ABSTRACT
Although immune checkpoint blockade have demonstrated promising results, their effects on gastric cancer (GC) are under investigation. Understanding the clinical significance of PD1 and its ligands' expression, together with T cell infiltration might provide clues for biomarkers screening in GC immunotherapy. Immunohistochemistry were performed on a tissue microarray including 1,014 GC specimens using PD1, PDL1 and PDL2 antibodies. T cell markers CD3 and CD8 were also stained and quantified by automated image analysis. Correlation with clinical features and outcome were analyzed after controlling for potential confounders including EBV infection, HER2, C-met and PCNA expression. 37.8% of the cases showed membranous PD-L1 expression in tumor cells and 74.9% in infiltrating immune cells. PDL1 expression rate was rather higher in patients without metastasis, in EBV positive group and those with C-met and PCNA expression. GC patients with high level PDL1 expression exhibited better survival. GC Patients with higher T cell infiltration also showed elevated PDL1, PDL2 and PD1 expression and predict favorable outcome, indicating an adaptive immune resistance mechanism may exist. The group of patients infiltrated with lower density CD3+ T cells also without PDL1 expression in tumor cells predict the worst outcome in the subgroup of different PTNM stage, which may suggest an inactive immune status. These results highlights the need to assess both PDL1 expression in all tumor context and the characterization of the GC immune microenvironment.
Introduction
Although declining in the last few decades, Gastric cancer (GC) still remains the third most common cancer worldwide.Citation1 It was postulated that GC, which harbored higher mutation frequency, would be more immunogenic and likely to benefit from immunotherapy, given its breakthrough effects on melanoma and lung cancer.Citation2,3,4
Cancer cells may exhibit immune inhibition to promote tumor progression and distant metastasis.Citation5 One key mechanism is the Programmed Cell Death 1 (PD1) / PD1 ligand 1 (PDL1) pathway. PD1 is typically expressed by activated lymphocytes, including CD8+ T cells, CD4+ T cells, natural killer (NK) T cells, B cells, activated monocytes and dendritic cells.Citation6 It is activated by its ligands PDL1 and PDL2 to suppress antigen-stimulated lymphocyte proliferation, migration and cytokine production, ultimately resulting in attenuating effector T cells function and immunological tolerance.Citation6 PDL1 is constitutively expressed on T and B cells, macrophages and dendritic cells, while PDL2 expression is much more typically restricted in activated DC and macrophages.Citation7 PDL1 have also been reported to be expressed in variety tumor cells including lung, colorectal, breast and melanoma.Citation8–11 To date, PD1 and its ligands in GC was only evaluated in small cohort of patients with controversial conclusions.Citation12–15 These results were probably confounded by the ethnic differences, the small sample size, and/or heterogeneous patient population.
In addition to PD1/PDL1, tumor infiltrating lymphocytes (TILs) reflects primary host immune response against solid tumors.Citation16,17 A strong histological lymphocytic reaction, high density of CD3+ and CD8+ T cells have been reported with better outcome in colorectal cancer and lung cancer, demonstrating an important role of T cell mediated host immunity in repressing tumor progression.Citation12,18,19 Considering the close relationship between PD1/PDL1 pathway and T cell activation, we used a tissue microarray (TMA) including 1,014 well annotated GC specimens to investigate the expression of PD1/PDL1, quantified tumor infiltrating CD3+ and CD8+ T cells density to determine their relationships with clinicopathological features and patients' prognosis.
Results
Patient characteristics
A total of 1014 surgically resected FFPE primary gastric cancer sampled in tissue microarrays could be assessed for the markers, including 739 males and 275 females without any preoperative therapy (). The median follow up is 39.0 months (range: 0.2–133.9 months) and the median age at diagnosis is 61 y (range: 22–89 y). 239 cases were located at the proximal part, including the gastro-esophageal junction, 259 cases in the corpus, 468 at the distal part including antrum and 5 cases disseminated to the whole gastric structures. All the tissue samples were identified as adenocarcinoma and 78% of samples were poorly or moderate differentiated, including 8% signet ring cell type. As shown in , 569 cases (56.1%) were intestinal subtype, while 242 (23.9%) were intestinal and 184 (18.1%) were mixed type. The vast majority of tumors were classified as clinical TNM stage 2(34.7%) and stage 3(44.1%). The 1-, 3-, 5-year overall survival is 0.846 ± 0.011, 0.613 ± 0.016, 0.521 ± 0.017, respectively.
Table 1. Clinicopathological and molecular features according to PDL1 expression.
PDL1, PDL2 and PD1 expression
PDL1 expression was evaluated in tumor and immune cells, respectively. Dichotomized by using an IRS of 2 as the cutoff value, 383 patients out of 1014 cases (37.8%) showed a membranous PDL1 expression in tumor cells. Also, the cytoplasmic expression of PDL1 were observed in 494 of 1014 cases (48.7%). Both cytoplasm and membrane expression were calculated while only membrane staining value was used in the further analysis considering it functions as a membrane ligand (Appendix table 1). PDL1 expression in tumor infiltrating immune cells was found in 759 of 1014 (74.9%) cases (, ).
Figure 1. PDL1, PDL2, PD1, CD3 and CD8 expression in gastric carcinoma by immunohistochemistry (× 200). PDL1 expression was evaluated based on staining in the cytoplasm and membrane of tumor cells and immune cells. A: Intense expression of PDL1 in tumor cells; B: Immune cells expressing PDL1 in the disseminated lymphocytes and macrophages; C: representative cytoplasmic expression of PDL2 in tumor tissues. D: PD1 expressed in the disseminated immune cells infiltrating the tumor tissues; E: The representative images of CD3+ T cells; F: The representative images of CD8+ T lymphocytes.
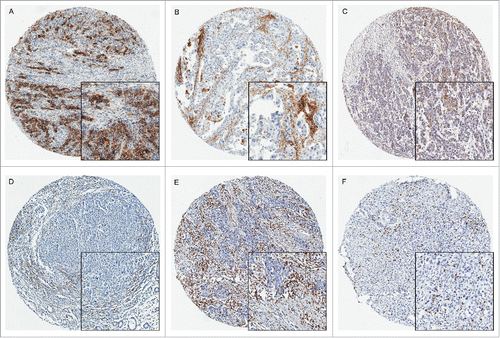
As shown in , PDL2 expression were mainly localized at cytoplasm. 506 of 1014 cases (49.9%) were found PDL2 expression in tumor and 203 (20%) cases in immune cells. PD1 diffusely distributed immune cells were present in 321(31.7%) GC cases. No distinct distribution pattern was found except accumulation in intratumoral lymph follicles. PD1 expression was also significantly associated with PDL1 and PDL2 expression in tumor cells and immune cells (Appendix table 2, online only).
Table 2. Survival analysis of 1014 gastric cancers.
Correlation of PDL1, PDL2, PD1 with clinic-pathological features
PDL1 membranous expression in tumor cells was negatively correlated with lymph node involvement, distant metastasis and pTNM stage (). Tumor PDL1 expression was more common in diffuse type GC. In addition, PDL1 expression rate was higher in the EBV positive group, C-met positive group comparing with the corresponding negative/deficient groups. In addition, higher intensity PCNA expression significantly correlated with positive PDL1 expression on both tumor and immune cells. Although PDL1 expression was more frequent in immune cells than in tumor cells, it showed the similar association with clinicopathological features ().
Similar to PDL1, PDL2 expression in tumor cells significantly correlated with pTNM, C-met, and PCNA expression. However, no significant association between PDL2 expressions in the infiltrating immune cells and clinical characteristics were observed after multiple testing. PD1 expression was positively related with T stage, tumor size, pTNM stage, and high level PCNA expression (Appendix table 3, online only).
Association of CD3 and CD8 with clinic-pathological parameters
The median percentage of CD3+ and CD8+ T cells among the total cells were 11.24 (range: 0.40–59.35) and 9.41 (range: 0.27–51.98). We also observed the significant positive relationship between CD3+, CD8+ cell density and PDL1, PDL2, PD1 expression, suggesting they may take interacting functions (Appendix table 2). CD3+ and CD8+ T cell density were both significantly higher in patients with younger age, smaller tumor size and with poor differentiation. They were also significantly increased in patients with lower potential to metastasis, such as negative vascular invasion, without lymph node metastasis and distant metastasis, or in the early pTNM stages. The density was significantly increased in EBV infectious patients and those with intense PCNA expression as well (Appendix table 4).
Prognostic significance
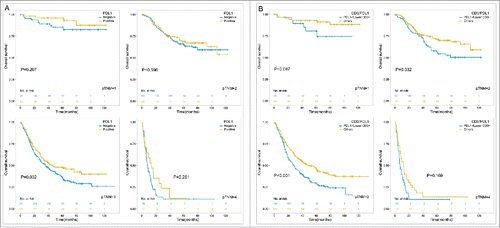
In the univariable survival analysis, differentiation, tumor size, vascular invasion, Lauren type, tumor invasion, lymph node status, distant metastasis and pathological TNM stage were significantly associated with patients' prognosis (). GC patients with PDL1 expression in tumor cells or immune cells were also showed better survival even in the subgroup of pathological stage analysis. (5-year OS, 0.609 ± 0.027 vs 0.475 ± 0.021, P<0.001; 0.546 ± 0.019 VS 0.447 ± 0.033, P<0.001 in tumor and immune cells respectively, , ). PDL2 expression in immune cells also showed significant positive association with survival (5-year OS, 0.588 ± 0.037 vs 0.504 ± 0.019, P = 0.015, ). While analyzing the impact of combined expression of PDL1, PDL2 and PD1 on survival, we found that patients expressing both PDL1 and PD1 showed higher survival than either PDL1 or PD1 positive patients ((5-year OS, 0.626 ± 0.041 vs 0.598 ± 0.035 vs 0.500 ± 0.042 vs 0.461 ± 0.024 for co-positive, PDL1 positive, PD1 positive, and co-negative respectively, p < 0.001, Appendix Fig 1, online only). Also, coexpression of PDL1 and PDL2 also showed the similar trends (5-year OS, 0.634 ± 0.035 vs 0.577 ± 0.041 vs 0.521 ± 0.031 vs 0.433 ± 0.028 for co-positive, PDL1 positive, PDL2 positive, and co-negative respectively, p < 0.001, Appendix Fig 1, online only), suggesting their interactions on immune reaction and further outcome (Supplementary ). As regarding to the tumor infiltrating immune cells, both higher CD3+ and CD8+ T cell densities showed better survival in a density dependent manner. (5-year OS, 0.614 ± 0.033 vs 0.570 ± 0.033 vs 0.508 ± 0.033 vs 0.391 ± 0.033 for lowest, second, third, highest CD3+ cell density respectively, p < 0.001, and 0.578 ± 0.033 vs 0.588 ± 0.032 vs 0.520 ± 0.033 vs 0.399 ± 0.033 for lowest, second, third, highest CD8+ cell density, p < 0.001, ).
Figure 2. Kaplan-Meire curves stratified by PDL1, CD3, and CD8 expression. PDL1 expression in tumor cells predicted better survival in GC patients using cut-off value of IRS for 2 and 5 as 3 groups (A, PDL1TU: PDL1 expression in tumor cells) or in the immune cells (B, PDL1IM: PDL1 expression in immune cells). CD3+ (C) or CD8+ (D) T cells both associated with better outcome in a density dependent manner.
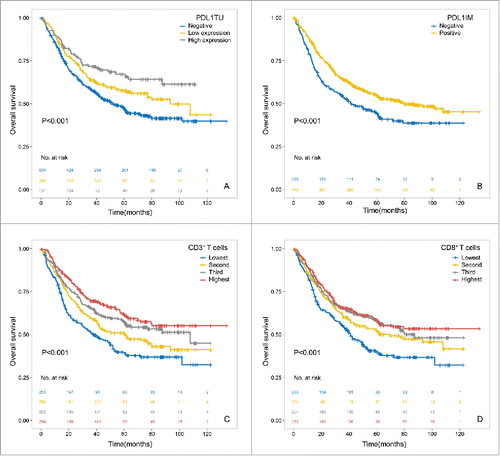
Figure 3. Prognostic significance of PDL1 expression (A) in tumor cells as well as the intersection of PDL1 expression and CD3+ T cell infiltration (B) in subgroups of different pTNM stages. Using the median of CD3 density as the cutoff value and PDL1 membrane expression in tumor cells, the patients were divided into 2 groups, one with CD3 density lower than median and PDL1 negative expression, and the other group consisted with the rest patients.
A Cox regression analysis including age, pTNM stage, differentiation, vascular invasion, tumor size, lauren type, PDL1 expression, PDL2 expression, CD3+ and CD8+ cell density showed tumor cells PDL1 expression remains significantly associated with better survival (HR = 0.786, 95%CI = 0.640–0.965, P = 0.021; ). While CD3+ cell density also remained as an independent significant favorable prognostic factor (HR: 0.978, 95%CI:0.967–0.989, P<0.001; AUC:0.763, 95%CI:0.732–0.794, P < 0.001, ).
Prognostic value of combination of CD3+ cell density and PDL1 expression
Considering the close relationship between CD3+ cell density and PDL1 expression with survival, we divided the patients into 2 subgroup, one with lower CD3+ infiltration together with PDL1 negative expression in tumor, the other group either have PDL1 expression or with higher CD3+ infiltration, Patients with low level of T cells infiltration and did not show PDL1 expression have the worst survival (5-year OS, 0.391 ± 0.028 vs 0.588 ± 0.020, p < 0.001), demonstrating lack or inactive immune reaction, and the other group indicating relatively active immune reaction and better survival.
In the subgroup analysis in different pTNM stages, the prognostic value of combination of CD3+ T cells density and PDL1 expression remained significant in stage 2 and stage 3, and showed the same tendency in stage 1and stage 4 (, panel B). It suggests distinguishing the prognosis of GC patients according to immune status within pTNM stages may be a more precise and applicable method in future.
Subgroup survival analysis stratified by EBV infection
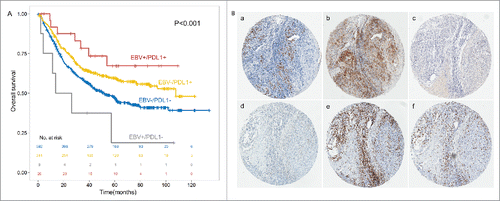
Interestingly, Survival curves stratified by PDL1 expression in the subgroup of 34 cases of EBV infected patients were significantly different. (5-year OS, 0.673 ± 0.104 vs 0.188 ± 0.158, P<0.001, ). That indicates although PDL1 showed the same tendency no matter with EBV infection or not, EBV infected GC patients with or without PDL1 expression may exhibited greatly different outcome.
Figure 4. PDL1 expression associated with better outcome in the subgroup of EBV+ and EBV- GC patients (A). PDL1, PDL2, PD1, CD3 and CD8 expression in the identical EBV infected gastric cancer tissue (× 200, B). In situ hybridization of EBER (a) was shown in the identical patients together with PDL1 (b), PDL2 (c) and PD1 (d) expression in GC tumor tissues and CD3 (e) and CD8 (f) in intratumoral lymphocytes.
TCGA database analysis and pathway
PDL1 mRNA high expressing patients in the TCGA database also have better outcome (HR: 0.824, 95%CI: 0.695–0.977, P = 0.026) in cox model adjusting for age, gender, pathological stage. PD1 also showed the same trend (HR: 0.780, 95%CI: 0.625–0.973, P = 0.028). A close relationship between PDL1, PDL2, PD1 expression and CD3D, CD3E, CD3G, CD3Z, CD8A, CD8B were also observed, consistent with our previous results (Appendix Fig 2). When combining the T cells density with PDL1 expression, Patients lack of PDL1 and CD3Z expression have worse outcome in the overall population and pTNM subgroup analysis (Appendix Fig 3).
A total of 679 differentially expressed genes were identified, including 238 genes overexpressed and 441 genes downregulated in the “PDL1-high” group (Appendix table 5). GO analysis revealed a major involvement in the regulation of immune response, notably the activation of T-cells. Many genes coded for proteins related to lymphocytes proliferation, leucocytes activation, response to virus, cytotoxic effector molecules, T cell differentiation, and chemokines related to T-cells activation and homing. In addition, several were MHC-related molecules, involved in the processing of endogenous antigens and presentation to cytotoxic and helper T-cells: mainly HLA-II molecules. Interestingly, CTLA4, which code for markers of T-cells exhaustion, were strongly overexpressed in the “PDL1- high” group. IDO, which code for cytokines synthesized in exhausted T-cells, were overexpressed in this group as well (Appendix table 6).
Discussion
Currently, more than 400 clinical trials focus on PDL1/PD1 immune checkpoint signaling, including 65 studies for gastrointestinal cancers.Citation20 Here we report PDL1 expression in a substantial amount of GCs either in tumor or immune cells, highlighting its association with T cell infiltration and further good outcome. Upon multivariable analysis, CD3+ T cells and PDL1 remained as the independent prognostic factor, suggesting that immunity is remarkably important for the outcome of GC patients. This is the largest and well-characterized cohort of GC to date to address several important findings.
Previous studies have reported conflicting results as to whether PDL1 expression is a favorable or adverse prognostic factor in gastric cancer.Citation13–15,21 Some small groups of study, including Asian and Western populations, showed a negative impact on patient survival. However, our results exhibiting a positive correlation with PDL1 expression and better outcome, are in keeping with a recent study in a Caucasian cohort of GC.Citation14 These divergences have often been related to the composition of study population, design and methods. Especially the absence of standardization of IHC, from the antibodies, to the cut-offs points. We here used the VENTANA SP142 as the primary antibody of PDL1 which have been validated and widely used including some clinical trials.Citation22–24 Furthermore, our analysis on TCGA database indicated that the PDL1 mRNA high expressing patients also associated with better outcome, which is consistent with our results on protein level. In fact, recent studies using validated IHC assays also showed favorable prognostic values of PDL1 in other cancers, opposite to those with non-validated antibodies.Citation8,25 The Blueprint Working Group, a cross-industry initiative, has been established to provide an analytical comparison of several PDL1 IHC assays for different antibody and evaluation criteria.Citation5
Another interesting finding is that PDL1 expression were significantly associated with EBV infection, together with T cells density. In the TCGA study, 15% EBV+ GCs were found possessed genomic amplification of chromosomal region 9p24.1, the locus of genes encoding PD1, PDL1 and PDL2.Citation26 Besides, Bass et.al reported that PDL1 expression was not restricted to GCs with 9p24.1 Alification, suggesting that EBV+ cancer may possess alternative multiple mechanisms such as IFN-r released by tumor infiltrating T cells.Citation27 Previous studies also found that EBV infection was also closely associated with CD8+ T cell infiltration.Citation13,28,29 These results indicated that EBV may mark a group of GCs with greater possibility to benefit from anti-PD1/PDL1 therapy, molecular pathways that participate in this distinct type of cancer may provide novel clues for the predictive marker screening.
Such improved prognostic value seems paradoxical given the known immunosuppressive role of PDL1 suggestive of antitumor escape mechanism.Citation5,30 In general, there are at least 2 mechanisms of PDL1 regulation in tumor: intrinsic immune resistance and adaptive resistance, the former one occurs when PDL1 upregulation was responded to oncogenic signaling, while the adaptive one happens secondary to the cytokine such as IFN-r.Citation31,32 Numerous studies have reported that TILs reflecting host immunity have been shown to be associated with favorable outcome.Citation19,33–35 In particular, GC infiltration by CD8+ lymphocytes has been suggested to be endowed with high prognostic value.Citation36 There are also reports that claiming CD8+ T cell density could be the best predictive marker of PD1 inhibitor therapy.Citation37 We hypothesize that PDL1 expression is rather a marker of adaptive immune resistance in response to engaged CD3+ and CD8+ TILs, and represents a negative feedback mechanism rather than a constitutive biomarker. This is supported by previous studies that suggested PDL1 expression is reflective of an active immune environment.Citation8,38–40 Indeed, we observed a robust immune response in the “PDL1-up” group in the TCGA database analysis. While associated with PD1 and PDL2 mRNA expression, together with other immunosuppressive molecules, such as IDO and CTLA4, “PDL1-up” group was highly suggestive of an activated profile of differentiated T-cells, suggesting cytokines secreted by TILs may participate in the PDL1 regulation. Actually, in our series PDL1 expression in immune cells was paradoxically higher in the group of MMR proficient GCs which supposed to have lower immunogenicity. Similar results were also previously reported in gastric and colorectal cancer.Citation8,9,14 That may support PDL1 upregulation may be the feedback of active immune reaction rather than the results of neoantigen expression. However, tumor-immune system interaction is high dynamic. We observed that PDL1 expression together with its partner PDL2 and PD1 gradually decreased from the early stage to the advanced stage, though a slight elevation in the very early stage (N0 to N1). Similar phenomenon also found on TILs with higher expression in younger patients and with smaller tumor burden. That indicates the dynamic immunoregulation during GC progression. Checkpoint inhibition may be more effective in the circumstance that pre-existing immunity is suppressed by PD1/PDL1. Actually, in anti-PD-L1 clinical trial samples there are emerging data that PD-L1 expression plus a T cell activation gene signature, may be associated with response.Citation41,42 which explains that recent study including 33 GCs showed that a tumor immune microenvironment dominated by IFN-r and T cell receptor signaling take benefit from pembrolizumab.Citation43
This study indicates PDL1 expression, and TILs in particular, are biologically important in GC. It remains to be seen whether TILs or PDL1 are effective biomarkers of PD1 blockade therapy response in GC, but our data highlight the need to assess PDL1 expression in all tumor compartments and also TILs including the subtypes of lymphocyte, as well as evidence for T-cell activation, to further understand the complexity of immune-oncology.
Materials and methods
Study population
Patients underwent curative gastrectomy for adenocarcinoma of the stomach or esophageo-gastric junction at Peking University Cancer Hospital between June 2003 and December 2012 were retrieved. The criteria for inclusion in this study were: (1) FFPE tissues were available; (3) Histologic identification of the adenocarcinoma. (3) Patients received no preoperative chemotherapy or radiotherapy. This study was approved by the institutional review boards, and appropriate written informed consent was obtained from all patients. pTNM stage was determined according to the 7th edition of the UICC guidelines.Citation44 Follow-up data were retrieved from hospital records. This study was approved by the institutional review boards, and appropriate written informed consent was obtained from all patients.
Immunohistochemistry
Immunostaining was performed using anti-PD-L1 antibody (SP142, Roche), anti-PD-L2 antibody (NBP1–88964, Novus Biologicals), anti-PD1 antibody(315M-96, Cell marque), anti-CD3 antibody (NCL−CD3–565, Leica), anti-CD8 antibody (CRM311C, Biocare Medical), anti-MutL homolog 1 (MLH1) antibody (IR079, DAKO), anti-mutS homolog 2 (MSH2) antibody (IR085, DAKO), anti-mutS homolog 6 (MSH6) antibody(IR086, DAKO), anti-PCNA antibody(CBL407, Millipore), anti-C-met antibody (clone SP44, Roche) and anti-HER2 antibody (clone 4B5, Roche) as primary antibodies. Normal IgG were used as the negative control. Details of the protocols and scoring schema for the HER2, PCNA, hMLH1, MSH2, MSH6, PD1, PDL1, PDL2, CD3 and CD8 were presented in supplementary documents.
EBER ISH
All the tissue slides were performed ISH using the BIO-HRP REMBRANDT® EBER RISH kit (PanPath, Netherlands) according to the instruction. A known EBV-positive Burkitt's lymphoma was used as a positive control, and hybridization without a probe was performed as a negative control.
Evaluation of immunohistochemistry staining
HER2 immunoreactivity was scored following the HER2 scoring scheme (scores 0, 1+, 2+ and 3+) according to HER2 overexpression assessment for gastric cancer.Citation45 For the evaluation of the PCNA expression in tumor cells, these averaged values were stratified into 5 scoring groups:-, not detected; ±, <10% positive cells; +, 10–20% weakly to moderately positive cells; ++, 10–20% intensely positive cells or 20–50% weakly positive cells; and +++, 20–50% positive cells with moderate to marked reactivity or >50% positive cells. Immunoreactivity of hMLH1, MSH2 and MSH6 was evaluated as follows: -, <10% of the tumor or epithelial cells showed positive immunoreactivity; +, ≥ 10% of these cells showed positive immunoreactivity. GCs were stratified according to DNA mismatch repair (MMR) status as following. Briefly, MMR-proficient tumors were defined as those simultaneously expressing MLH1, MSH2 and MSH6, while MMR-deficient tumors were defined as those with loss of MLH1 and loss of both MSH6 and MSH2 expression.
PDL1, PDL2 and PD1
Intensity and percentage of stained cells was evaluated separately for tumor and immune cells by 3 pathologists. For the evaluation of PD-L1 expression, both the membranous and cytoplasmic staining was evaluated and the following immunoreactivity scoring system (IRS) was applied: Category A rated the percentage of immunoreactive cells and was graded as 0 (negative, or <1 positive), 1 (1–10% positive), 2 (10–50%) and 3 (> 50%). Category B documented the intensity of immunostaining as 0 (no immunostaining), 1 (weak), 2 (moderate), or 3 (strong). The addition of category A and B resulted in an IRS ranging from 0 to 614. PDL2 evaluation was identical with PDL1, only the localization is cytoplasm. The immunostaining of PD-1 in immune cells was rated as present or absent. The final interpretation was determined by consensus. There was a high level of consistency among the 3 pathologists, and in the few discrepant cases (< 5%) a consensus was reached after joint review.
CD3 and CD8
Stained slides were scanned at x20 magnification using an Aperio XT digital slide scanner and subjected to automated image analysis to detect and quantify immunoreactivity. An in-house developed software system, TMAi was used to discriminate brown (immunopositive) pixels, blue (immunonegative) pixels and white (empty space) pixels. All cores were reviewed after the image analysis process by a senior GI histopathologist to confirm that (a) the detection of the brown DAB staining had been performed accurately by the software and (b) to exclude all cores which contained tumor cells. The percentage immunoreactivity (Positive cells/ (positive cells +negative cells)*100) from all available cores per case was averaged and used as a surrogate for the extent of immune cell infiltration.
TCGA database analysis
Differentially expressed genes between PDL1-high and PDL1-nohigh GCs were identified using TCGA mRNA sequencing data of 415 cases of GCs (http://gdac.broadinstitute.org/).Citation26 The lower quantiles was used as cutoff to define the PDL1-no high group and the PDL1-high group. Gene expression analyses were done using the R package “limma,” with adjustment for false discovery rate. Top differentially expressed genes were used for hierarchical clustering using heatmap.2 function available in the “gplots” R package (version 3.1.1). Gene Ontological analysis of the resulting gene list was based on GO biologic processes of the Database for Annotation, Vizualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/).
Statistical analyses
Correlations analysis was assessed by Fisher's exact or Cochran-Mantel-Haensel χ2 test. Overall Survival was calculated using the Kaplan-Meier method, with Log rank test to determine significance of differences. Hazard ratios of variables were calculated by univariale Cox regression model and those having p-values up to 0.05 were included in a multivariable Cox regression, combined with iterative backward LR method to identify independent prognostic variables. All statistical tests were 2-sided at the 5% level of significance. False discovery rate was controlled by applying the explorative Simes (Benjamini-Hochberg) procedure group-wise for each biomarker.Citation46 We wrote the article in accordance with the criteria specified in the reporting recommendations for tumor marker prognostic studies (REMARK).Citation47 Statistical analyses were performed with SPSS 21.0 (IBM Corporation).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KONI_A_1356144_supplementary_materials.zip
Download Zip (741.5 KB)Acknowledgments
The authors thank all patients who participated in this study.
Additional information
Funding
References
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. doi:10.3322/caac.21262. PMID:25651787
- Drake CG, Lipson EJ, Brahmer JR. Breathing new life into immunotherapy: review of melanoma, lung and kidney cancer. Nat Rev Clin Oncol. 2014;11:24-37. doi:10.1038/nrclinonc.2013.208. PMID:24247168
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214-8. doi:10.1038/nature12213. PMID:23770567
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr., Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546-58. doi:10.1126/science.1235122. PMID:23539594
- Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275-87. doi:10.1038/nrc.2016.36. PMID:27079802
- Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568-71. doi:10.1038/nature13954. PMID:25428505
- Latchman Y, Wood CR, Chernova T, Chaudhary D, Borde M, Chernova I, Iwai Y, Long AJ, Brown JA, Nunes R, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2:261-8. doi:10.1038/85330. PMID:11224527
- Droeser RA, Hirt C, Viehl CT, Frey DM, Nebiker C, Huber X, Zlobec I, Eppenberger-Castori S, Tzankov A, Rosso R, et al. Clinical impact of programmed cell death ligand 1 expression in colorectal cancer. Eur J Cancer. 2013;49:2233-42. doi:10.1016/j.ejca.2013.02.015. PMID:23478000
- Masugi Y, Nishihara R, Yang J, Mima K, da Silva A, Shi Y, Inamura K, Cao Y, Song M, Nowak JA, et al. Tumour CD274 (PD-L1) expression and T cells in colorectal cancer. Gut. 2016;66:1463-73. doi:10.1136/gutjnl-2016-311421. PMID:27196573
- Sabatier R, Finetti P, Mamessier E, Adelaide J, Chaffanet M, Ali HR, Viens P, Caldas C, Birnbaum D, Bertucci F. Prognostic and predictive value of PDL1 expression in breast cancer. Oncotarget. 2015;6:5449-64. doi:10.18632/oncotarget.3216. PMID:25669979
- Ohigashi Y, Sho M, Yamada Y, Tsurui Y, Hamada K, Ikeda N, Mizuno T, Yoriki R, Kashizuka H, Yane K, et al. Clinical significance of programmed death-1 ligand-1 and programmed death-1 ligand-2 expression in human esophageal cancer. Clin Cancer Res. 2005;11:2947-53. doi:10.1158/1078-0432.CCR-04-1469. PMID:15837746
- Lee HE, Chae SW, Lee YJ, Kim MA, Lee HS, Lee BL, Kim WH. Prognostic implications of type and density of tumour-infiltrating lymphocytes in gastric cancer. Br J Cancer. 2008;99:1704-11. doi:10.1038/sj.bjc.6604738. PMID:18941457
- Thompson ED, Zahurak M, Murphy A, Cornish T, Cuka N, Abdelfatah E, Yang S, Duncan M, Ahuja N, Taube JM, et al. Patterns of PD-L1 expression and CD8 T cell infiltration in gastric adenocarcinomas and associated immune stroma. Gut. 2017;66(5):794-801. doi:10.1136/gutjnl-2015-310839. PMID:26801886
- Boger C, Behrens HM, Mathiak M, Kruger S, Kalthoff H, Rocken C. PD-L1 is an independent prognostic predictor in gastric cancer of Western patients. Oncotarget. 2016;7:24269-83. doi:10.18632/oncotarget.8169. PMID:27009855
- Eto S, Yoshikawa K, Nishi M, Higashijima J, Tokunaga T, Nakao T, Kashihara H, Takasu C, Iwata T, Shimada M. Programmed cell death protein 1 expression is an independent prognostic factor in gastric cancer after curative resection. Gastric Cancer. 2016;19:466-71. doi:10.1007/s10120-015-0519-7. PMID:26210691
- Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298-306. doi:10.1038/nrc3245. PMID:22419253
- Teng MW, Ngiow SF, Ribas A, Smyth MJ. Classifying cancers based on T-cell infiltration and PD-L1. Cancer Res. 2015;75:2139-45. doi:10.1158/0008-5472.CAN-15-0255. PMID:25977340
- Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960-4. doi:10.1126/science.1129139. PMID:17008531
- Ogino S, Nosho K, Irahara N, Meyerhardt JA, Baba Y, Shima K, Glickman JN, Ferrone CR, Mino-Kenudson M, Tanaka N, et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15:6412-20. doi:10.1158/1078-0432.CCR-09-1438. PMID:19825961
- de Guillebon E, Roussille P, Frouin E, Tougeron D. Anti program death-1/anti program death-ligand 1 in digestive cancers. World J Gastrointest Oncol. 2015;7:95-101. doi:10.4251/wjgo.v7.i8.95. PMID:26306141
- Zhang L, Qiu M, Jin Y, Ji J, Li B, Wang X, Yan S, Xu R, Yang D. Programmed cell death ligand 1 (PD-L1) expression on gastric cancer and its relationship with clinicopathologic factors. Int J Clin Exp Pathol. 2015;8:11084-91. doi:10.1186/s12967-016-0925-6. PMID:26617827
- Gaule P, Smithy JW, Toki M, Rehman J, Patell-Socha F, Cougot D, Collin P, Morrill P, Neumeister V, Rimm DL. A quantitative comparison of antibodies to programmed cell death 1 ligand 1. JAMA Oncol. 2017;3(2):256-259. doi:10.1001/jamaoncol.2016.3015. PMID:27541827
- Schats KA, Van Vre EA, De Schepper S, Boeckx C, Schrijvers DM, Waelput W, Fransen E, Vanden Bempt I, Neyns B, De Meester I, et al. Validated PD-L1 immunohistochemistry assays (E1L3N & SP142) reveal similar immune cell staining patterns in melanoma when using the same sensitive detection system. Histopathology. 2017 Jan;70(2):253-263. doi:10.1111/his.13056. PMID:27496355
- Sun WY, Lee YK, Koo JS. Expression of PD-L1 in triple-negative breast cancer based on different immunohistochemical antibodies. J Transl Med. 2016;14:173. doi:10.1186/12967-016-0925-6. PMID:27286842
- McDermott DF, Sosman JA, Sznol M, Massard C, Gordon MS, Hamid O, Powderly JD, Infante JR, Fassò M, Wang YV, et al. Atezolizumab, an anti-programmed death-ligand 1 antibody, in metastatic renal cell carcinoma: long-term safety, clinical activity, and immune correlates from a phase ia study. J Clin Oncol. 2016;34:833-42. doi:10.1200/JCO.2015.63.7421. PMID:26755520
- Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-9. doi:10.1038/nature13480. PMID:25079317
- Derks S, Liao X, Chiaravalli AM, Xu X, Camargo MC, Solcia E, Sessa F, Fleitas T, Freeman GJ, Rodig SJ, et al. Abundant PD-L1 expression in Epstein-Barr Virus-infected gastric cancers. Oncotarget. 2016;7:32925-32. doi:10.18632/oncotarget.9076. PMID:27147580
- Koji H, Yazawa T, Nakabayashi K, Fujioka Y, Kamma H, Yamada A. CD8-positive T-cell lymphoproliferative disorder associated with Epstein-Barr virus-infected B-cells in a rheumatoid arthritis patient under methotrexate treatment. Mod Rheumatol. 2016;26:271-5. doi:10.3109/14397595.2013.850613. PMID:24386983
- Hara S, Hoshino Y, Naitou T, Nagano K, Iwai M, Suzuki K, Yamamoto K, Nagasaka T, Morishima T, Kimura H. Association of virus infected-T cell in severe hepatitis caused by primary Epstein-Barr virus infection. J Clin Virol. 2006;35:250-6. doi:10.1016/j.jcv.2005.07.009. PMID:16181807
- Postow MA, Callahan MK, Wolchok JD. Immune checkpoint blockade in cancer therapy. J Clin Oncol. 2015;33:1974-82. doi:10.1200/JCO.2014.59.4358. PMID:25605845
- Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, Gajewski TF. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. 2013;5:200ra116. doi:10.1126/scitranslmed.3006504. PMID:23986400
- Taube JM, Young GD, McMiller TL, Chen S, Salas JT, Pritchard TS, Xu H, Meeker AK, Fan J, Cheadle C, et al. Differential expression of immune-regulatory genes associated with PD-L1 display in melanoma: implications for PD-1 pathway blockade. Clin Cancer Res. 2015;21:3969-76. doi:10.1158/1078-0432.CCR-15-0244. PMID:25944800
- Savas P, Salgado R, Denkert C, Sotiriou C, Darcy PK, Smyth MJ, Loi S. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat Rev Clin Oncol. 2016;13:228-41. doi:10.1038/nrclinonc.2015.215. PMID:26667975
- Schalper KA, Brown J, Carvajal-Hausdorf D, McLaughlin J, Velcheti V, Syrigos KN, Herbst RS, Rimm DL. Objective measurement and clinical significance of TILs in non-small cell lung cancer. J Natl Cancer Inst. 2015;3:107-9. doi:10.1093/jnci/dju435. PMID:25650315
- Eerola AK, Soini Y, Paakko P. A high number of tumor-infiltrating lymphocytes are associated with a small tumor size, low tumor stage, and a favorable prognosis in operated small cell lung carcinoma. Clin Cancer Res. 2000;6:1875-81. PMID:10815910
- Feichtenbeiner A, Haas M, Buttner M, Grabenbauer GG, Fietkau R, Distel LV. Critical role of spatial interaction between CD8(+) and Foxp3(+) cells in human gastric cancer: the distance matters. Cancer Immunol Immunother. 2014;63:111-9. doi:10.1007/s00262-013-1491-x. PMID:24170095
- Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, Chen L, Pardoll DM, Topalian SL, Anders RA, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20:5064-74. doi:10.1158/1078-0432.CCR-13-3271. PMID:24714771
- Velcheti V, Schalper KA, Carvajal DE, Anagnostou VK, Syrigos KN, Sznol M, Herbst RS, Gettinger SN, Chen L, Rimm DL. Programmed death ligand-1 expression in non-small cell lung cancer. Lab Invest. 2014;94:107-16. doi:10.1038/labinvest.2013.130. PMID:24217091
- Bertucci F, Finetti P, Colpaert C, Mamessier E, Parizel M, Dirix L, Viens P, Birnbaum D, van Laere S. PDL1 expression in inflammatory breast cancer is frequent and predicts for the pathological response to chemotherapy. Oncotarget. 2015;6:13506-19. doi:10.18632/oncotarget.3642. PMID:25940795
- Guo L, Li W, Zhu X, Ling Y, Qiu T, Dong L, Fang Y, Yang H, Ying J. PD-L1 expression and CD274 gene alteration in triple-negative breast cancer: implication for prognostic biomarker. Springerplus. 2016;5:805. doi:10.1186/s40064-016-2513-x. PMID:27390646
- Ribas A, Robert C, Hodi SF, Wolchok JD, Joshua AM, Hwu W, Weber JS, Zarour HM, Kefford R, Loboda A, Albright A, et al. Association of response to programmed death receptor 1 (PD-1) blockade with pembrolizumab (MK-3475) with an interferoninflammatory immune gene signature. J Clin Oncol. 2015;33, no. 15_suppl.3001. doi:10.1200/jco.2015.33.15_suppl.3001. PMID:25667273
- Seiwert TY, Burtness B, Weiss J, Eder JP, Yearley J, Murphy E, Nebozhyn M, McClanahan T, Ayers M, Lunceford JK, et al. Inflamed-phenotype gene expression signatures to predict benefit from the antiPD-1 antibody pembrolizumab in PD-L1+ head and neck cancer patients. J Clin Oncol. 2015;33, no. 15_suppl.6017. doi:10.1200/jco.2015.33.15_suppl.6017
- Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Albright A, Cheng J, Kang P, Ebbinghaus S, Yearley J, et al. Relationship between immune gene signatures and clinical response to PD-1 blockade with pembrolizumab (MK-3475) in patients with advanced solid tumors. J Immunother Cancer. 2015;3:80. doi:10.1186/2051-1426-3-S2-P80. PMCID:PMC4645163
- Washington K. 7th edition of the AJCC cancer staging manual: stomach. Ann Surg Oncol. 2010;17:3077-9. doi:10.1245/s10434-010-1362-z. PMID:20882416
- Ruschoff J, Dietel M, Baretton G, Arbogast S, Walch A, Monges G, Chenard MP, Penault-Llorca F, Nagelmeier I, Schlake W, et al. HER2 diagnostics in gastric cancer-guideline validation and development of standardized immunohistochemical testing. Virchows Arch. 2010;457:299-307. doi:10.1007/s00428-010-0952-2. PMID:20665045
- Benjamini Y, Cohen R. Weighted false discovery rate controlling procedures for clinical trials. Biostatistics. 2017 Jan;18(1):91-104. doi:10.1093/biostatistics/kxw030. PMID:27445132
- McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, Statistics Subcommittee of NCI-EORTC Working Group on Cancer Diagnostics. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst. 2005;97:1180-4. doi:10.1093/jnci/dji237. PMID:16106022
