably higher in HNSCC than in oral mucosa (). We further analyzed the clinical pathological characteristic of HNSCC, and found that the CD8+/CTLA4 ratio, but not the expression of CTLA4 and CD8+ was significantly decreased in metastatic lymphocyte nodes compared with in negative nodes (Fig. S2A) in a pathology independent manner (Fig. S2B). Further, Kaplan–Meier survival analysis indicated that CTLA4 and CD8+ exhibited no correlation with overall survival in patient with HNSCC, while the CD8+/CTLA4 ratio exhibited a significant positive correlation with overall survival in patients with HNSCC (). High CD8+/CTLA4 ratio in tumor indicates an improved prognosis of patients with HNSCC. Meanwhile, the results of Cox proporational hazard model analysis confirmed the validity of the CD8+/CTLA4 ratio as a positive prognostic marker (Table S2). These data indicated that CTLA4 was upregulated in the TIL (tumor-infiltrating lymphocyte) of HNSCC and the high CD8+/CTLA4 ratio was correlated with improved prognosis.
Figure 1. CD8+/CTLA4 protein expression ratio is high in HNSCC and correlated with prognosis. (A) Immunohistochemical analysis of CD8+ and CTLA4 protein expression in HNSCC. Representative pictures are presented at different staining intensities (Low and High). Scale bar, 100 μm. (B) Quantification of CD8+, CTLA4 and CD8+/CTLA4 ratio protein expression histoscore in oral mucosa, dysplasia (Dys), and head neck squamous cell carcinoma (HNSCC). (Graph Pad One way ANOVA with post Tukey test. *p < 0.05; **p < 0.01; ***p < 0.001). (C) Kaplan–Meier survival analysis by CD8+, CTLA4 and CD8+/CTLA4 ratio in HNSCC patients (*p < 0.05).
CD8+/CTLA4 ratio was correlated with suppressive myeloid cells in human HNSCC
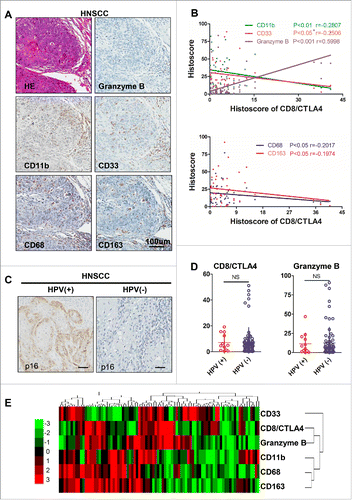
Cumulative studies demonstrated that suppressive myeloid cells, including MDSCs and M2 macrophages, play an indispensable role in tumorigenesis and tumor progression.Citation17,18 This finding promoted us to detect the correlation between the CD8+/CTLA4 ratio and suppressive myeloid cells. We analyzed the CD11b, CD33, CD68, CD163, and Granzyme B expression by immunohistochemical staining in human HNSCC as shown in . The expression of CD8+/CTLA4 ratio was a negatively correlated with CD11b and CD33 (markers for human MDSCs) and positively correlated with the expression of Granzyme B (). Similarly, the expression of CD68 and CD163 (markers for M2 type macrophages) was negatively correlated with the CD8+/CTLA4 ratio (). HPV is recognized as the causative agent of a growing subset of HNSCC.Citation19 Using p16 immunohistochemical staining () and HPV DNA in situ hybridization technique (data not shown) to monitor HPV infection, we found that there was no statistically significant difference in the CD8+/CTLA4 ratio and Granzyme B expression between HPV-positive and HPV-negative human HNSCC (). To further confirm whether CD8+/CTLA4 is associated with suppressive myeloid cells, we used a hierarchical clustering approach to evaluate the CD11b, CD33, CD68, CD163, and Granzyme B expression in human HNSCC (). Above data verified that the CD8+/CTLA4 ratio is correlated with suppressive myeloid cells in human HNSCC but not associated with HPV infection.
Figure 2. CD8+/CTLA4 protein expression ratio is correlated with immature suppressive myeloid cells in HNSCC. (A) Representative pictures of hematoxylin and eosin (HE) staining and immunohistochemical staining for Granzyme B, CD11b, CD33, CD68 and CD163 in HNSCC serial section. Scale bar, 100 μm. (B) The protein expression histoscore of CD8+/CTLA4 ratio exhibits a positive correlation with Granzyme B and a negative correlation with CD11b, CD33, CD68 and CD163 in HNSCC. (C) Representative immunohistochemistry staining of p16 to identify HPV-positive and HPV-negative HNSCC. Scale bar, 50 μm. (D) Quantification the CD8+/CTLA4 ratio and Granzyme B histoscore of HPV-positive and HPV-negative in HNSCC. NS, No Significant. (E) Hierarchical clustering of CD8+/CTLA4 ratio, CD11b, CD33, CD68, CD163, and Granzyme B histoscore results in human HNSCC.
Accumulation of MDSCs and M2 macrophages in the Tgfbr1/Pten 2cKO mouse HNSCC model
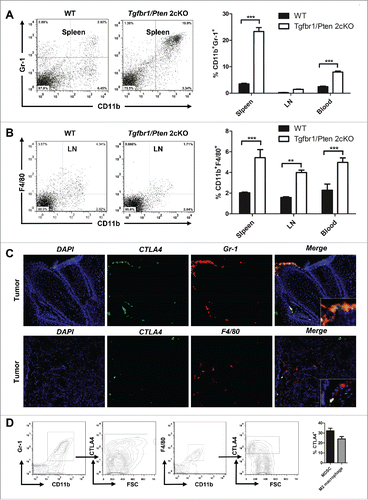
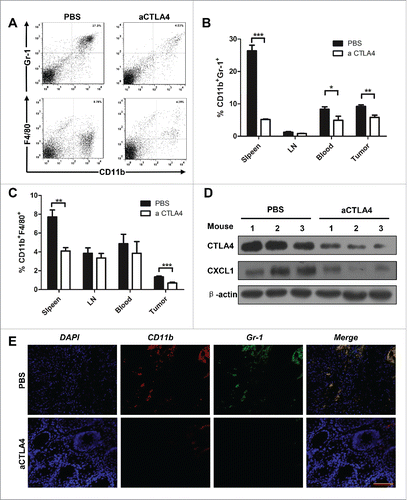
By using next-generation “omic” technologies, PTEN loss has been confirmed as a frequent molecular event in HNSCC.Citation20 Loss of the tumor suppressor SMAD4, leading to abrogation of TGF-β signaling, has been reported in HPV- HNSCC.Citation21 By combining deletion of important tumor suppressors, Pten and Tgfbr1, we constructed a spontaneous immune competent HNSCC mouse model (HPV-).Citation22 In tumor bearing mice, significant increases in CD11b+ and Gr1+ cell populations were found in spleen and blood (). In addition, significant increases in CD11b+ and F4/80+ cell populations were found in the spleen, lymph node, and blood of tumor bearing mice (). Considering the negative role of CTLA4 activation in antitumor immunity, we subsequently analyzed the co-expression of CTLA4 in MDSCs and M2 macrophages. We found that CTLA4 was partly expressed in MDSCs as indicated by the co-location of CTLA4 and Gr-1 (). In addition, CTLA4 may be expressed in the tumor infiltrating M2 macrophage, as indicated by the co-location of F4/80 (M2 macrophage) and CTLA-4 (). Further, quantification of percentage of CD11b+Gr-1+CTLA4+ and CD11b+F4/80+CTLA4+ cells in tumor of Tgfbr1/Pten 2cKO mice by flow cytometry (). Given these data, we hypothesized that CTLA4 may regulates tumor infiltrating immature myeloid cell of HNSCC and exhibits a potential immune suppression role of HNSCC tumorigenesis.
Figure 3. The accumulation of MDSCs and M2 macrophages in HNSCC mouse model. (A) Representative dot plots of CD11b+Gr-1+ MDSC in the spleen of wide type mice and Tgfbr1/Pten 2cKO mice (left) and bar graph shows quantitative analysis the number of CD11b+Gr-1+ MDSCs in spleen, draining lymphocyte node (LN), and blood of wide type mice and Tgfbr1/Pten 2cKO mice (right, t test, ***p < 0.001). (B) Representative dot plots of CD11b+F4/80+ M2 macrophages in the DLN of wide type mice and Tgfbr1/Pten 2cKO mice (left) and bar graph shows quantitative analysis the number of CD11b+F4/80+ M2 macrophages in spleen, LN and blood of wide type mice and Tgfbr1/Pten 2cKO mice (right, t test, **p < 0.01; ***p < 0.001). (C) Tumor sections were analysis by immunofluorescence for CTLA4+Gr-1+ and CTLA4+F4/80+ cells. Representative images were shown. (D) Quantification of percentage of CD11b+Gr-1+CTLA+ and CD11b+F4/80+CTLA4+ cells in tumor of Tgfbr1/Pten 2cKO mice.
CTLA4 blockade delayed tumorigenesis in Tgfbr1/Pten 2cKO mouse HNSCC model
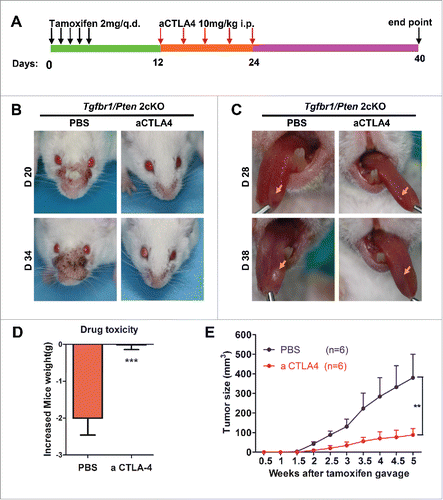
To test the hypothesis, we used immunocompetent Tgfbr1/Pten 2cKO mouse HNSCC models. The mice received a CTLA4 blocking antibody (9D9, 10 mg/kg; n = 6 mice) or PBS intraperitoneal injection every 3 d (). Tumor growth was assessed every day after tamoxifen gavage. We observed that blockade of CTLA4 was sufficient to reduce in the tumor burden of head neck () and tongue (). Meanwhile, CTLA4 blockade caused no additional cytotoxicity as compared with the PBS treatment group (). In addition, the tumor growth curve of the head and neck illustrated that CTLA4 blockade effectively decreased the tumor burden ().
Figure 4. aCTLA4 prevents tumorigenesis in HNSCC mouse model. (A) Schematic representation of the spontaneous immunocompetent HNSCC mouse model. (B) Representative images of tumor in mouse model with or without aCTLA4 treatment at day 20 and day 34 after tamoxifen gavage. (C) Representative images of tongue cancer in mouse model with or without aCTLA4 treatment at day 28 and day 38 after tamoxifen gavage. (D) Body weight of mice model was used to reflect drug toxicity in each group. (t test, ***p < 0.001). (E) Tumor volumes over time. On week five, p < 0.01 between mice treated aCTLA4 and PBS group (n = 6 in each group).
CTLA4 blockade reduced MDSCs and M2 macrophages in the Tgfbr1/Pten 2cKO mouse HNSCC model
Many studies have indicated that CTLA4 is a pivotal molecule in Treg.Citation37 Indeed, we also found blockade of CTLA4 significantly reduced the Treg cell population in our mouse model (Fig. S3). Considering that MDSCs and M2 macrophages can function as a negative regulator of antitumor immune response in HNSCC,Citation17,18 We focused on whether therapeutic blockade of CTLA4 could decrease MDSCs and M2 macrophages in this mouse model. The single cell suspense from the spleen, LN, blood, and tumor were stained by antibody for CD11b, Gr-1, and F4/80 (). Indeed, CTLA4 blockade remarkably decreased CD11b+Gr1+ MDSCs in the spleen, blood and tumor (). Similarly, CD11b+F4/80+ M2 macrophages were significantly reduced in the spleen and tumor in the treatment group as compared with the control (). Western bolt analysis supports that blockade of CTLA4 could effectively attenuate the recruitment of MDSCs and M2 macrophage by reducing chemokine CXCL1 in the tumor (). Immunofluorescence staining further confirmed a significant decrease of the Gr1+CD11b+ double positive MDSCs cell population in CTLA4 blockade group as compared with the control group ().
Figure 5. aCTLA4 reduced MDSCs and M2 macrophages in HNSCC mouse model. (A) Representative dot plots of CD11b+Gr-1+ MDSCs and CD11b+F4/80+ M2 macrophages in mouse model from spleen with or without aCTLA4 treatment. (B) The percentage of CD11b+Gr-1+ MDSCs was quantified in mouse model from spleen, draining lymphocyte node (LN), blood and tumor with or without aCTLA4 treatment (t test, *p < 0.05; **p < 0.01; ***p < 0.001). (C) The percentage of CD11b+F4/80+ M2 macrophage was quantified in mouse model from spleen, LN, blood and tumor with or without aCTLA4 treatment (t test, **p < 0.01; ***p < 0.001). (D) Western blot analysis the protein expression of CTLA4 and CXCL1 in mice bearing tumor with or without aCTLA4 treatment. β-actin was used as loading control. (E) Tumor sections were analysis by immunofluorescence for CD11b+Gr-1+ MDSCs. Representative images were shown from aCTLA4 and PBS treatment group. Scale bar, 100 μm.
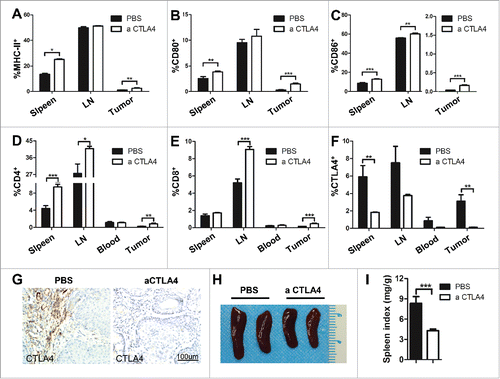
CTLA4 blockade improved T cell effector function in Tgfbr1/Pten 2cKO mouse HNSCC model
Among the costimulatory factors widely studied in the immune system is the CD28/CTLA4 - CD80/CD86 pathway, which critically controls the nature and duration of the T cell response.Citation23 This finding promoted us to detect the co-stimulating molecular MHC-II, CD80, and CD86 by flow cytometry. We found that mice treated with aCTLA4 enhanced the MHC-II cell population in the spleen and tumor (). Meanwhile, single-color analysis of the same cells revealed that CTLA4 blockade increased the CD80 positive and CD86 positive cell population in spleen, LN, and tumor (). Nevertheless, the increase of in the CD80 positive cell population has not reached statistically significant difference in LN. In general, these data suggested that CTLA4 blockade improved the condition of immunosuppressive status of the mouse model. Immunosuppression of effector T cells in the tumor microenvironment is mediated by increased expression of co-inhibitory receptor, including CTLA4.Citation24 To determine whether aCTLA4 had an broader role in activating T cells, single cell suspense from the spleen, LN, blood, and tumor were stained with CD4+, CD8+, and CTLA4 antibody. Quantitative the flow cytometry results demonstrated that CTLA4 blockade significantly increased the CD4+ and CD8+ T cells and reduced CTLA4+ cell population in the aCTLA4 treatment mice as compared with the control (). Further, immunohistochemical staining analyzed the expression of CTLA4 in the tumor of Tgfbr1/Pten 2cKO mouse HNSCC model with or without aCTLA4 treatment (). Additionally, the reverse of the immune suppressive status could be indicated by the reduction in the spleen size and weight observed in the aCTLA4 treatment group as compared with the control (). The combined results suggested that CTLA4 blockade significantly enhanced the T cells antitumor effect.
Figure 6. CTLA4 blockade improved T cell effector function in Tgfbr1/Pten 2cKO mouse HNSCC model. (A–C) Frequencies of MHC-II+(A), CD80+(B) and CD86+(C) cell population from mice spleen, draining lymphocyte node (LN) and tumor tissue with aCTLA4 and PBS treatment were summarized by flow cytometry. (t test, *p < 0.05; **p < 0.01; ***p < 0.001). (D–F) Quantification the percentage of CD4+(D) , CD8+(E), and CTLA4+(F) cell population from mice spleen, LN and tumor tissue with aCTLA4 and PBS treatment by flow cytometry. (t test, *p < 0.05; **p < 0.01; ***p < 0.001). (G) Immunohistochemistry was conducted on mice bearing tumor with or without aCTLA4 treatment for CTLA4 protein expression. Scale bar, 100 μm. (H) Representative photos of spleen from mice with or without aCTLA4 treatment were shown. (I) Spleen index (spleen weight: body weight ratio) was calculated in aCTLA4 and PBS treatment group (t test, ***p < 0.001).
Discussion
Our current study revealed the CD8+/CTLA4 ratio as a prognostic factor for patients with HNSCC and showed that the pathogenic role of CTLA4 in HNSCC via the recruitment of immature myeloid cells, including MDSCs and M2 macrophages. The present study demonstrated the important role of host immunity and cancer cell educated immunosuppression in HNSCC progression. Moreover, we used an immune competent transgenic HNSCC mouse model to elucidate the role of CTLA4 in HNSCC tumorigenesis. Furthermore, we showed that blockade of CTLA4 inhibited tumor growth by reducing MDSCs and M2 macrophages, as well as promoting antigen presentation and the activation of T cell in the mouse model.
The CD8+ T cell can predict disease recurrence and prognosis to a certain extent in HNSCC.Citation25 The expression of CTLA4 as a negative regulator is upregulated in the activator CD8+ T cell.Citation26 Herein, we evaluated the expression of CTLA4 and CD8+ in 86 patients with HNSCC. We found a positive correlation between the CD8+/CTLA4 ratio and the overall survival time of patients with HNSCC. Additionally, our study, consistent with previous study, demonstrated that immunosuppressive myeloid cells, including MDSCs and M2 macrophages, correlated with poor prognosis in patients with HNSCC.Citation12,27 On the basis of this finding, we analyzed the correlation between the CD8+/CTLA4 ratio and the expression of CD11b and CD33 (MDSCs markers in human) as well as CD68 and CD163 (M2 macrophages markers in human) in 86 patients with HNSCC. Remarkably, we observed a significant correlation between the CD8+/CTLA4 ratio and the immunosuppressive cell population. HPV infection is considered one of the most important etiologic cases of HNSCC.Citation28 A recent study indicated that different patterns of intratumor immune cell infiltrates in HPV+ HNSCC compared with HPV− HNSCC.Citation29 Interestingly, we found that CD8+/CTLA4 remained correlated with the immunosuppressive cell population in HPV− HNSCC.
MDSCs and M2 macrophages are identified as important immunosuppressive cells in the tumor environment.Citation30,31 Significantly increased MDSCs and M2 macrophages exist in tumor bearing transgenic mice as compared with wide type mice.Citation32 CTLA4 is a key inhibitory receptor that affects T cell function. CTLA4 is usually found in the intracellular compartment of resting T cell.Citation33 In our study, we found that part of MDSCs and M2 macrophages expressed CTLA4. The results indicated that CTLA4 may regulate the development of MDSCs and M2 macrophages in the tumor microenvironment. In line with our results, a recent study showed that exhausted T cells are associated with increased CTLA-4 expression in MDSCs in melanoma.Citation34 However, it remained unclear whether CTLA-4 inhibits tumor growth in a MDSCs and M2 macrophage dependent manner. In our mouse model, we observed that blockade of CTLA4 reduced the number of MDSCs and M2 macrophages in the tumor and the immune organ. In addition to regulating immunosuppressive cell population, we wanted to know whether CTLA4 affected other immune components such as the co-stimulatory molecule. In physiological status, T cell activation, cytokine production, proliferation, and differentiation need two signal models: MHC-peptide complex by the TCR and recognition of CD80 or CD86 by CD28.Citation35 CD28 and CTLA4 are the critical receptors that determine the early outcome of stimulation through banding of CD80 or CD86.Citation36 CD28 is constitutively expressed on the T cell surface, whereas the expression of CTLA4 is increased after T cell activation.Citation26 CTLA4 can function as a negative regulator of T cell activation and proliferation.Citation37 Consistent with our observations, the expression of CTLA4 is upregulated in tumor patients.Citation38 Moreover, in our study, the low of CD8+/CTLA4 ratio is associated with poor prognosis in patients with HNSCC. Blockade of CTLA4 expression could promote T cell activation via the CD28/B7 signal pathway.Citation39 In our in vivo study, upregulation of the expression levels of MHC-II (signal 1) and CD80/CD86 (signal 2) by blockade of CTLA4 promoted T cell activation and proliferation.
Chronic inflammation is a common framework of HNSCC. Secretion of IL-1β stimulates cells to produce the chemokine CXCL1 in HNSCC.Citation40 CXCL1 is one of the ligands of the chemokine receptor CXCR2, which is a crucial chemokine for the recruitment of MDSCs to sites of inflammation.Citation41 Our previous study demonstrated the high level of CXCL1 in tongue SCC as compared with normal tongue.Citation32 In our current study, we detected a decrease in MDSCs with the abatement of CXCL1 by inhibiting CTLA4. CXCL1 was reported to promote macrophage infiltration in addition to its recruitment of MDSCs.Citation42 This finding coincides with the current result that M2 macrophages were recruited to the site of tumor in our mouse model.
Though there are not completed clinical of ipilimumab (an lgG1 mAb targeting the CTLA4 receptor) in HNSCC patients, ipilimumab has been shown to improve overall survival in patients with advanced melanoma.Citation43,44 Moreover, our study suggested that targeting CTLA4 successfully reversed the immunosuppressive status by inhibiting MDSCs and M2 macrophage recruitment as well as promoting T cell activation. Nevertheless, targeting CTLA4 may still have an effect on other cells populations such as Treg.Citation45 Additionally, a recent study indicated that ipilimumab enhances cetuximab-mediated ADCC by eliminating intratumor Treg in HNSCC.Citation38
In summary, our results characterized CD8+/CTLA4 as a potential biomarker with clinical prognostic value for HNSCC. By using immune competent transgenic mouse HNSCC models, we directly observed that targeting CTLA4 decreased immature myeloid cells (MDSCs and M2 macrophages) and promote T cell activation in tumor microenvironment and macro-environment. Thus, our study provides important information for future application in patients with HNSCC.
Materials and methods
Human HNSCC tissue array
This study was approved by School and Hospital of Stomatology of Wuhan University Medical Ethics Committee, and informed consent was obtained from the patients before they underwent surgery. Custom made tissue array were used for immunohistochemistry staining, including 86 HNSCC, 12 oral epithelial Dys, and 32 normal oral mucosa tissue samples as previously described.Citation46
Immunocompetent and spontaneous HNSCC mouse model
The mice were fed in specific pathogen-free condition. All experiment procedures were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Wuhan University. Time inducible tissue-specific Tgfbr1/Pten 2cKO mouse (K14-CreERtam+/−; Tgfbr1flox/flox; Ptenflox/flox) were maintained and genotyped according to published protocols.Citation47 All the mice were bred in the FVBN/CD1/129/C57 mixed background.
CTLA4 antibody treatment
The mAb anti-CTLA4 antibody (9D9) was purchased from BioXcell (West Lebanon, NH, USA) and was stored at 4° C at a concentration of 5.52 mg/mL. The working solution was diluted in with a final concentration of 1 ug/uL immediately before use. The PBS was used as a negative control for tumorigenesis experiments. After oral gavage of tamoxifen for 5 consequent days, the Tgfbr1/Pten 2cKO mice were randomly divides into vehicle group (PBS 100 μL, i.p; n = 6 mice), treatment group (10 mg/kg 9D9, i.p; n = 6 mice). For all animals, general inspection and monitoring were performed every day. The tumor sizes were measured using a micrometer caliper and by taking photographs every other day. The endpoint was determined according to a systematic evaluation by the veterinarian. The mice were euthanized at the end of the studies, and the tumors were fixed in paraffin overnight or frozen at −80°C for subsequent immunostaining or Western blot.
Flow cytometry analysis
The single cell suspense was obtained from spleen, lymphocyte node (LN), blood, and tumor of HNSCC mouse model. The following anti-mouse antibody were used for staining: FITC-conjugated CD4+, CD8+, and CD11b, PE-conjugated CTLA4 and Gr-1 (all from Becton Dickinson, Mountain View, CA); Percp-Cy5.5-conjugated F4/80, PE-conjugated MHC-II, CD80 and CD86 (all from eBioscience, San Diego, CA); and isotype-matched IgG controls (eBioscience, San Diego, CA). The cells were analyzed using FlowJo (Tree Star, Ashland, OR) and gated by the side scatter and forward scatter filters. Death cells were excluded by staining 7AAD (Invitrogen,).
Western blot analysis
Tumor from HNSCC mouse model were carefully dissected (n = 6, respectively). A total amount of 30 µg protein from each sample was denatured and then subjected to 12% SDS-polyacrylamide gel electrophoresis followed by transfer onto polyvinylidene fluoride membranes (Millipore Corporation, Bil-lerica, MA). Next, the blots were stained using an enhanced chemiluminescence detection kit (West Pico, Thermo). The following antibodies were used for Western blot analysis: CTLA4 (Santa Cruz Biotechnology), CXCL1 (GeneTex, Irine CA). β-actin was used as a loading control.
Immunofluorescence
Tumor sections were hydrated in alcohol, washed three times in PBS, retrieved using sodium citrate in a pressure cooker, blocked with 2.5% bovine serum album in PBS buffer for 1 h at 37°C and were then incubated with primary antibody or isotype-matched IgG controls overnight at 4°C. The next day, slides were incubated with fluorochrome conjugated secondary antibodies (Alexa 594 anti-rabbit and Alexa 488 anti-mouse; Invitrogen) and mounted in Vectashield with 4', 6-diamidino-2-phenylindole (DAPI; Vector Laboratories). Fluorescence images were then captured using a CLSM-310, Zeiss fluorescence microscope.
Immunohistochemistry
The tissue array sections were stained as previous protocol.Citation46 In brief, the sections were incubated overnight at 4°C with antibody for CTLA4 (Santa Cruz Biotechnology, California, USA), CD8 (Zymed, China), CD68 (Zymed, China), CD163 (CWBiotech, China), CD11b (Abcam, Cambridge, UK), CD33 (Zymed, China), p16 (Zymed, China), Granzyme B (Zymed, China) or isotype-matched IgG controls. Then, the sections were incubated with a secondary biotinylated immunoglobulin G antibody solution and an avidin-biotin-peroxidase reagent. At last, after being washed three times with phosphate buffer saline, the sections lightly counterstained with Mayer's haematoxylin (Invitrogen, USA).
Scoring system, hierarchical clustering, and data visualization
Whole slices were scanned using an Aperio ScanScope CS scanner (Vista, CA, USA) with a background substrate for each slice, and quantified using Aperio Quantification software (Version 9.1) for membrane, nuclear, or pixel quantification. An area of interest was selected either in the epithelial or the cancerous area for scanning and quantification. The histoscore of membrane and nuclear staining was calculated as a percentage of different positive cells using the formula (3+)×3+(2+)×2+(1+)×1.Citation46 The histoscore of pixel quantification was calculated as the total intensity/total cell number. The threshold for scanning of different positive cells was set according to the standard controls provided by Aperio.Citation47 Hierarchical clustering was performed as our previous produce.Citation46
Statistical analysis
Data analyses were performed using Graph Pad Prism version 5.0 for Windows (Graph Pad Software Inc., La Jolla, CA). One-way ANOVA followed by the post-Tukey multiple comparison tests and unpaired t test was used to analyze significant difference. The Kaplan–Meier was used to analyze survival curve and the log-rank test to detect the differences of overall survival. Two-tailed Pearson's statistics was used for the correlation. Dates are represented as the mean ± SEM. ***p values of < 0.05 were considered statistically significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KONI_A_1151594_supplementary_material.zip
Download Zip (239.1 KB)Funding
This work was supported by National Natural Science Foundation of China 81272963, 81472528 (Z.J S.), 81272964, 81472529 (W.F.Z). Z.J.S was supported by program for new century excellent talents in university (NCET-13-0439), Ministry of Education of China.
References
- Hashibe M, Brennan P, Chuang SC, Boccia S, Castellsague X, Chen C, Curado MP, Dal Maso L, Daudt AW, Fabianova E et al. Interaction between tobacco and alcohol use and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. Cancer Epidemiol Biomarkers Prev 2009; 18:541-50; PMID:19190158; http://dx.doi.org/10.1158/1055-9965.EPI-08-0347
- Tong CC, Kao J, Sikora AG. Recognizing and reversing the immunosuppressive tumor microenvironment of head and neck cancer. Immunol Res 2012; 54:266-74; PMID:22454102; http://dx.doi.org/10.1007/s12026-012-8306-6
- Quan J, Johnson NW, Zhou G, Parsons PG, Boyle GM, Gao J. Potential molecular targets for inhibiting bone invasion by oral squamous cell carcinoma: a review of mechanisms. Cancer Metastasis Rev 2012; 31:209-19; PMID:22101806; http://dx.doi.org/10.1007/s10555-011-9335-7
- Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 2015; 27:450-61; PMID:25858804; http://dx.doi.org/10.1016/j.ccell.2015.03.001
- Tivol EA, Borriello F, Schweitzer AN, Lynch WP, Bluestone JA, Sharpe AH. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity 1995; 3:541-7; PMID:7584144; http://dx.doi.org/10.1016/1074-7613(95)90125-6
- Lenschow DJ, Herold KC, Rhee L, Patel B, Koons A, Qin HY, Fuchs E, Singh B, Thompson CB, Bluestone JA. CD28/B7 regulation of Th1 and Th2 subsets in the development of autoimmune diabetes. Immunity 1996; 5:285-93; PMID:8808683; http://dx.doi.org/10.1016/S1074-7613(00)80323-4
- Hathcock KS, Laszlo G, Pucillo C, Linsley P, Hodes RJ. Comparative analysis of B7-1 and B7-2 costimulatory ligands: expression and function. J Exp Med 1994; 180:631-40; PMID:7519245; http://dx.doi.org/10.1084/jem.180.2.631
- Peggs KS, Quezada SA. Ipilimumab: attenuation of an inhibitory immune checkpoint improves survival in metastatic melanoma. Expert Rev Anticancer Ther 2010; 10:1697-701; PMID:21080797; http://dx.doi.org/10.1586/era.10.144
- Galdiero MR, Bonavita E, Barajon I, Garlanda C, Mantovani A, Jaillon S. Tumor associated macrophages and neutrophils in cancer. Immunobiology 2013; 218:1402-10; PMID:23891329; http://dx.doi.org/10.1016/j.imbio.2013.06.003
- Ochando JC, Chen SH. Myeloid-derived suppressor cells in transplantation and cancer. Immunol Res 2012; 54:275-85; PMID:22535241; http://dx.doi.org/10.1007/s12026-012-8335-1
- Serafini P, Borrello I, Bronte V. Myeloid suppressor cells in cancer: recruitment, phenotype, properties, and mechanisms of immune suppression. Semin Cancer Biol 2006; 16:53-65; PMID:16168663; http://dx.doi.org/10.1016/j.semcancer.2005.07.005
- Weed DT, Vella JL, Reis IM, De la Fuente AC, Gomez C, Sargi Z, Nazarian R, Califano J, Borrello I, Serafini P. Tadalafil reduces myeloid-derived suppressor cells and regulatory T cells and promotes tumor immunity in patients with head and neck squamous cell carcinoma. Clin Cancer Res 2015; 21:39-48; PMID:25320361; http://dx.doi.org/10.1158/1078-0432.CCR-14-1711
- Liu Y, Cao X. The origin and function of tumor-associated macrophages. Cell Mol Immunol 2015; 12:1-4; http://dx.doi.org/10.1038/cmi.2014.83
- Sica A, Larghi P, Mancino A, Rubino L, Porta C, Totaro MG, Rimoldi M, Biswas SK, Allavena P, Mantovani A. Macrophage polarization in tumour progression. Semin Cancer Biol 2008; 18:349-55; PMID:18467122; http://dx.doi.org/10.1016/j.semcancer.2008.03.004
- Ostuni R, Kratochvill F, Murray PJ, Natoli G. Macrophages and cancer: from mechanisms to therapeutic implications. Trends Immunol 2015; 36:229-39; PMID:25770924; http://dx.doi.org/10.1016/j.it.2015.02.004
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007; 9:166-80; PMID:17356713; http://dx.doi.org/10.1593/neo.07112
- Trellakis S, Bruderek K, Hutte J, Elian M, Hoffmann TK, Lang S, Brandau S. Granulocytic myeloid-derived suppressor cells are cryosensitive and their frequency does not correlate with serum concentrations of colony-stimulating factors in head and neck cancer. Innate Immun 2013; 19:328-36; PMID:23160385; http://dx.doi.org/10.1177/1753425912463618
- Lyford-Pike S, Peng S, Young GD, Taube JM, Westra WH, Akpeng B, Bruno TC, Richmon JD, Wang H, Bishop JA et al. Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res 2013; 73:1733-41; PMID:23288508; http://dx.doi.org/10.1158/0008-5472.CAN-12-2384
- Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol 2011; 29:4294-301; PMID:21969503; http://dx.doi.org/10.1200/JCO.2011.36.4596
- Sepiashvili L, Bruce JP, Huang SH, O'Sullivan B, Liu FF, Kislinger T. Novel insights into head and neck cancer using next-generation “omic” technologies. Cancer Res 2015; 75:480-6; PMID:25589349; http://dx.doi.org/10.1158/0008-5472.CAN-14-3124
- Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer 2011; 11:9-22; PMID:21160525; http://dx.doi.org/10.1038/nrc2982
- Bian Y, Hall B, Sun ZJ, Molinolo A, Chen W, Gutkind JS, Waes CV, Kulkarni AB. Loss of TGF-β signaling and PTEN promotes head and neck squamous cell carcinoma through cellular senescence evasion and cancer-related inflammation. Oncogene 2012; 31:3322-32; PMID:22037217; http://dx.doi.org/10.1038/onc.2011.494
- Rudd CE, Taylor A, Schneider H. CD28 and CTLA-4 coreceptor expression and signal transduction. Immunol Rev 2009; 229:12-26; PMID:19426212; http://dx.doi.org/10.1111/j.1600-065X.2009.00770.x
- Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 2011; 331:1565-70; PMID:21436444; http://dx.doi.org/10.1126/science.1203486
- Czystowska M, Gooding W, Szczepanski MJ, Lopez-Abaitero A, Ferris RL, Johnson JT, Whiteside TL. The immune signature of CD8(+)CCR7(+) T cells in the peripheral circulation associates with disease recurrence in patients with HNSCC. Clin Cancer Res 2013; 19:889-99; PMID:23363813; http://dx.doi.org/10.1158/1078-0432.CCR-12-2191
- Walunas TL, Lenschow DJ, Bakker CY, Linsley PS, Freeman GJ, Green JM, Thompson CB, Bluestone JA. CTLA-4 can function as a negative regulator of T cell activation. Immunity 1994; 1:405-13; PMID:7882171; http://dx.doi.org/10.1016/1074-7613(94)90071-X
- He KF, Zhang L, Huang CF, Ma SR, Wang YF, Wang WM, Zhao ZL, Liu B, Zhao YF, Zhang WF et al. CD163+ tumor-associated macrophages correlated with poor prognosis and cancer stem cells in oral squamous cell carcinoma. Biomed Res Int 2014; 2014:838632; PMID:24883329; http://dx.doi.org/10.1155/2014/838632.
- Martin D, Abba MC, Molinolo AA, Vitale-Cross L, Wang Z, Zaida M, Delic NC, Samuels Y, Lyons JG, Gutkind JS. The head and neck cancer cell oncogenome: a platform for the development of precision molecular therapies. Oncotarget 2014; 5:8906-23; PMID:25275298; http://dx.doi.org/10.18632/oncotarget.2417
- Partlova S, Boucek J, Kloudova K, Lukesova E, Zabrodsky M, Grega M, Fucikova J, Truxova I, Tachezy R, Spisek R et al. Distinct patterns of intratumoral immune cell infiltrates in patients with HPV-associated compared to non-virally induced head and neck squamous cell carcinoma. Oncoimmunology 2015; 4:e965570; PMID:25949860; http://dx.doi.org/10.4161/21624011.2014.965570
- Flemming A. Cancer: Re-educating tumour-associated macrophages. Nat Rev Drug Discov 2011; 10:177; PMID:21358735; http://dx.doi.org/10.1038/nrd3399
- Talmadge JE, Gabrilovich DI. History of myeloid-derived suppressor cells. Nat Rev Cancer 2013; 13:739-52; PMID:24060865; http://dx.doi.org/10.1038/nrc3581
- Yu GT, Bu LL, Huang CF, Zhang WF, Chen WJ, Gutkind JS, Kulkarni AB, Sun ZJ. PD-1 blockade attenuates immunosuppressive myeloid cells due to inhibition of CD47/SIRPalpha axis in HPV negative head and neck squamous cell carcinoma. Oncotarget 2015; 6:42067-80; PMID:26573233; http://dx.doi.org/10.18632/oncotarget.5955.
- Buchbinder E, Hodi FS. Cytotoxic T lymphocyte antigen-4 and immune checkpoint blockade. J Clin Invest 2015; 125:3377-83; PMID:26325034; http://dx.doi.org/10.1172/JCI80012
- Chevolet I, Speeckaert R, Schreuer M, Neyns B, Krysko O, Bachert C, Hennart B, Allorge D, van Geel N, Van Gele M et al. Characterization of the immune network of IDO, tryptophan metabolism, PD-L1, and in circulating immune cells in melanoma. Oncoimmunology 2015; 4:e982382; PMID:25949897; http://dx.doi.org/10.4161/2162402X.2014.982382
- Kaufmann DE, Walker BD. PD-1 and CTLA-4 inhibitory cosignaling pathways in HIV infection and the potential for therapeutic intervention. J Immunol 2009; 182:5891-7; PMID:19414738; http://dx.doi.org/10.4049/jimmunol.0803771
- Egen JG, Kuhns MS, Allison JP. CTLA-4: new insights into its biological function and use in tumor immunotherapy. Nat Immunol 2002; 3:611-8; PMID:12087419; http://dx.doi.org/10.1038/ni0702-611
- Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science 1996; 271:1734-6; PMID:8596936; http://dx.doi.org/10.1126/science.271.5256.1734
- Jie HB, Schuler PJ, Lee SC, Srivastava RM, Argiris A, Ferrone S, Whiteside TL, Ferris RL. CTLA-4(+) Regulatory T Cells Increased in Cetuximab-Treated Head and Neck Cancer Patients Suppress NK Cell Cytotoxicity and Correlate with Poor Prognosis. Cancer Res 2015; 75:2200-10; PMID:25832655; http://dx.doi.org/10.1158/0008-5472.CAN-14-2788
- Ewing MM, Karper JC, Abdul S, de Jong RC, Peters HA, de Vries MR, Redeker A, Kuiper J, Toes RE, Arens R et al. T-cell co-stimulation by CD28-CD80/86 and its negative regulator CTLA-4 strongly influence accelerated atherosclerosis development. Int J Cardiol 2013; 168:1965-74; PMID:23351788; http://dx.doi.org/10.1016/j.ijcard.2012.12.085
- Lee CH, Chang JS, Syu SH, Wong TS, Chan JY, Tang YC, Yang ZP, Yang WC, Chen CT, Lu SC et al. IL-1beta promotes malignant transformation and tumor aggressiveness in oral cancer. J Cell Physiol 2015; 230:875-84; PMID:25204733; http://dx.doi.org/10.1002/jcp.24816
- Oppenheim JJ, Zachariae CO, Mukaida N, Matsushima K. Properties of the novel proinflammatory supergene “intercrine” cytokine family. Annu Rev Immunol 1991; 9:617-48; PMID:1910690; http://dx.doi.org/10.1146/annurev.iy.09.040191.003153
- Remus EW, Sayeed I, Won S, Lyle AN, Stein DG. Progesterone protects endothelial cells after cerebrovascular occlusion by decreasing MCP-1- and CXCL1-mediated macrophage infiltration. Exp Neurol 2015; 271:401-8; PMID:26188381; http://dx.doi.org/10.1016/j.expneurol.2015.07.010
- Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010; 363:711-23; PMID:20525992; http://dx.doi.org/10.1056/NEJMoa1003466
- Swanson MS, Sinha UK. Rationale for combined blockade of PD-1 and CTLA-4 in advanced head and neck squamous cell cancer-review of current data. Oral Oncol 2015; 51:12-5; PMID:25459157; http://dx.doi.org/10.1016/j.oraloncology.2014.10.010
- Simpson TR, Li F, Montalvo-Ortiz W, Sepulveda MA, Bergerhoff K, Arce F, Roddie C, Henry JY, Yagita H, Wolchok JD et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J Exp Med 2013; 210:1695-710; PMID:23897981; http://dx.doi.org/10.1084/jem.20130579
- Yu GT, Bu LL, Zhao YY, Liu B, Zhang WF, Zhao YF, Zhang L, Sun ZJ. Inhibition of mTOR reduce Stat3 and PAI related angiogenesis in salivary gland adenoid cystic carcinoma. Am J Cancer Res 2014; 4:764-75; PMID:25520866
- Sun ZJ, Zhang L, Hall B, Bian Y, Gutkind JS, Kulkarni AB. Chemopreventive and chemotherapeutic actions of mTOR inhibitor in genetically defined head and neck squamous cell carcinoma mouse model. Clin Cancer Res 2012; 18:5304-13; PMID:22859719; http://dx.doi.org/10.1158/1078-0432.CCR-12-1371
