ABSTRACT
Microorganisms mainly exist in the form of biofilm in nature. Biofilm can contaminate food and drinking water system, as well as cause chronic wound infections, thereby posing a potential threat to public health safety. In the last two decades, researchers have made efforts to investigate the genetic contributors control different stages of biofilm development (adherence, initiation, maturation, and dispersal). As an opportunistic pathogen, C. albicans causes severe superficial or systemic infections with high morbidity and mortality under conditions of immune dysfunction. It has been reported that 80% of C. albicans infections are directly or indirectly associated with biofilm formation on host or abiotic surfaces including indwelling medical devices, resulting in high morbidity and mortality. Significantly, the outcome of C. albicans biofilm development includes enhanced invasion, exacerbated inflammatory responses and intrinsic resistance to antimicrobial chemotherapy. Thus, this review aimed at providing a comprehensive overview of the regulatory network controls microbial biofilm development, with C. albicans as a representative, served as reference for therapeutic targets.
1. Microbial biofilms in various applications
Microbial biofilm was firstly discovered as a stack of small moving living things from plaque sample by microscopy in the 17th century [Citation1]. Biofilm is defined as a complex net structural mixture that composed of microorganisms and their autocrine extracellular matrix after adhering to contact surface [Citation2–4]. The majority of microbial cells exist as biofilm in nature to resist adverse living conditions including nutritional deficiency, extreme temperature, acid and alkali, ultraviolet rays, disinfectants, and antibiotics [Citation1,Citation5]. A variety of research on microbial biofilm have been conducted mainly to prevent its harmfulness to human or to take advantage of its beneficial aspects, extensively involved in clinical, food, environment, drinking water system and many other fields. Microbial biofilms have potential threat to public health. It causes chronic wound infections resulting in recurrences of inflammation [Citation6]. Medical device implants including artificial heart valves and joint prostheses are prone to be contaminated by microbial biofilms, imposing burden on the patients and medical expenses for treatment [Citation7,Citation8]. It has been estimated that biofilm is related to 65%-80% of microbial infections [Citation9,Citation10]. Microbial biofilm can also corrode food processing equipment and contaminate food, increasing the difficulty of equipment cleaning and the risk of foodborne illness [Citation11–15]. In drinking water systems, microorganisms form biofilm after attaching to distribution pipes. Under shear force of the water flow, biofilms diffuse into water and enter thousands of households through the tap water distribution system, further affect the health of residents [Citation16–18]. In the United States, water-borne infections alone cause more than 40,000 hospitalizations every year [Citation19] Due to the adverse aspect of microbial biofilm, researchers have conducted various studies on their characteristics and prevention methods. Meanwhile, some scholars have also studied how to develop and take advantage of microbial biofilms. Due to the strong viability and high density, biofilm cells can well accumulate the secreted enzymes and fermentation products such as acid and alcohol, thus be used to continuously and efficiently produce biochemicals [Citation20]. Besides, factories also utilize biofilm reactors to treat sewage. The mixed biofilm composed of various aerobic bacteria, anaerobic bacteria and fungi with a continuous adsorption and decomposition function on organic matter, nitrogen and phosphorus in sewage can be used to purify sewage [Citation21].
2. Structure and heterogeneity of microbial biofilms
The distribution of components, subgroups and species in biofilms is heterogeneous, leading to different metabolic and vitality characteristics from planktonic cell. The composition and structure of the microbial biofilm are changeable in different stages, mainly including various biofilm cells and extracellular components. With the secretion of the extracellular substances by attached cells and the development of the biofilms, the proportion of the extracellular components can increase to 80%-85% [Citation22]. For maturation biofilm, the water content could even reach 97%, and the water channels throughout the extracellular matrix are conducive to material exchange for cells [Citation23]. The basic structural unit in biofilm is micro-community encapsulated by extracellular matrix. The density of extracellular components close to the cells are intensive [Citation24,Citation25]. In addition to the extracellular adhesion components actively secreted by cells, the matrix components also include cell metabolism and lysis products, absorbed nutrients and their further decomposition products by enzymes [Citation26,Citation27]. The main skeletal structure connecting cells is polysaccharides, which are the abundant components of extracellular matrix except for water. Secondly, polysaccharides and lipids also play a role in enhancing the stability of the skeletal structure. Studies have shown that extracellular adhesive proteins can be combined with polysaccharides and are an important component of the skeleton [Citation28]. Lipids and polysaccharides interaction form lipopolysaccharides which have the properties of surfactants and contribute to structural stability [Citation29]. Apart from polysaccharides, both ends of eDNA chain link to different cell surfaces, thereby increasing intercellular adhesion [Citation30]. In addition, small molecules may participate in partial network cross-linking. The various components in the extracellular matrix and their interactions determine the internal chemical environment, physical structure, and strength of the biofilms, and provide various residents with completely different living conditions from planktonic cells.
However, the microenvironment of each single cell in maturated biofilm is different. The heterogeneity is formed with the development of biofilm which is related to the external conditions, the species and metabolism activity of microorganisms, as well as the shape and structure of biofilm. The initially adhered cells can be fully in contact with external nutrients and oxygen. The environment for each single cell is similar at this stage. With the increase of the thickness and width of biofilm and the penetration or dissolution of nutrients and oxygen, the gradient of oxygen and nutrients in biofilm gradually increase. The maturated biofilm has a highly hydrated environment, and the diffusion rate of oxygen is approximately 60% of that in water [Citation31]. In addition, the cells at outer layer will preferentially acquire and consume nutrients and oxygen resulting in a lack of oxygen and nutrients in the inner layer. In addition, the difference in the metabolic activity of microorganisms causes the heterogeneity in biofilms. For example, microorganisms produce the acetic acid by anaerobic respiration altering partial pH. At present, relatively few studies have been reported on the structure and heterogeneity of anaerobic bacteria biofilms, but it is necessary as they cause infections including cholecystitis, otitis media, and endocarditis. We can infer the heterogeneity of the anaerobic bacteria biofilm based on the formation mechanism of the heterogeneity. Under anaerobic conditions in vivo, there is a general lack of oxygen at different depths of the biofilm. Anaerobic bacteria prevalent undergo anaerobic fermentation, so the heterogeneity of the biofilm is relatively weak. The actual situation under this model needs further verification.
The heterogeneity of the biofilm environment promotes the differentiation of metabolic subpopulations to improve their adaptability. In general, the surface cells at outer layer and the planktonic cells that have just adhered to the matrix can obtain sufficient nutrients and oxygen, thus have stronger metabolic activity, and larger size. Even if the cells at inner layer obtain a certain amount of nutrients and oxygen through the water channels in the biofilm, the preferential acquisition of metabolic substrates by the cells at outer layer makes the inner cells undergo relatively slow metabolic activity, and the cells closer to the center even stop growing or die [Citation32].
Cells with different metabolic activities in the community cooperate to improve survival ability. A typical way of cooperation is cross-feeding. For example, E. coli may not completely convert the substrate into carbon dioxide in low-oxygen environment, but produce an intermediate metabolite acetate, which can be completely oxidized to carbon dioxide by the cells at outer layer. The cross-feeding reduce accumulation of intermediate metabolites and maintain osmotic pressure stability in biofilm [Citation33]. For P. aeruginosa biofilm, cells ferment in hypoxic zone and produce acetic acid used as carbon source by cells at aerobic outer layer, thereby contributing to maintain acid-base balance in biofilm [Citation34,Citation35]. In addition, if biofilm adheres to the nutrient substrate and the substrate at the bottom is the only source of nutrients, cross-feeding provide metabolic substrate for the cells at upper aerobic layer with no nutrition resource.
3. Representative biofilm former-Candida albicans
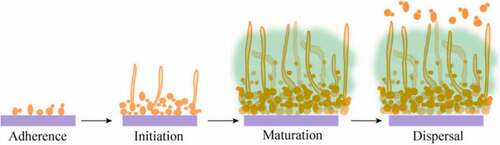
Aside from common composition and structure, biofilms formed by various species differ. Here we selected Candida albicans as a representative biofilm former to further discuss the regulatory network of biofilm formation during four stages. C. albicans is a commensal fungus that is frequently a benign member of the skin and mucosal flora. The growth of C. albicans is limited due to immunity and the presence of other commensal microorganisms [Citation36]. However, it would be regarded as pathogenic causing superficial and systemic infection in immunocompromised individuals. In recent years, fungal infections have become more prevalent due to the increase in the number of transplant devices and the increase in the abuse of antifungal drug and immunosuppressive patients. Candidiasis is usually associated with indwelling medical devices (such as catheter, heart valves, vascular bypass grafts, dental implants, etc.), which is difficult to cure and create huge challenge in medicine. Vulvovaginal candidiasis is common and may affect up to 75% of women at least once in a lifetime. A small percentage of women (5–10%) have experienced long-term recurrent episodes that have seriously affected their quality of life. In addition, acute pseudomembranous infection of the mouth or vagina is some of the most common diseases. Under the current antifungal treatment conditions, mortality in patients with candidiasis is increasing and as high as 40%, especially in those with bloodstream infections [Citation37,Citation38]. Fungal infections are usually caused by Candida, one of the most common fungi associated with the disease [Citation39]. In fact, C. albicans may cause many diseases mainly because it with some virulence factors including biofilm formation, phenotyping changes and production some harmful substance to cells, such as hemolysins, phospholipases and proteases. The secretion of aspartic protease from C. albicans is one of the main factors determining its toxicity by promoting invasion and counteracting the role of host defense systems in pathogenicity. C. albicans can grow in yeast and hyphae forms (pseudohyphal and hyphae) [Citation40,Citation41]. C. albicans in hyphae form has strong adhesion and invasive ability to the host. Generally, the yeast form disseminates into the bloodstream, spreading the organism to different host niches, and after that the hyphal form is contributed to penetrate epithelial and endothelial cells, then colonize [Citation42]. The transition from yeast form to hyphal form is the infection process of C. albicans. The more mycelial infection, the higher the infection rate. The transformation of C. albicans phenotype plays an important role in the pathogenesis of C. albicans and biofilm formation [Citation43–45]. Eighty percent of the infections caused by C. albicans are directly or indirectly related to the formation of biofilm. Compared with planktonic state, C. albicans biofilm is the more common form of C. albicans growth in nature [Citation46]. Like other microorganisms, the formation of C. albicans biofilm consists of four steps: adherence, initiation, maturation and dispersal (). The biofilm of C. albicans is an aggregated population including a bacterial cell population and an extracellular polymer produced by C. albicans in contact with a surface of biotic or abiotic material. It is surrounded by extracellular polysaccharides, proteins, lipids and DNA [Citation8] Biofilms are a population of organic cells regulated by signaling molecules, not just random aggregation of many individual cells [Citation47].
Figure 1. Four steps of C. albicans biofilm development. In the adherence step, yeast-form cells adhere to the substrate. In the initiation step, cells form microcolonies and hyphae begin to form. In the maturation step, the biofilm biomass expands. In the dispersal step, yeast-form cells are released to surrounding environment.
According to the National Institutes of Health statistics, most human infections are caused by biofilms [Citation48]. The formation of biofilm could protect the pathogen from host defenses and plays a key role in C. albicans infection [Citation49,Citation50]. C. albicans biofilms are surface-associated structured communities of yeast, hyphae and pseudohyphal cells surrounded by extracellular matrices. C. albicans biofilms often cause life-threatening systemic infections and are particularly difficult to eradicate due to their high resistance to antifungal drug. Biofilm of C. albicans is a form of survival that adapts to environmental changes. It is resistant to antifungal drugs and can evade immune host responses. Once the biofilm is formed, drug resistance will rise sharply. The minimum inhibitory concentration (MIC) of C. albicans is increased by 30 to 20,000 times compared to planktonic cells. In addition, the biofilm at the dispersal stage continuously releases the fungal cells into environment, and the fungal cells migrate to other parts of the body to cause repeated infection. This greatly increases the difficulty of completely curing fungal infections [Citation51,Citation52]. Even worse, because C. albicans biofilm is strongly embedded to the indwelling medical devices, the removal device is necessary to treat C. albicans infection during treatment [Citation53].
It is necessary to understand the molecular mechanism of biofilm formation in order to reduce and prevent infections caused by C. albicans. C. albicans biofilms were the first described on medical devices, however, research had been impeded because the lack of genetic and molecular tools. Most studies on fungal biofilms are in vitro. In recent years, some scholars have used animal models such as intubation-related infections and oral infections to conduct in vivo studies of biofilms. During biofilm formation, genes play an important role in biofilm formation in C. albicans and they participate in every process from adhesion to dispersion. The research of molecular biology of C. albicans biofilm can make us have a deeper understanding of the biofilm formation mechanism and its resistance mechanism. In addition, it can provide new ideas and references for the study of deep fungal infections. In this review, we mainly focus on biofilm formation and regulatory mechanisms of C. albicans.
4. Biofilm adherence
4.1 General adherence in microbial biofilms
Adhesion is the first and most important step in the formation of biofilms. The adhesion phase includes a reversible adhesion phase and an irreversible adhesion phase. Adhesion often leads to the formation of biofilms [Citation39]. After approaching a relatively stable settlement, microorganisms reversibly adhere to the contact surface by nonspecific interaction including hydrophobic bond and ionic bond [Citation54,Citation55]. The interaction between microorganisms and surface is affected by chemical characteristics of both sides and related to the shape and roughness of contact surface. The mechanism of how these attached cells aggregate into community is not clear. These settled microbial cells can reproduce and form micro-communities with suitable nutritional conditions. Studies have shown that microbial cells can regulate aggregation through quorum sensing (QS) [Citation56]. QS is a way to transfer information between cells. As the cell density increases, signal molecules reach a certain threshold to promote intraspecies or interspecies communication. These signal molecules affect the expression of specific genes through a series of transmission processes, thereby regulating the microbial community behavior. At present, QS signal molecules including self-induction peptide (AIP) of Gram-positive bacteria, N-acetylus deridate (AHL) of Gram-negative bacteria, and AI-2 which generally exist in microbial cells are well documented [Citation57]. QS signal molecules may promote the aggregation of microbial cells on the surface by affecting twitching of flagella or other movement organs [Citation58,Citation59]. Regarding how QS signal affects the direction of movement has not been studied clearly. In addition, microbial cells can secrete adhesin to make the interaction with surface more stable [Citation60]. This effect is more obvious in vivo as microbial cells may secrete adhesin to specifically bind to host surface proteins [Citation61,Citation62]. For example, the fibrinogen-binding proteins AB (FnbAB) of Staphylococcus aureus can combine with the fibronectin of tissue cells [Citation63,Citation64] The adhered single cells eventually form a monolayer cell population through replication and mutual attraction. Microbial cells can fully contact nutrients and oxygen similar to planktonic cells, with relatively strong proliferation and metabolic activities.
4.2 Biofilm adherence in C. albicans
C. albicans typically adheres to the surface of the host and/or medical device and causes infection. The ability to form biofilms is related to microbial species, host, and surface properties [Citation65,Citation66]. In general, the rougher the surface of the adhesive material, the easier it is to form a biofilm. In addition, the chemical composition, critical tension, surface energy, hydrophilic/hydrophobic, surface charge and other factors on the surface of the adhesive material have a great influence on microorganism adhesion. In the process of forming biofilms by C. albicans, they generally aggregate and adhere to each other on a biological or abiotic surface in the form of yeast. In this process, adhesion proteins, adhesins, are key inducers of cell adhesion and colonization, and they play an important role in the formation of biofilms [Citation67]. Adhesion is the interaction between pathogen cells and the host surface. Thus, the cell wall structure of the pathogen, host cell surface receptors and the external environment can regulate the adhesion process [Citation68].
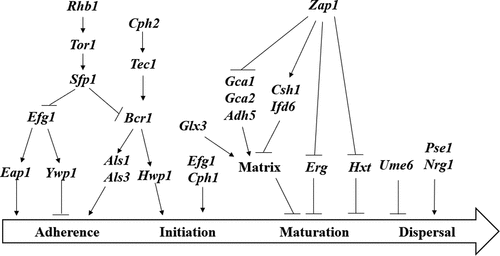
As a class of cell wall glycoproteins, Als protein family is one of the most studied protein molecules in C. albicans that regulate cell adhesion and biofilm formation. It consists of eight proteins with the same N-terminal secretory signal sequence and C-terminal glycosylphosphatidylinositol, namely Als1-7 and Als9 [Citation69]. In the Als protein family, Als1, Als3 and Als5 play an important role in the adhesion of C. albicans to host cells. Among them, Als1 and Als3 proteins mainly adhere to host endothelial cells and epithelial cells, while Als5 proteins are mainly bound to host extracellular matrix proteins. ALS1 is the first identified gene in C. albicans gene family. Als1 has a highly similar amino acid sequence to saccharomyces cerevisiae cell surface adhesion protein alpha lectin, and it is speculated that Als1 is an important cell adhesion protein in C. albicans [Citation70]. Als3 protein plays a key role in the biofilm formation of C. albicans. When the gene Als3 was knocked out, the mutant Als3/Als3 could not form a normal biofilm on biomaterials in vitro [Citation70]. In addition, the protein expression levels of Als1 and Als3 are related to cell morphology. Als1 can be expressed in yeast and mycelial cells, while Als3 is a mycelial cell-specific protein [Citation71].
Tec1p is required for hyphal formation. Biofilm produced by the Tec1 null mutant (Δ/Δ Tec1) strain was rudimentary, less than 20 µm deep, and composed exclusively of yeast cells, while its parental strain formed a biofilm 250–450 µm deep that included many hyphal filaments [Citation72]. In addition, the zinc finger protein Bcr1 plays a necessary role in formation of biofilms by C. albicans. Als3 is a key target for Bcr1 action [Citation73]. The ability of the Als3/Als3 mutant to form biofilms is diminished. Moreover, overexpression of ALS3 can compensate for defects in the formation of biofilm by the bcr1/bcr1 mutant. Overexpression of Als1 or Hwp1 in the Bcr1/Bcr1 mutant contributes to the formation of biofilms [Citation74]. RTA3 with a 7-transmembrane domain topology is similar to the G-protein-coupled receptor and is a fungal community. Rta3 regulates the expression of Bcr1 target genes involved in cell surface properties, adhesion and hyphae growth. They found that the Rta3/Rta3 mutant is a biofilm defect in a rat model of venous catheter infection, and that Bcr1 overexpression rescues this defect, suggesting that Bcr1 acts downstream of Rta3 to mediate biofilm formation in C. albicans [Citation75].
The Efg1 gene is an important regulatory factor with some function during the growth of C. albicans, such as involved in the colonization process by regulate the expression of Eap1 and Ywp1 genes. Its expression would be modified by the immune system of the host. In addition, it confers to C. albicans the capacity to transition from commensal microorganism to opportunistic pathogen status [Citation76]. Mutant defective in the enhanced filamentous growth transcriptional factor (Efg1), a major activator of hyphal development, presented impaired formation of a monolayer of cells on polystyrene surfaces. This defect in biofilm development may occur because of altered surface-protein composition and adherence properties of the Efg1 null mutant (Δ/Δ efg1) [Citation73]. In addition, the lack of functioning Efg1 in C. albicans strains yielded only pseudohyphae on solid media and without growth in liquid media [Citation77]. The Efg1 gene, an important regulatory factor of transcription in biofilm, is involved in the colonization process by C. albicans, modifying its expression according to the immune system of the host and conferring to C. albicans the capacity to transition from commensal microorganism to opportunistic pathogen status. Ywp1p is also a GPI-CWP protein, which is mainly expressed on the surface of yeast form cells at the end of logarithmic growth phase and is not found in hyphae and pseudomycelium. Ywp1 wild type strains can only form a monolayer of cell adhesion, while Ywp1 deletion mutants are more likely to adhere to the surface of polystyrene or other substrates and form biofilm. Therefore, it can be considered that Ywplp is a negative regulatory protein that exerts an inhibitory effect on biofilm formation.
C. albicans Sfp1 functions as an activator to regulate ribosomal gene expression. The deletion of the Sfp1 gene enhanced cell adhesion and biofilm formation in comparison with the wild-type strain. Interestingly, the Sfp1 deleted mutant also exhibited an increase in the expression of the Als1, Als3 and Hwp1 genes, which encode adhesin proteins. The deletion of Bcr1 or Efg1 in the Δ/ΔSfp1 background significantly reduced the expression levels of Als1, Als3 and Hwp1 genes. Sfp1 may regulate adhesin genes and biofilm formation through Bcr1 and Efg1. Reduced expression of adhesin genes was also detected after Sfp1 overexpression in the Δ/ΔSfp1 and Δ/ΔRhb1 strains. Sfp1 appears to function downstream of the Rhb1-Tor1 signaling pathway [Citation78].
Sphingolipids are a class of sphingosine backbones or long chain bases (LCB) containing lipids [Citation79]. They are widely present in eukaryotic cell membranes [Citation79,Citation80]. When sphingolipids coexist with sterols and glycerophospholipids, they confer plasma membrane integrity and plasticity [Citation81]. CaFEN1 and CaFEN12 have important regulatory effects on cell wall integrity, hyphae and biofilm formation because they are involved in sphingolipid biosynthesis. Deletion of these two genes results in strains that are highly sensitive to amphotericin B, which most likely due to their weak cell walls and the inability to form biofilms.
MP65 gene encodes β-glucanase mannoprotein and plays an important role in the host-fungus relationship, morphogenesis and pathogenicity. The mp65/mp65 mutant is sensitive to a variety of cell degrading agents and wall-perturbing. Mp65 promotes the formation of biofilm by inducing the formation of hyphae. The mp65/mp65 mutant showed an activation of two MAPKs (Mkc1p and Cek1p). The mitogen-activated protein kinase (MAPK) signaling pathway is the first one associated with the morphogenesis of C. albicans and is highly conserved in the evolution of different species [Citation82]. Mkc1 is an MAPK protein kinase, which is a part of the intact cell wall structure of C. albicans and is involved in the regulation of cell surface contact [Citation83]. When Mkc1 is knockout, the mkc1/mkc1 mutant forms lots of biofilms with hyphal defects [Citation84]. Cek1 is an extracellular signal-regulated kinase that is also an important member of the MAPK family and plays a role in cell surface contact reactions [Citation85]. Like MKC1, CEK1 is also an important regulator of the formation of normal biofilms in C. albicans, but the specific regulatory mechanisms of Mkc1 and Cek1 for biofilm formation are not clearly [Citation86]. Silvia Sandini et al found that the Mp65/Mp65 mutant compared to the wild type demonstrated a marked reduction in adhesion to biliary epithelial cells (BEC) and Caco-2 cells and severe defects in biofilm formation [Citation87] ().
5. Biofilm initiation
In the initiation stage, the cells propagate to form microcolonies, and germ tubes form to yield hyphae. The extension of hyphae causes the microcolony to dissolve with each other. Cells secrete extracellular polymers, bind to individual fungal cells and form fungal masses. Biofilm composed mainly of extracellular matrix begins to be generated and surrounds the microcolony. At the same time, the extracellular matrix is produced. The growth of mycelium causes small colonies to fuse to form a monolayer matrix. This matrix is mainly composed of polysaccharide.
Hwp1 is a GPI (glycosyl phosphatidyl inositol)-anchored cell wall proteins. Hwp1 is a mycelial-specific cell wall protein that plays a role in regulating the formation of cell buds and hyphae in biofilms. In addition, Hwp1 promotes the contact and adhesion of C. albicans to host cells. Hwp1 is an important cell surface protein required for the formation of biofilms by C. albicans in vivo [Citation88].
The three-dimensional network of extracellular matrices (ECM) formed outside the biofilm prevents or delays the penetration of antibiotics. In addition, the target of antifungal drugs such as echinococcal antifungal and pneumocandin is β-1,3-glucan. In biofilms, the content of β-1.3-glucan is much higher than that of planktonic. In addition, the extracellular matrix plays an important role in protecting the cells inside the biofilm [Citation89]. Extracellular DNA (eDNA) has been identified as part of the ECM. The mechanisms of eDNA release include cell lysis, quorum sensing, and excretion of vesicle-containing DNA [Citation90]. To determine whether eDNA plays a role in biofilm formation, Margarida Martins et al. [Citation91] studied the relationship between eDNA content and biofilm formation in C. albicans ECM. DNase and exogenous DNA were added to the biofilm to monitor biofilm development as an indicator of eDNA action. The involvement of eDNA is required in the early stage of biofilm formation [Citation92,Citation93]. The biofilm biomass is decreased if the biofilm is treatment with deoxyribonuclease I (DNAse). The proteins in the extracellular matrix of biofilm play an important role in the surface adhesion, morphological transformation, biofilm maturity, dissociation and structural stability of C. albicans cells. Proteins associated with adhesion of C. albicans Bcr1, Als3, Hwp1 etc., can regulate the adhesion of cells to different host substrates, which is the basis of biofilm formation. The protein Bgl2, which is involved in the morphological transformation of C. albicans, regulates its transformation from the yeast form to the hyphae phase and plays a key role in formation of biofilms.
Quorum sensing is a phenomenon in which microbes communicate and regulate their behavior through secret signaling molecules, which is the basis of biofilm formation [Citation94]. The fungal QS signaling molecules currently found include farnesol and tyrosol found in C. albicans [Citation95,Citation96], as well as phenylethanol and chromitol found in Saccharomyces cerevisiae [Citation97]. Farnesol is currently the most studied QS signal molecule. Experiments have confirmed that farnesol can inhibit the transformation of C. albicans from yeast to hyphae form, and can also affect the adhesion process of biofilm cells, the structure of mature biofilms and organisms. Dispersion of cells in the envelope, thereby inhibiting the formation and growth of C. albicans biofilm [Citation98]. Davis et al. [Citation99], showed that farnesol inhibits hyphae growth by inhibiting the Ras1-Cdc35-PKA signaling pathway, and affects other cellular metabolic processes including stress response, metabolism, and drug resistance (). Another study of the fungal QS signaling molecule is tyrosol, which promotes the formation of germ tubes, thereby promoting the transformation of C. albicans from the yeast form to the hyphae form [Citation100]. The regulation of biofilm by QS system is complicated and closely related to the regulation of some drug resistance genes [Citation101]. However, the mechanism remains unclear and deserves further study and discussion.
6. Biofilm maturation
6.1 General maturation in microbial biofilms
After microbial cells attached to surface, extracellular polymeric substances (EPS) including polysaccharides, proteins, lipids, nucleic acids and other components were secreted to enhance cell-to-cell interaction [Citation102]. Studies have verified the activation of ica gene contribute to the secretion of polysaccharide intercellular adhesion (PIA), which is a significant factor for biofilm formation [Citation103]. However, research have shown biofilm can be formed in an ica-independent manner. A variety of adhesive proteins play a substitute role for PIA [Citation104–106]. In addition, extracellular enzymes alter the composition and structure of EPS. Petra Tielen et al [Citation107]. found that the extracellular esterase EstA and proteolytic elastic protease LasB produced during mucoid Pseudomonas aeruginosa biofilm maturation increased the concentration of rhamnolipids and decreased the alginate concentration, eventually enhanced the hydrophobicity and viscosity of EPS. The chemical composition, structure and morphology of the biofilm formed by the same species under various conditions or by different species differ, resulting in diverse cell metabolic activities. eDNA can strengthen cell-to-cell interaction to show positive effect on biofilm formation [Citation108]. For example, cid can mediate the lysis of S. aureus cells and the release of DNA, thereby promoting biofilm maturation [Citation109]. Meanwhile, QS system and ion-mediated electrical signals promote communication between cells, thereby enhance the overall adaptability of the biofilm [Citation110]. Biofilms can also attract distant cells by electrical signal to adhere EPS. Jacqueline Humphries et al. [Citation111] found that electrical signals mediated by potassium ion channel produced by the Bacillus subtilis biofilm can attract microbial cells to move to themselves by changing the membrane potential of distant cells. As the attached cells upward movement and new microbial cell attachment to EPS, eventually forming a tower-shaped maturation morphology [Citation112].
6.2 Biofilm maturation in C. albicans
The mature biofilm of C. albicans is a dense mesh system composed of large numbers of extracellular polymer-encapsulating yeast cells, hyphae and pseudomycelium. Observation by electron microscopy, the biofilm morphology of C. albicans is a three-dimensional structure with hyphae intertwined. It is not a simple cell accumulation, but a highly structured microbial community. The degree of fungal resistance and biofilm formation are also consistent, mature biofilms are more resistant. During the maturation, the amount of extracellular material increased with incubation time until C. albicans communities were completely encased within this material.
The glyoxalase Glx3 is an abundant proteomic component of the biofilm extracellular matrix. The glx3/glx3 mutant showed decreased fitness and formed less biofilm as compared to wild type and a reintegrate strain [Citation113]. Zap1 is a zinc-sensitive transcription factor. In mature biofilms, Zap1 acts as a negative regulator in several pathways. Zap1 inhibits the production of the extracellular matrix of the biofilm. Zap1 downstream target genes Csh1, Ifd6, Gca1 and Adh5 can regulate β-1,3 glucan production. Among them, Csh1 and Ifd6 can inhibit the formation of extracellular matrix under the control of Zap1, while Gca1, Gca2 and Adh5 can promote the formation and accumulation of extracellular matrix under the control of Zap1. In addition, Zap1 inhibits the expression of the Erg gene and the Hxt gene, and the expression of these two genes inhibits the maturation of the biofilm [Citation114]. ().
7. Biofilm dispersal
7.1 General dispersal in microbial biofilms
Microbial biofilm cannot grow indefinitely. When the cell population is extraordinary intensive, the nutrients are insufficient, or under the stress of internal and external environments, biofilm will enter the final stage of its life cycle – – dispersal [Citation115]. The biofilm dispersal is divided into self-initiated dispersal and passive dispersal. The former is characterized as gene expression is regulated to promote the dispersal, which is mainly manifested in inhibiting the production of extracellular matrix, producing enzymes which can degrade extracellular components, and enhancing cell motility [Citation116–118]. As the increase of biofilm biomass, the gradient of oxygen and nutrients in biofilm increase. The closer to the inner layer, the less nutrients and oxygen concentration will be, and the metabolic processes of cells may be affected. The spontaneous dispersal initiates from partial biofilm rather than holistic. For P. aeruginosa, c-di-GMP decreases in the inner layer, reducing the production of EPS and matrix adhesion but increasing cell movement [Citation119]. The consumption of glucose or self-inducing peptide AIP activate S. aureus agr QS system which is inhibited during the biofilm attachment and maturation. The activation of agr system increases the activity of proteases such as serine proteases, thereby degrading the proteins in extracellular matrix and promoting S. aureus biofilm dispersal [Citation109,Citation120]. For the staphylococcal biofilm, thermonuclease reduces the stability of EPS by degrading its eDNA [Citation109]. Lysis of dead cells release enzymes contributing to EPS degradation. The movement of fimbriae is inhibited during biofilm maturation, but enteropathogenic Escherichia coli restore the motility of fimbriae by altering the conformation of its type IV bundle-forming pili, resulting in accelerate of biofilm dispersal [Citation121,Citation122]. The latter passive dispersal does not rely on the regulation of gene expression. The most typical case is that the shear force of fluid physically detach biofilm [Citation123]. The dispersed biofilm cells gradually transform into planktonic and look for the next settlement site. This step may lead to the expansion of microbial contamination/infection [Citation124].
7.2 Biofilm dispersal in C. albicans
The dispersion process is the steps in which bacteria change from biofilm to planktonic lifestyle. The dispersion of cells in mature biofilms is an important component of the biofilm life cycle. Some of cells in the C. albicans biofilms diffuses into the surrounding environment in the form of yeast to form new biofilms. Studies have shown that dispersed cells have stronger adhesion, toxicity and drug resistance. Dispersion occurs throughout the growth cycle of the biofilm. The dispersed C. albicans cells showed a single yeast morphology with greater adherence and increased pathogenicity in the infected mouse model [Citation125,Citation126].
At present, there are few regulatory mechanisms for the diffusion phase of C. albicans biofilm. Only Ume6, Pes1 and Nrg1 regulatory mechanisms have been reported. The Ume6, Pes1 and Nrg1 genes play a role in regulating the dispersion of cells in the biofilm. Overexpression of the Ume6 gene can reduce the release of cells from the biofilm, whereas overexpression of Pes1 and Nrg1 genes enhances cell dispersion. Changes in the expression or activity of Ume6 or Pes1 during biofilm maturation are responses to quorum sensing molecule accumulation, which determines cell dispersion ().
8. Conclusion
The drug resistance produced by biofilms is closely related to many challenges of modern medical treatment, and the complexity of its formation and drug resistance mechanisms makes it extremely difficult to clinically treat biofilm-associated infections. In the last two decades, researchers have made efforts to investigate the genetic contributors control different stages of biofilm development (adherence, initiation, maturation, and dispersal). Many studies have revealed the existence of the biofilm of C. albicans and the specific process of biofilm formation, confirming that the formation of its biofilm can increase the resistance of the fungal host to defense mechanisms and decrease the susceptibility to antifungal drugs. There has been some progress in the study of mechanisms. The biofilm formation of fungi has an important influence on its pathogenic mechanism. The complexity of its formation and resistance mechanisms determines the clinical treatment of biofilm-associated infections is extremely difficult. The formation of Candida biofilm on human medical devices is a major source of diffuse C. albicans infection. Multiple links will affect the formation of biofilm, and adhesion may be the most important stage. These studies will help people deepen understanding the biological characteristics and molecular regulation mechanisms of C. albicans biofilms, provide theoretical basis for prevention and treatment of C. albicans infections, and provide new ideas about the development of new antifungal drugs.
Highlights
Structure and heterogeneity of microbial biofilms are in depth reviewed.
The genetic contributors control biofilm development from adherence to dispersal are reviewed.
This review provides biological characteristics and molecular regulation mechanisms of C. albicans biofilms,
The review yields theoretical basis for prevention and treatment of C. albicans infections.
Data Availability
All data generated or analyzed during this study are included in this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15(2):167–193.
- Hurlow J, Couch K, Laforet K. Clinical biofilms: a challenging frontier in wound care. Adv Wound Care (New Rochelle). 2015;4(5):295–301.
- Cortés ME, Bonilla JC, Sinisterra RD. Biofilm formation, control and novel strategies for eradication. Political Science. 2011;2:896–905.
- Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8(9):881.
- Shirtliff ME, Mader JT, Camper AK. Molecular interactions in biofilms. Chem Biol. 2002;9(8):859–871.
- Marrie TJ, Nelligan J, Costerton JW. A scanning and transmission electron microscopic study of an infected endocardial pacemaker lead. Circulation. 1982;66(6):1339–1341.
- Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358(9276):135–138.
- Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol. 2009;11(7):1034–1043.
- Jamal M, Ahmad W, Andleeb S, et al. Bacterial biofilm and associated infections. J Chin Med Assoc. 2018;81(1):7–11.
- Van Acker H, Van Dijck P, Coenye T. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol. 2014;22(6):326–333.
- Chmielewski RAN, Frank JF. Biofilm formation and control in food processing facilities. Compr Rev Food SCI F. 2003;2(1):22–32.
- Chmielewski RAN, Frank JF. A predictive model for heat inactivation of Listeria monocytogenes biofilm on buna-N rubber. LWT. 2006;39(1):11–19.
- Saitou K, Furuhata K, Kawakami Y, et al. Biofilm formation abilities and disinfectant-resistance of Pseudomonas aeruginosa isolated from cockroaches captured in hospitals. Food Rev Int. 1992;8:573–603.
- Simões M, Simões LC, Vieira MJ. A review of current and emergent biofilm control strategies. LWT. 2010;43(4):573–583.
- Mattila‐Sandholm T, Wirtanen G. Biofilm formation in the industry: areview. Food Rev Int. 1992;8(4):573–603.
- Flemming HC, Percival SL, Walker JT. Contamination potential of biofilms in water distribution systems. Water Sci. Technol. 2018;2:271–280.
- Helmi K, Skraber S, Gantzer C, et al. Interactions of Cryptosporidium parvum, Giardia lamblia, vaccinal poliovirus type 1, and bacteriophages Φ174 and MS2 with a drinking water biofilm and a wastewater biofilm. Appl Environ Microbiol. 2008;74(7):2079–2088.
- Ashbolt NJ. Microbial contamination of drinking water and disease outcomes in developing regions. Toxicology. 2004;198(1–3):229–238.
- Collier SA, Stockman LJ, Hicks LA, et al. Direct healthcare costs of selected diseases primarily or partially transmitted by water. Epidemiol Infect. 2012;140(11):2003–2013.
- Jiang Y, Liu Y, Zhang X, et al. Biofilm application in the microbial biochemicals production process. Biotechnol Adv. 2021;48:107724.
- Biase AD, Kowalski MS, Devlin TR, et al. Moving bed biofilm reactor technology in municipal wastewater treatment: a review. J Environ Manage. 2019;247:849–866.
- Kokare CR, Chakraborty S, Khopade AN, et al. Biofilm: importance and applications. Indian J Biotechnol. 2009;8:159–168.
- Nazir R, Zaffar MR, Amin I. Bacterial biofilms: the remarkable heterogeneous biological communities and nitrogen fixing microorganisms in lakes. Freshwater Microbiology. 2019: 307–340.
- Salama Y, Chennaoui M, Mountadar M, et al. Influence of support media on COD and BOD removal from domestic wastewater using biological treatment in batch mode. Desalin Water Treat. 2015;54(1):37–43.
- Sheng GP, Yu HQ. Characterization of extracellular polymeric substances of aerobic and anaerobic sludge using three-dimensional excitation and emission matrix fluorescence spectroscopy. Water Res. 2006;40(6):1233–1239.
- Salama Y, Chennaoui M, Sylla A, et al. Characterization, structure, and function of extracellular polymeric substances (EPS) of microbial biofilm in biological wastewater treatment systems: a review. Desalin Water Treat. 2015;57(35):16220–16237.
- Costa OYA, Raaijmakers JM, Kuramae EE. Microbial Extracellular Polymeric Substances: ecological Function and Impact on Soil Aggregation. Front Microbiol. 2018;9:1636.
- Sutherland IW. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology. 2001;147(1):3–9.
- Matsuyama T, Nakagawa Y. Surface-active exolipids analysis of absolute chemical structures and biological functions. J Microbiol Methods. 1996;25(2):165–175.
- Yang L, Barken KB, Skindersoe ME, et al. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiol. 2007;153(5):1318–1328.
- Stewart PS. Diffusion in biofilms. J Bacteriol. 2003;185(5):1485–1491.
- Ramey BE, Koutsoudis M, Bodman SBV, et al. Biofilm formation in plant-microbe associations. Curr Opin Microbiol. 2004;7(6):602–609.
- Nikolic N, Barner T, Ackermann M. Analysis of fluorescent reporters indicates heterogeneity in glucose uptake and utilization in clonal bacterial populations. BMC Microbiol. 2013;13(1):258.
- Lin YC, Cornell WC, Jo J, et al. The Pseudomonas aeruginosa complement of lactate dehydrogenases enables use of d- and l-lactate and metabolic cross-feeding. mBio. 2018;9(5). 10.1128/mBio.00961-18.
- Schiessl KT, Hu F, Jo J, et al. Phenazine production promotes antibiotic tolerance and metabolic heterogeneity in Pseudomonas aeruginosa biofilms. Nat Commun. 2019;10(1):762.
- Mathe L, Van DP. Recent insights into Candida albicans biofilm resistance mechanisms. Curr Genet. 2013;59(4):251–264.
- Warnock DW. Trends in the epidemiology of invasive fungal infections. Nihon Ishinkin Gakkai Zasshi. 2007;48(1):1–12.
- Barnes RA. Early diagnosis of fungal infection in immunocompromised patients. J Antimicrob Chemoth. 2008;61(Supplement 1):i3–6.
- Chandra J, Kuhn DM, Mukherjee PK, et al. Biofilm Formation by the Fungal Pathogen Candida albicans Development, Architecture, and Drug Resistance. J Bacteriol. 2001;183(18):5385–5394.
- Berman J. Morphogenesis and cell cycle progression in Candida albicans. Curr Opin Microbiol. 2006;9(6):595–601.
- Guo DD, Yue HZ, Wei YJ, et al. Genetic regulatory mechanisms of Candida albicansbiofilm formation. Chinese Journal of Biotechnology. 2017;33(9):1567–1581.
- Dalle F, Wachtler B, L’Ollivier C, et al. Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell Microbiol. 2010;12(2):248–271.
- Gil-Bona A, Parra GC, Hernáez M, et al. Candida albicanscell shaving uncovers new proteins involved in cell wall integrity, yeast to hypha transition, stress response and host-pathogen interaction. J Proteomics. Vol. 127. 2015. p. 340–351.
- Shao J, Wang T, Yan Y, et al. Matrine reduces yeast-to-hypha transition and resistance of a fluconazole-resistant strain of Candida albicans. J Appl Microbiol. 2014;117(3):618–626.
- Ramage G, Saville SP, Wickes BL, et al. Inhibition of Candida albicans Biofilm Formation by Farnesol, a Quorum-Sensing Molecule. Appl Environ Microbiol. 2002;68(11):5459–5463.
- Fanning S, Mitchell AP. Fungal biofilms. PLoS Pathog. 2012;8(4):e1002585.
- Aggarwal S, Stewart P, Hozalski R. Biofilm Cohesive Strength as a Basis for Biofilm Recalcitrance: are Bacterial Biofilms Overdesigned? Fibertas Academica. 2018;8(2):29–32.
- Nett J, Lincoln L, Marchillo K, et al. Putative role of beta-1,3 glucans in Candida albicansbiofilm resistance. Antimicrob Agents Ch. 2007;51(2):510–520.
- Lazzell AL, Chaturvedi AK, Pierce CG, et al. Treatment and prevention of Candida albicans biofilms with caspofungin in a novel central venous catheter murine model of candidiasis. J Antimicrob Chemoth. 2009;64(3):567–570.
- Hawser SP, Douglas LJ. Biofilm formation by Candida species on the surface of catheter materials in vitro. Infect Immun. 1994;62(3):915–921.
- Hawser SP, Douglas LJ. Resistance of Candida albicans biofilms to antifungal agents in vitro. Antimicrob Agents Ch. 1995;39(9):2128–2131.
- Ramage G, Vande WK, Wickes BL, et al. Standardized Method for In Vitro Antifungal Susceptibility Testing of Candida albicans Biofilms. Antimicrob Agents Ch. 2001;45(9):2475–2479.
- Kojic EM, Darouiche RO. Candida Infections of Medical Devices. Clin Microbiol Rev. 2004;17(2):255–267.
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95–108.
- Emily J, Marsh HL, Wang H. A three-tiered approach to differentiate Listeria monocytogenes biofilm-forming abilities. FEMS Microbiol Lett. 2003;228(2):203–210.
- Shrout JD, Tolker-Nielsen T, Givskov M, et al. The contribution of cell-cell signaling and motility to bacterial biofilm formation. MRS Bull. 2011;36(5):367–373.
- Nahar S, Mizan MFR, Ha AJ, et al. Advances and future prospects of enzyme-based biofilm prevention approaches in the food industry. Compr Rev Food Sci Food Saf. 2018;17(6):1484–1502.
- Bradle DE. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can J Microbiol. 1980;26(2):146–154.
- O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54(1):49–79.
- Ryu JH, Beuchat LR. Biofilm formation by Escherichia coli O157: h7on stainless steel: effect of exopolysaccharide and Curli production on its resistance to chlorine. Appl Environ Microbiol. 2005;71(1):247–254.
- Vazquez V, Liang X, Horndahl JK, et al. Fibrinogen is a ligand for the Staphylococcus aureus microbial surface components recognizing adhesive matrix molecules (MSCRAMM) bone sialoprotein-binding protein (Bbp). J Biol Chem. 2011;286(34):29797–29805.
- Paterson GK, Orihuela CJ. Pneumococcal microbial surface components recognizing adhesive matrix molecules targeting of the extracellular matrix. Mol Microbiol. 2010;77(1):1–5.
- Foster TJ, Geoghegan JA, Ganesh VK, et al. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12(1):49–62.
- Kang M, Ko YP, Liang X, et al. Collagen-binding microbial surface components recognizing adhesive matrix molecule (MSCRAMM) of Gram-positive bacteria inhibit complement activation via the classical pathway. J Biol Chem. 2013;288(28):20520–20531.
- Henriques M, Gasparetto K, Azeredo J, et al. Experimental methodology to quantify Candida albicans cell surface hydrophobicity. Biotechnol Lett. 2002;24(13):1111–1115.
- Chaffin WL. Candida albicanscell wall proteins. Microbiol Mol Biol Rev. 2008;72:495–544.
- Silva S, Negri M, Henriques M, et al. Adherence and biofilm formation of non-Candida albicansCandida species. Trends Microbiol. 2011;19(5):241–247.
- Wang YC, Huang SH, Lan CY, et al. Prediction of phenotype-associated genes via a cellular network approach: a Candida albicans infection case study. PLoS One. 2012;7(4):e35339.
- Tronchin G, Pihet M, Lopes-Bezerra LM, et al. Adherence mechanisms in human pathogenic fungi. Med Mycol. 2008;46(8):749–772.
- Cota E, Hoyer LL. The Candida albicans agglutinin-like sequence family of adhesins: functional insights gained from structural analysis. Future Microbiol. 2015;10(10):1635–1648.
- Nobile CJ, Schneider HA, Nett JE, et al. Complementary adhesin function in Candida albicans biofilm formation. Curr Biol. 2008;18(14):1017–1024.
- Ramage G, Vande WK, López-Ribot JL, et al. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol Lett. 2002;214(1):95–100.
- Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the Candida albicans transcription factor Bcr1p. Curr Biol. 2005;15(12):1150–1155.
- Nobile CJ, Andes DR, Nett JE, et al. Critical role of Bcr1-dependent adhesins in Candida albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006;2(7):e63.
- Srivastava A, Sircaik S, Husain F, et al. Distinct roles of the 7-transmembrane receptor protein Rta3 in regulating the asymmetric distribution of phosphatidylcholine across the plasma membrane and biofilm formation in Candida albicans. Cell Microbiol. 2017;19(12):E12767.
- Pierce VJ, Kumamoto AC, Lorenz M. Variation in Candida albicans EFG1 Expression Enables Host-Dependent Changes in Colonizing Fungal Populations. MBio. 2012;3(4):e00117–12.
- Stoldt VR, Sonneborn A, Leuker CE, et al. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. Embo J. 1997;16(8):1982–1991.
- Chen HF, Lan CY, Coste AT. Role of SFP1 in the Regulation of Candida albicans Biofilm Formation. PLoS One. 2015;10(6):e0129903.
- Dickson RC. Thematic review series: sphingolipids. New insights into sphingolipid metabolism and function in budding yeast. J Lipid Res. 2008;49(5):909–921.
- Patton JL, Lester RL. The phosphoinositol sphingolipids of Saccharomyces cerevisiae are highly localized in the plasma membrane. Journal Bacteriol. 1991;173(10):3101–3108
- Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat Rev Mol Cell Bio. 2008;9(2):125–138.
- Moosa MYS, Alangaden GJ, Manavathu E, et al. Resistance to amphotericin B does not emerge during treatment for invasive aspergillosis. J Antimicrob Chemother. 2002;49(1):209–213.
- Navarro GF, Alonso MR, Rico H, et al. A role for the MAP kinase gene MKC1 in cell wall construction and morphological transitions in Candida albicans. Microbiology. 1998;144(2):411–424.
- Nobile CJ, Mitchell AP. Genetics and genomics of Candida albicansbiofilm formation. Cell Microbiol. 2006;8(9):1382–1391.
- Mollinedo F. Lipid raft involvement in yeast cell growth and death. Front Oncol. 2012;2(140). DOI:10.3389/fonc.2012.00140
- Csank C, Schröppel K, Leberer E, et al. Roles of the Candida albicansmitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect Immun. 1998;66(6):2713–2721.
- Sandini S, Stringaro A, Arancia S, et al. The MP65 gene is required for cell wall integrity, adherence to epithelial cells and biofilm formation in Candida albicans. BMC Microbiol. 2011;11(1):106.
- Nobile CJ, Nett JE, Andes DR, et al. Function of Candida albicansadhesin Hwp1 in biofilm formation. Eukaryot Cell. 2006;5(10):1604–1610.
- Nieto MC, Telleria O, Cisterna R. Sentinel surveillance of invasive candidiasis in Spain: epidemiology and antifungal susceptibility. Diagn Microbiol Infect Dis. 2015;81(1):34–40.
- Allesen-Holm M, Barken KB, Yang L, et al. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59(4):1114–1128.
- Martins M, Henriques M, Lopez-Ribot JL, et al. Addition of DNAse improves the in vitro activity of antifungal drugs against Candida albicans biofilms. Mycoses. 2012;55(1):80–85.
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, et al. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295(5559):1487.
- Izano EA, Amarante MA, Kher WB, et al. Differential Roles of Poly-N-Acetylglucosamine Surface Polysaccharide and Extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis Biofilms. Appl Environ Microbiol. 2008;74(2):470–476.
- Hornby JM, Jensen. EC, Lisec AD, et al. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microb. 2001;67(7):2982–2992.
- Yu LH, Wei X, Ma M, et al. Possible inhibitory molecular mechanism of farnesol on the development of fluconazole resistance in Candida albicans biofilm. Antimicrob Agents Ch. 2012;56(2):770–775.
- Chen H, Fujita M, Feng QH, et al. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc Natl Acad Sci U S A. 2004;101(14):5048–5052.
- Chen H, Fink GR. Feedback control of morphogenesis in fungi by aromatic alcohols. Gene Dev. 2006;20(9):1150–1161.
- Deveau A, Hogan DA. Linking quorum sensing regulation and biofilm formation by Candida albicans. Methods Mol Biol. 2011;692:219–233.
- Davis HA, Piispanen AE, Stateva LI, et al. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol Microbiol. 2008;67(1):47–62.
- Rde AC, Teixeira CE, Brilhante RS, et al. Exogenous tyrosol inhibits planktonic cells and biofilms of Candida species and enhances their susceptibility to antifungals. FEMS Yeast Res. 2015;15(4) :fov012
- Cao YY, Cao YB, Xu Z, et al. cDNA microarray analysis of differential gene expression in Candida albicans biofilm exposed to farnesol. Antimicrob Agents Ch. 2005;49(2):584–589.
- Armbruster CR, Parsek MR. New insight into the early stages of biofilm formation. Proc Natl Acad Sci U S A. 2018;115(17):4317–4319.
- Kot B, Sytykiewicz H, Sprawka I. Expression of the biofilm-associated genes in methicillin-resistant Staphylococcus aureus in biofilm and planktonic conditions. Int J Mol Sci. 2018;19(11):3478.
- Pozzi C, Waters EM, Rudkin JK, et al. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog. 2012;8(4):e1002626.
- Rohde H, Burandt EC, Siemssen N, et al. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials. 2007;28(9):1711–1720.
- Kogan G, Sadovskaya I, Chaignon P, et al. Biofilms of clinical strains of Staphylococcus that do not contain polysaccharide intercellular adhesin. FEMS Microbiol Lett. 2006;255(1):11–16.
- Tielen P, Rosenau F, Wilhelm S, et al. Extracellular enzymes affect biofilm formation of mucoid Pseudomonas aeruginosa. Microbiology (Reading). 2010;156(7):2239–2252.
- DeFrancesco AS, Masloboeva N, Syed AK, et al. Genome-wide screen for genes involved in eDNA release during biofilm formation by Staphylococcus aureus. Proc Natl Acad Sci U S A. 2017;114(29):5969–5978.
- Mann EE, Rice KC, Boles BR, et al. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4(6):e5822.
- Prindle A, Liu J, Asally M, et al. Ion channels enable electrical communication in bacterial communities. Nature. 2015;527(7576):59–63.
- Humphries J, Xiong L, Liu J, et al. Species-independent attraction to biofilms through electrical signaling. Cell. 2017;168(1–2):200–209.
- Klausen M, Aaes‐Jørgensen A, Molin S, et al. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol Microbiol. 2003;50(1):61–68.
- Laura C, Estefania GH, Jordan FP, et al. Deletion of GLX3 in Candida albicans affects temperature tolerance, biofilm formation and virulence. Fems Yeast Res. 2019;19(2):124.
- Garcia SS, Aubert S, Iraqui I, et al. Candida albicans Biofilms: a Developmental State Associated With Specific and Stable Gene Expression Patterns. Eukaryot Cell. 2004;3(2):536–545.
- Kaplan JB, Ragunath C, Ramasubbu N, et al. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J Bacteriol. 2003;185(16):4693–4698.
- Rumbaugh KP, Sauer K. Biofilm dispersion. Nat Rev Microbiol. 2020;18(10):571–586.
- Williamson KS, Richards LA, Perez-Osorio AC, et al. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J Bacteriol. 2012;194(8):2062–2073.
- Serra DO, Hengge R. Stress responses go three dimensional - the spatial order of physiological differentiation in bacterial macrocolony biofilms. Environ Microbiol. 2014;16(6):1455–1471.
- An S, Wu J, Zhang LH. Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-Di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl Environ Microbiol. 2010;76(24):8160–8173.
- Boles BR, Horswill AR. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4(4):e1000052.
- Cleary J, Lai LC, Shaw RK, et al. Enteropathogenic Escherichia coli (EPEC) adhesion to intestinal epithelial cells: role of bundle-forming pili (BFP), EspA filaments and intimin. Microbiol. 2004;150(3):527–538.
- Stuart Knutton RKS, Anantha RP, Donnenberg MS, et al. The type IV bundle-forming pilus of enteropathogenic Escherichia coli undergoes dramatic alterations in structure associated with bacterial adherence, aggregation and dispersal. Mol Microbiol. 1999;33(3):499–509.
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: acommon cause of persistent infections. Sci. 1999;284(5418):1318–1322.
- Uppuluri P, Acosta Zaldívar M, Anderson MZ, et al. Candida albicansdispersed cells are developmentally distinct from biofilm and planktonic cells. mBio. 2018;9(4). DOI:10.1128/mBio.01338-18.
- Vila TVM, Rozental S. Biofilm Formation as a Pathogenicity Factor of Medically Important Fungi. InTech. 2016;1–24.
- Finkel JS, Mitchell AP. Genetic control of Candida albicansbiofilm development. Nat Rev Microbiol. 2011;9(2):109–118.