ABSTRACT
The anterior cingulate cortex (ACC) is a core brain region processing pain emotion. In this study, we performed RNA sequencing analysis to reveal transcriptomic profiles of the ACC in a rat chronic constriction injury (CCI) model. A total of 1628 differentially expressed genes (DEGs) were identified by comparing sham-operated rats with rats of 12 hours, 1, 3, 7, and 14 days after surgery, respectively. Although these inflammatory-related DEGs were generally increased after CCI, different kinetics of time-series expression were observed with the development of neuropathic pain affection. Specifically, the expression of Ccl5, Cxcl9 and Cxcl13 continued to increase following CCI. The expression of Ccl2, Ccl3, Ccl4, Ccl6, and Ccl7 were initially upregulated after CCI and subsequently decreased after 12 hours. Similarly, the expression of Rac2, Cd68, Icam-1, Ptprc, Itgb2, and Fcgr2b increased after 12 hours but reduced after 1 day. However, the expression of the above genes increased again 7 days after CCI, when the neuropathic pain affection had developed. Furthermore, gene ontology analysis, Kyoto Encyclopedia of Genes and Genomes pathway enrichment and interaction network analyses further showed a high connectivity degree among these chemokine targeting genes. Similar expressional changes in these genes were found in the rat spinal dorsal horn responsible for nociception processing. Taken together, our results indicated chemokines and their targeting genes in the ACC may be differentially involved in the initiation and maintenance of neuropathic pain affection. These genes may be a target for not only the nociception but also the pain affection following nerve injury.
1. Introduction
Neuropathic pain caused by the injury or disease of the somatosensory nervous system is a pathological condition that places a heavy burden on patients. According to the data of the International Association for the Study of Pain in 2019, approximately 10% of the world’s population suffered from neuropathic pain[Citation1]. Therefore, improving our understanding of the pathogenesis of neuropathic pain and developing effective treatments is crucial for the wellbeing of these patients.
The anterior cingulate cortex (ACC) is located in the forebrain and is an important part of the limbic system. Evidence has shown that the ACC is a core brain region that processes pain emotion. Clinical studies have revealed significant changes in the structure and function of the ACC in patients with chronic pain change significantly [Citation2–4]. Synaptic plasticity of the ACC is a key mechanism underlying the development of chronic pain, which has been supported by numerous animal experiments [Citation5,Citation6]. Peripheral inflammatory pain or neuropathic pain causes considerable changes in the synaptic transmission and morphological plasticity of ACC neurons [Citation7–9], and blocking this abnormal plasticity of the ACC induces a significant analgesic effect [Citation10,Citation11].
To date, RNA sequencing analyses of the ACC have been conducted to reveal the mechanisms underlying psychiatric disorders [Citation12–14], diurnal rhythms[Citation15] and cognitive dysfunction[Citation16], however, those for neuropathic pain remain scarce. A recent study revealed the entire transcriptome of the spinal cord, ACC, and amygdala following spinal nerve ligation (SNI) and compared between sham-surgery mice and mice of 7 days after SNI[Citation17]. In order to undertake a more comprehensive investigation of the ACC cells under neuropathic pain conditions, we performed RNA sequencing analysis to distinguish DEGs between sham-operated rats and rats of 12 hours and 1, 3, 7, and 14 days after chronic constriction injury (CCI). We expected to further understand the molecular mechanisms underlying different stages of neuropathic pain and provide therapeutic targets for neuropathic pain. We performed gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment, protein-protein interaction (PPI) network analyses and Short time-series expression miner analysis (STEM) to investigate the mechanism underlying the initiation and maintenance of neuropathic pain. In addition, we compared gene expression patterns of chemokines in ACC and spinal dorsal horn to further explore the role of these genes in pain.
2. Methods
2.1. Animals
Male Sprague–Dawley rats weighing 220 g − 250 g were purchased form Hunan SLAC Laboratory Animal Co., Ltd. (Changsha, China). Rats were raised in groups of three per cage with a free access to food and water under a 12 h light/dark cycle in a suitable environment for temperature and humidity. All procedures were conducted in accordance with the National Institutes of Health guidelines for the care and use of Laboratory animals and approved by the Institutional Ethics Committee of Central South University. A total of 60 rats subject to CCI or sham-operated surgery were included in this study. 48 rats were randomly divided into 6 groups for the measurements of paw withdrawal mechanical threshold (PWMT), paw withdrawal thermal latency (PWTL) and sucrose preference test (SPT). Due to time limitation, we cannot finish all the behavioral assessments of the CCI-7d rats in one day. Another 12 rats were randomly divided equally into sham and CCI group for the experiment of place escape/avoidance paradigm (PEAP).
2.2. CCI model
CCI was performed in rats according to the method of Bennett and Xie[Citation18]. After the rats were anesthetized with isoflurane, the left sciatic nerve was exposed and tied using around by four snug ligatures (4–0) with equal tightness at consistent intervals. After ligation, the nerve was repositioned. In sham surgery rats, the left sciatic nerve was just exposed without ligation.
2.3. Behavioral assessment
PWMT and PWTL of the rats were measured on 1 day before CCI and from 12 hours to 14 days after CCI. The measurement before CCI was used as the basic threshold (BL). All assessments were carried out after the rats were acclimated to specific individual chambers for at least 30 min. The PWMT was measured using von Frey filaments (North Coast Medical, San Jose, CA, USA) that ranged from 0.4–15 g, as described in our previous study [Citation19,Citation20]. Briefly, the stimuli were applied vertically to the plantar surface of the left hind paw, and the minimal force needed to induce three consistent withdrawal responses (lifting or licking) was defined as the PWMT. A thermal pain test instrument (Tes7370, Ugo Basile, Comerio, Italy) [Citation19,Citation20] was used to assess the PTWL. Briefly, after the rats were habituated in the cage for 30 min, a continuous heat stimuli were applied to the plantar surface of the hindpaw to evoke withdrawal responses, and a timer recorded the response latency. The cutoff time was set to 30 s, and each rat was tested for three times with a 5 min interval. The three latencies were averaged to provide the PTWL.
SPT was performed according to a previous study[Citation21]. Rats were given 1% sucrose solution for 3–5 days. Subsequently, the experiment was carried out 1 day before CCI (BL) and 3, 7, and 14 days after CCI. During the test, rats were fed in a single cage and given two bottles of water (1% sucrose solution and water). The positions of the sucrose solution and water were exchanged every 12 hours. After 24 hours, the consumption of sucrose solution and water was measured, and the percentage of sucrose preference was calculated.
PEAP was performed by rats as described previously[Citation22]. Rats were placed in a 30 × 30 × 30 cm chamber and were allowed to move freely for 30 minutes before the test. The chamber was placed on top of a mesh floor. One half of the chamber was painted black (dark area), and the other half was the light area. A 60 g von Frey filament was applied to the plantar surface of the hind paw every 15 s for 30 min. The injured hind paw was stimulated within the dark area, and the hind paw contralateral to the CCI was stimulated within the light area. The time spent in the light area throughout the test was recorded and converted to a percentage to reflect the level of pain affect. All behavioral tests were performed by investigators who were blinded to the experimental groups.
2.4. Total RNA extraction and purification
Rats were sacrificed under deep anesthesia at different time points. The ACC and spinal cord were extracted from each rat after perfusion with phosphate-buffered saline, immediately frozen in dry ice, and stored at −80°C. Total RNA was extracted using the RNeasy Micro Kit (Cat# 74,004, Qiagen) following manufacturer instructions and the RIN number was checked to inspect RNA integrity using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, US). Qualified total RNA was further purified using the RNAClean XP Kit (Cat A63987, Beckman Coulter, Inc. Kraemer Boulevard Brea, CA, USA) and the RNase-Free DNase Set (Cat#79,254, QIAGEN, GmBH, Germany).
2.5. RNA sequence and data analysis
The preparation of the cDNA library of all ACC and spinal cord samples and the sequencing were performed by Shanghai Biotechnology Corporation and KangChen Biotechnology Corporation, respectively. On average, 6 G of raw data per sample were obtained. Unqualified sequencing reads were removed, and genome mapping was performed for clean reads using the HISAT2 (version:2.0.4) tool[Citation23]. The reads were mapped to rn6. To achieve gene expression standardization across different genes and samples, clean reads were transformed into fragments per kilobase of exon model per million mapped reads (FPKM)[Citation24]. Fold change was calculated according to the FPKM value.
GO and KEGG pathway analyses of gene functional annotation clustering were performed using the OmicStudio tools [Citation25,Citation26](https://www.omicstudio.cn/tool). Genes with a fold change > 1.5 and p < 0.05 were considered differentially expressed.
STEM was used to classify the input genes according to their expression trends and was combined with functional enrichment analysis to mine the data for internal patterns more effectively. STEM (Version: 1.3.11) was applied to cluster time-series gene expression data[Citation27].
The PPI network analysis was performed using the STRING online software (http://string-db.org/). Next, the interaction data in tsv format was downloaded and then analyzed by MCC algorithms of Plug-in cytoHubba for Cytoscape (Version: 3.7.1) software in JAVA platform [Citation26,Citation28,Citation29].
The ACC and spinal cord sequencing results were submitted to the Gene Expression Omnibus (GEO) repository and assigned GEO accession numbers: GSE172133 and GSE175760, respectively.
2.6. Statistical analysis
The GraphPad Prism 7 was used for statistical analysis. Data were analyzed using a two-way repeated-measures analysis of variance, followed by Sidak’s multiple comparisons test. All data are presented as means ± standard deviations and statistical significance was set at p < 0.05.
3. Results
In this study, we aimed to reveal more comprehensive transcriptomic profiles of the ACC in CCI rats to further understand the molecular mechanisms underlying different stages of neuropathic pain and provide therapeutic targets for neuropathic pain. We performed RNA sequencing analysis and a total of 1628 DEGs were identified by comparing sham-operated rats with rats of 12 hours, 1, 3, 7, and 14 days after surgery, respectively. GO and KEGG analysis revealed that most of the DEGs were involved in inflammatory and immune process. Although these DEGs were generally increased after CCI, they demonstrated different kinetics in time-series expression. We screened out chemokines and their targeting genes through STEM, and PPI network analysis showed high connectivity among them. Similar expressional changes in these genes were also exhibited in the rat spinal dorsal horn. Thus, these genes may be a target for not only nociception but also pain affection following nerve injury.
3.1. Behavioral characterization of rats following CCI
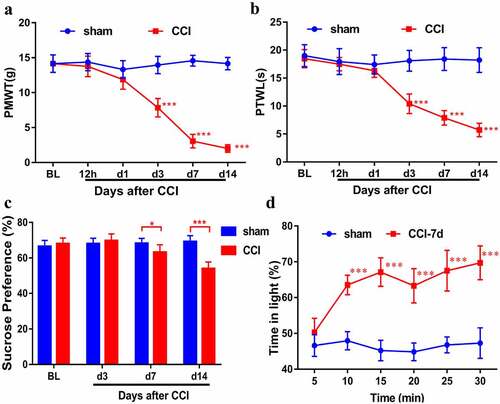
We performed CCI in rats and then measured behavioral characterization of them. Compared with the sham rats, the PWMT and PWTL of the CCI rats decreased from 3 days to 14 days, indicating that CCI rats developed allodynia and thermal hyperalgesia ( and B, n = 8). SPT showed a significant decrease in the intake of sucrose in CCI rats, suggesting that CCI rats developed depression (, n = 8). CCI rats spent more time than sham rats in the light area to escape the stimulation during the PEAP test, whereas sham rats were willing to stay in the dark area (a preferred area) (, n = 6). These results indicated that the neuropathic pain model was successfully established.
Figure 1. Behavioral characterization of rats following CCI. Mechanical allodynia (a) and thermal hyperalgesia (b) were induced after CCI surgery (n = 8). (c). Sucrose preference test (n = 8). (d). CCI rats spent more time in the light area than sham rats during the PEAP test (n = 6). Results are represented as means ± SD. Two-way ANOVA; *p< 0.05 vs. sham group, ***p< 0.001 vs. sham group.
3.2. The regulation of transcripts in the ACC of the rat following CCI
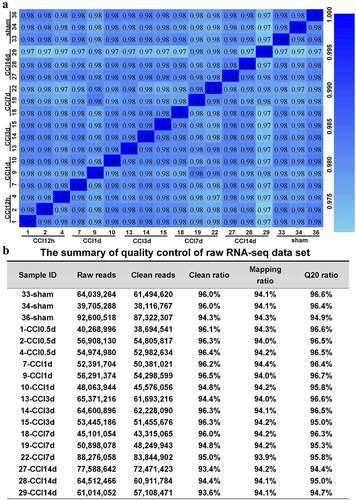
We performed RNA sequencing analysis to reveal transcriptomic profiles of the ACC in CCI rats (n = 3). The transcriptome data were acquired from CCI rats of 12 hours and 1, 3, 7, and 14 days after surgery and sham-operated rats. shows the quality control of sequencing data for each sample. On average, approximately 60 million clean reads were collected, and a mapping rate of approximately 93%–97% was achieved. Q20 ratios were all above 94%. We then calculated the correlation value between every two samples based on normalized expression results and drew a correlation heat map ().
Figure 2. Quality control of the raw RNA sequencing dataset of the ACC. (a). Heatmap of the correlations between each sample using the Pearson test. (b). The summary of quality control of the raw RNA sequencing dataset showing raw reads, clean reads, clean ratio, mapping ratio and Q20 (Phred quality Q scores) of the 18 samples.
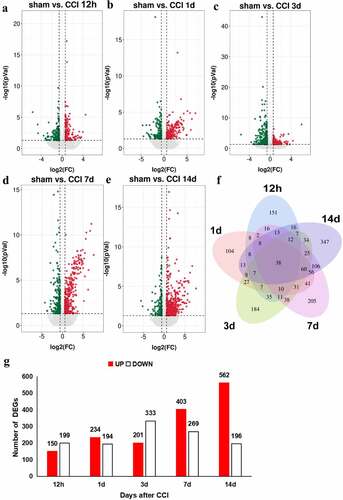
Our data revealed 1628 DEGs with cutoffs of fold changes > 1.5 and p < 0.05. The representative distributions of up- or down-regulated genes are shown in the volcano plot in . As shown in the Venn diagram (), 38 DEGs were shared across five time points. Twelve hours after CCI, 349 genes were differentially expressed: 150 were up-regulated and 199 were down-regulated. On days 1, 3, 7, and 14, the ratio of up-regulated DEGs to all DEGs were 234/428, 201/524, 403/672, and 562/748, respectively, and the ratio of down-regulated DEGs to all DEGs were 194/428, 333/524, 269/672, and 196/748 (). Overall, during the 14 days, the number of up-regulated genes in the ACC after CCI followed an upward trend, reaching a peak at 14 days after nerve injury, and only slightly decreasing on the third day after nerve injury, while the number of down-regulated genes was largely unchanged 1 day after nerve injury, increased significantly 3 days after injury, and subsequently decreased gradually. The top 5 up- and down-regulated DEGs in each time points were listed in Table S1 and S2. These findings revealed that a large number of unique DEGs are expressed during different stages of neuropathic pain development.
Figure 3. Regulation of transcripts in the ACC of rats following CCI. (a-e). Volcano plots of all the DEGs of the sham group compared with those of the CCI groups. Log2(fold change) is plotted as the abscissa, and -log10(p-value) is plotted as the ordinate. Up-regulated genes are red and down-regulated genes are green. Gray dots represent genes with no significant difference. (f). Venn diagram showing the number of unique and shared DEGs with a fold change > 1.5 and p-value < 0.05 at each time point. (g). Histogram showing the statistics of up- and down-regulated DEGs at each time point.
3.3. GO analysis of DEGs
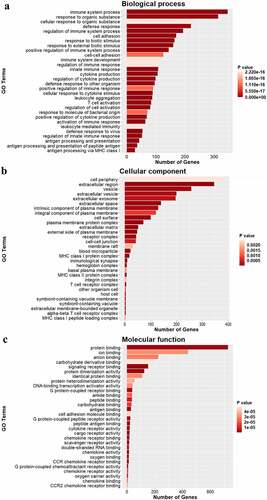
To further understand the specific functions of DEGs in the ACC of rats following CCI, we performed GO analysis to analyze the biological processes, cellular components, and molecular functions of DEGs. The GO analysis revealed that the biological processes were enriched with ‘immune system processes,’ ‘defense responses,’ ‘regulation of immune system processes,’ ‘cell adhesion,’ and ‘cytokine production,’ which suggested that strong immune and inflammatory responses occurred in the ACC following CCI. The cellular components were mainly ‘cell periphery,’ ‘vesicles,’ ‘the intrinsic component of the plasma membrane,’ ‘the integral component of the plasma membrane,’ and ‘the plasma membrane protein complex,’ which indicated that multiple membrane components are involved in neuropathic pain. The molecular function terms were focused on ‘protein binding,’ ‘signaling receptor binding,’ ‘G protein-coupled receptor binding,’ ‘antigen binding,’ and ‘CCR chemokine receptor binding.’ Taken together, the GO analysis revealed that immune and inflammatory responses are key physiological processes involved in the development of neuropathic pain ().
3.4. Analysis of the KEGG pathways
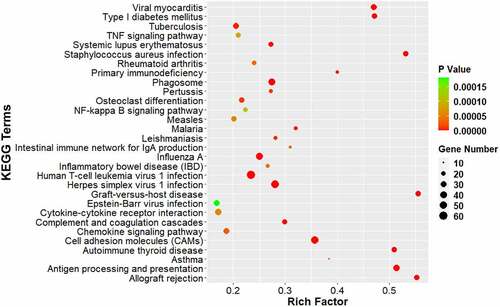
We analyzed the DEGs using KEGG pathway enrichment analysis to identify the major signaling pathways involved in neuropathic pain (). Of the top 30 pathways, DEGs were enriched in the TNF signaling pathway, phagosome, Nuclear factor-kappa B signaling pathway, cytokine–cytokine receptor interaction, complement and coagulation cascades, chemokine signaling pathway, cell adhesion molecules (CAMs), and antigen processing and presentation, which indicated that the cytokines and complements participate in the development of neuropathic pain. In addition, several pathways involved in infection and immune system diseases were also enriched, such as viral myocarditis and Type I diabetes mellitus. These results suggest that immune and inflammatory responses are important processes that occur in the ACC after CCI.
Figure 5. KEGG pathway analysis of DEGs. The comparison of pathway enrichment in the ACC of rats following CCI. The top 30 significantly enriched KEGG pathways are shown. The KEGG terms are plotted as the ordinate and the rich factor is plotted as the abscissa. The size of the dots represented the gene number.
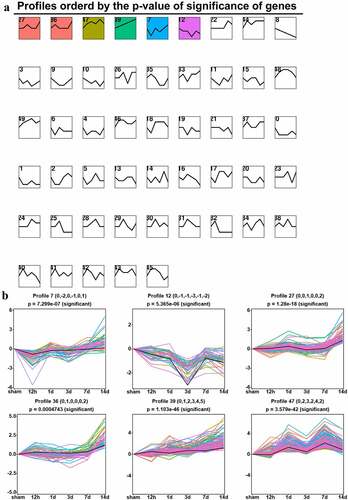
Different kinetics of time-series expression of DEGs in the ACC after CCI Except for several common DEGs, a large number of unique DEGs exist during different stages of neuropathic pain development. We classified all DEGs into 50 clusters based on time series (sham, 12 hours, and 1, 3, 7, and 14 days after CCI). We identified six clusters of significantly different gene expression profiles: 7, 12, 27, 36, 39, and 47. Apart from profile 12, all the other five profiles showed an overall upward trend. Surprisingly, we found that profiles 27 and 36 were up-regulated in sequence. Profile 36 DEGs were up-regulated to reach a small peak at 12 hours and returned on day 1, whereas the DEGs of profile 27 began to up-regulate at 12 hours, reached a small peak on day 1, and returned on day 3. Both of these profiles continued to increase 7 days after CCI. In addition, the profile 39 DEGs continued to increase ().
Figure 6. Short time-series expression miner analysis of DEGs in the ACC following CCI. (a). Model profiles of time series gene expression. Data were sampled at six time points: 0 days (sham), 12 hours, and 1, 3, 7 and 14 days after CCI. The profile ID number is in the top left-hand corner, and the profiles with statistical differences are shown in color. (b). Significant model profiles provided detailed information on the K-means cluster.
To further understand the functions of DEGs in profile 27, 36, 39, we performed GO analysis to identify the biological processes, molecular functions, and cellular components against them. As shown in Figure S1A, the enriched biological processes of profile 36 were those representing immune processes, which included ‘immune response,’ ‘response to cytokines,’ ‘response to interferon (IFN)-gamma,’ and ‘cellular response to IFN-gamma.’ The role of chemokines in enriched molecular function terms was particularly prominent and included ‘cytokine receptor binding,’ ‘cytokine activity,’ ‘chemokine receptor binding,’ ‘chemokine activity,’ and ‘CCR chemokine receptor binding.’ The enriched cellular components were focused mainly on the nucleus, such as ‘nuclear chromosome,’ ‘chromosome,’ and ‘chromatin.’ For profile 27, enriched biological processes were those representing immune processes, such as ‘immune system processes,’ ‘cell activation,’ ‘lymphocyte aggregation,’ ‘leukocyte aggregation,’ ‘mononuclear cell proliferation,’ ‘lymphocyte proliferation,’ ‘leukocyte proliferation,’ and ‘T cell proliferation.’ The enriched molecular function terms were ‘protein binding,’ ‘enzyme binding,’ ‘kinase binding,’ and ‘MHC class II protein complex binding.’ The enriched cellular components were extranuclear and included ‘membrane,’ ‘cell periphery,’ and ‘plasma membrane.’ As for the DEGs of profile 39, the biological processes were ‘immune system processes,’ ‘immune response,’ ‘positive regulation of immune system processes,’ and ‘cell activation.’ The molecular function terms were ‘anion binding,’ ‘carbohydrate derivative binding,’ and ‘ribonucleoside binding,’ and the cellular components were ‘protein-containing complex,’ ‘plasma membrane protein complex,’ ‘chromosome,’ and ‘nuclear chromosome.’
To identify the major signaling pathways involved in the formation and development of neuropathic pain, we further analyzed the DEGs of profiles 27, 36, and 39 using KEGG pathway enrichment analysis. Results showed that significantly enriched DEGs were in the classifications of ‘cytokine–cytokine receptor interactions,’ ‘chemokine signaling pathway,’ ‘CAMs,’ and other cell-defense-related pathways (Figure S1B). Among these, cytokine-related pathways were predominant in profiles 36 and 39, whereas immune cell-related pathways were predominant in profile 27, such as ‘Fc gamma R-mediated phagocytosis,’ ‘B cell signaling pathway,’ ‘leukocyte transendothelial migration,’ and ‘T cell receptor signaling pathway.’ These results indicated that the initial increase of these DEGs may reflect the early response of the ACC to nerve injury, whereas the later increase of these genes may reflect their involvement in the development of neuropathic pain affection.
3.5. PPI network analysis of DEGs
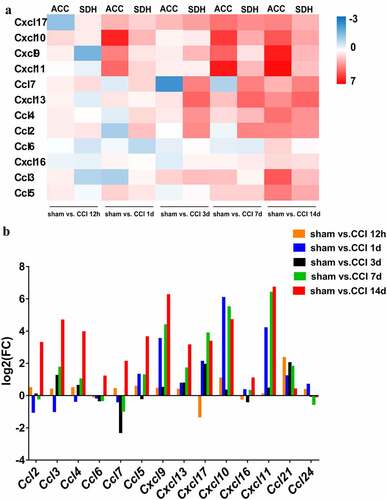
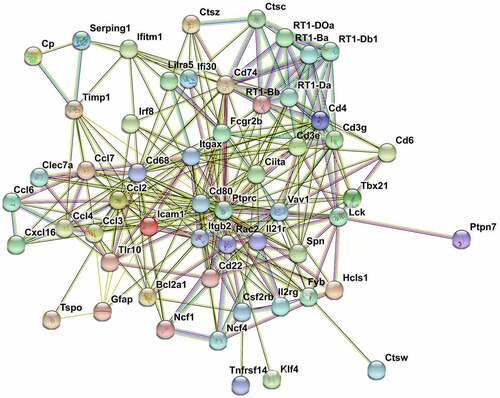
We performed function analysis of the DEGs from profiles 27, 36, and 39 using literature retrieval and found that more than half were from the chemokine family (). Ccl5, Cxcl9, and Cxcl13 were in profile 39, and Ccl2, Ccl3, Ccl4, Ccl6, and Ccl7 were in profile 36. In profile 27, there were several inflammatory and immune-related genes, such as Rac2, Cd68, Icam-1, Ptprc, and Itgb2. Many of these genes have known functions in neuroinflammation or nerve injury. Because the genes in profiles 27 and 36 changed in sequence, we performed PPI network analysis to investigate the underlying relationship between the two profiles. The PPI network analysis revealed complex interactions among genes and contained 62 nodes and 282 edges (p< 0.001). The following genes showed high connectivity: Ptprc (degree = 35), Cd68 (degree = 26), Ccl2 (degree = 19), Vav1 (degree = 17), Rac2 (degree = 17), Itgb2 (degree = 17), Fcgr2b (degree = 17), Icam-1 (degree = 15), Ccl3 (degree = 12), Ccl4 (degree = 11), Ccl7 (degree = 10) (). These results suggested that chemokines and chemokine-targeting genes in the ACC are closely related. Dynamic analysis of the ACC transcriptome profiles revealed that there were temporal differences in the expression of chemokines and their targeting genes. The above results indicated that these genes are differentially involved in the initiation and maintenance of neuropathic pain affection.
Table 1. Function analysis of DEGs for profiles 27, 36, and 39. Gene symbols of DEGs are shown
Figure 7. PPI network nalysis of DEGs. STRING analysis for the PPI networks of DEGs for profiles 27 and 36. Network nodes represent proteins. Edges represent protein-protein associations, which include known interactions, predicted interactions, text mining, co-expression and protein homology. PPI enrichment p-value< 0.001.
3.6. Similar expressional changes of chemokines in the ACC and spinal cord
To determine the specific expression patterns of each chemokine in the ACC, a bar chart was generated to visualize their expression levels during the different stages. As shown in , all chemokines varied in their expression patterns. Chemokines in the spinal cord are responsible for processing nociception following nerve injury[Citation30]. Therefore, we compared gene expression patterns of chemokines in the ACC with those in the spinal cord. The quality control of the sequencing data of each spinal cord sample is shown in Table S3. As shown in the heatmap in , all chemokines showed generally upward trends and exhibited similar expressional changes, which suggested that DEGs are differentially involved in the initiation and maintenance of neuropathic pain affection. Thus, these genes may be a target for not only nociception but also pain affection following nerve injury.
4. Discussion
The pain experience includes pain perception and emotional affection. The ACC is a key brain region that is involved in the regulation of pain. Pain emotions include anxiety, depression, aversion, fear et al. In this study, CCI rats developed pain perception and pain-related affection, manifesting as a decrease in threshold and latency to mechanical and thermal stimuli, a reduction in intake of sugar due to depression, and aversion to noxious stimuli. We performed RNA sequencing of the ACC in CCI rats and found 1628 DEGs that were primarily involved in inflammatory and immune processes.
ACC activation is closely related to the inflammatory response. Harrison found that ACC activation was associated with increased IL-6 accompanied by fatigue and confusion, in healthy individuals who received typhoid vaccination[Citation31]. ACC activation also occurs during stress-induced inflammatory responses[Citation32]. Moreover, microglial activation has been detected using labeling translocator protein labeling with positron emission tomography in patients with major depressive disorder, whose ACC showed elevated TSPO levels [Citation32,Citation33]. It is well established that neuropathic pain is associated with profound neuroinflammation and immune responses, which involve chemokines to some extent. Chemokines were found up-regulated in dendritic cells that infiltrated into tumors compared with dendritic cells from juxtatumor tissues in the model of cancer pain[Citation34]. In our study, chemokines showed different kinetics of time-series expression in ACC and spinal dorsal horn from CCI rats with the development of neuropathic pain. Evidence has shown that chemokines function in the dorsal root ganglion (DRG) and spinal cord under chronic pain conditions[Citation30]. However, few studies on chemokines in the brain have been conducted. Our results showed that several chemokines, such as Ccl5, Cxcl9, and Cxcl13, were persistently up-regulated in the ACC during the development and maintenance of neuropathic pain. Recent studies have revealed that increases in Cxcl13 and its receptor, C-X-C chemokine receptor type 1 (Cxcr5), triggers neuropathic pain-related conditioned place aversion. Moreover, cxcl13 mRNA was found to gradually increase in the ACC from 1 to 10 days following surgery, which is consistent with our results. Cxcll13/Cxcr5 are also up-regulated in the trigeminal ganglion, DRG, and spinal cord under pain conditions. Furthermore, pain hypersensitivity is attenuated by inhibiting Cxcl13/Cxcr5 signaling[Citation30]. Under pathological conditions, Ccl5, which predominantly recruits and activates T lymphocytes and natural killer (NK) cells, is localized in infiltrating lymphocytes of the blood-brain barrier, whereas Cxcl9 is localized in cerebral microvessels and glial cells[Citation35]. Moreover, Ccl5 is closely related to pain in the DRG, spinal cord,[Citation36]and the injured nerve[Citation37]. Intraperitoneal injection of a Ccl5 receptor antagonist or knocking out the Ccl5 gene in rats reduces the infiltration of macrophages and the release of pro-inflammatory cytokines, such as TNFα, IL-1β, IL-6 and IFN-γ following nerve injury [Citation37,Citation38]. Cxcl9 is up-regulated in spinal astrocytes after SNL, whereas intrathecal injection of Cxcl9 or inhibition of spinal Cxcl9 is ineffective for pain relief[Citation39]. Therefore, further study of Ccl5 and Cxcl9 in the ACC is crucial to clarify their roles in neuropathic pain.
Previous reports have suggested that the participation of chemokines in the recruitment of immune cells is dependent on time and context. For example, Ccl2/Ccr2, and not Ccl5/Ccr5, have been shown to recruit monocytes, which exacerbates inflammation in osteoarthritis[Citation40]. In atherosclerosis, Ccr2− monocytes rely on Ccr5, whereas Ccr2+ monocytes are dependent on Cx3cr1 to enter plaques[Citation41]. We found that C-C chemokine receptor type 1 (Ccr1) ligands (i.e., Ccl2, Ccl3, Ccl4, Ccl6, Ccl7, but not Ccl5) were up-regulated in the ACC at two time points: before 12 hours and after 7 days after surgery. Studies have reported that the levels of Ccl2, Ccl3, Ccl4, Ccl6, and Ccl7 significantly increased in the spinal cord after CCI. In addition, Ccl2, Ccl6, and Ccl7 were up-regulated in the DRG in CCI rats, and single and multiple intrathecal injections of J113863 (a Ccr1 antagonist) relieves mechanical and thermal pain. Moreover, repeated intrathecal injections of J11386 can reduce the activation and infiltration of microglia, monocytes, neutrophils, lymphocytes, etc., thereby regulating the levels of pronociceptive (IL-1b, IL-6 and IL-18) and antinociceptive properties (IL-1RA)[Citation42].
Another profile of DEGs was also up-regulated twice and the first time occurred 12 hours to 1 day after CCI. In contrast to the DEGs of the above profile, the GO and KEGG pathway enrichment analyses of profile 27 showed that 1) biological processes involved in the activation, proliferation, aggregation, and adhesion of various immune cells were particularly prominent; 2) cellular components included the ‘membrane,’ ‘cell surface,’ ‘external side of the plasma membrane,’ and ‘cell periphery,’ rather than the nucleus; 3) molecular functions were ‘protein binding,’ ‘enzyme binding,’ ‘MHC class II protein binding,’ and ‘MHC class II protein complex binding’; 4) the signaling pathways of the T cell receptor signaling pathway, NK cell-mediated cytotoxicity, leukocyte transendothelial migration, Fc gamma R-mediated phagocytosis, and B cell receptor signaling pathway were mainly immune-related. Results suggested that the expression of these DEGs in profile 27 in the ACC after CCI was inseparable from the role of chemokines. In profile 27, the DEGs included Rac2, Cd68, Icam-1, Ptprc, and Itgb2. Rac2 is a member of the Rho Small GTPase family and regulates the cytoskeletal dynamics, cell shape, migration, adhesion, gene transcription and signal transduction[Citation43]. A previous study identified Rac2 as a crucial gene that participates in the pathological process of neuropathic pain [Citation44,Citation45], which may be related to its regulation of inflammation and immune response [Citation46,Citation47]. Cd68 is a lysosome marker associated with phagocytosis expressed in activated microglia[Citation48]. In the early stages of systemic inflammation, microglia are attracted to vessels to protect the blood-brain barrier integrity by Ccl5-Ccr5 signaling. Once the blood-brain barrier becomes damaged, microglia transformed into a reactive phenotype expressing Cd68, which amplify the neuroinflammation[Citation49]. Icam-1 was identified as a cell surface glycoprotein that regulates inflammation and injury resolution[Citation50]. After SNI, Icam-1 is up-regulated in small extracellular vesicles[Citation51]. Moreover, Icam-1 expressed in the vascular endothelium recruits opioid-containing immune cells to promote analgesia[Citation52]. Futhermore, Icam-1 is closely related to chemokines. After Ccl7 treatment, Icam-1 was significantly increased in human umbilical endothelial cells[Citation53].However, Icam-1 can also inhibit the expression of Ccl2 by up-regulating miR-124, thereby promoting macrophage polarization[Citation54]. Recombinant Protein Tyrosine Phosphatase Receptor Type C (Ptprc) acts as a positive regulator of T-cell coactivation. Ptprc up-regulates in microglia isolated from LPS-injected mice[Citation55]. Itgb2 (Cd18) is a transmembrane glycoprotein that aggravates injury following spinal cord injury[Citation56].Taken together, the initial increase in these DEGs may reflect an early response of the ACC to nerve injury, whereas the later increase of these genes may reflect their involvement in the development of neuropathic pain affection. These results indicated that chemokines and their targeting genes in the ACC are differentially involved in the initiation and maintenance of neuropathic pain affection. Thus, these genes may be a target for not only the nociception but also the pain affection following nerve injury.
We demonstrated that the expression pattern of chemokines in the ACC was similar to that of the spinal cord after CCI. The connection between the ACC and spinal cord is complex and remains poorly understood. Chen injected the retrograde tracer Fluoro-Gold into the cervical spinal cord of mice, and found that labeled neurons were primarily located in layer V of the ACC. After injecting phaseolus vulgaris leucoagglutinin, an anterograde neuronal tracer, and a gene-edited rabies virus respectively into the ACC, labeled neurons were located primarily in lamina I–III of the spinal cord. Moreover, the expression of GluA1 and potentiated AMPA receptor (AMPAR)-mediated postsynaptic responses were increased in the ACC-spinal cord projecting neurons following nerve injury. Further research found that Ca2+-permeable AMPAR antagonist NASPM reverses the potentiation of postsynaptic responses and pain sensitization [Citation57–59]. However, further investigations are needed to clarify the connection between the ACC and spinal cord, and ascertaining the role of chemokines between the ACC and spinal cord may offer profound insight into the mechanisms underlying neuropathic pain.
The main reasons for the high prevalence of neuropathic pain are the lack of clear pathogenesis and effective treatments. Unlike opioids and nonsteroidal anti-inflammatory drugs, which effectively relieve nociceptive pain, drugs currently used for the treatment of neuropathic pain, such as tricyclic antidepressants, 5-hydroxytryptamine/norepinephrine reuptake inhibitors, and ion channel modulators/blockers, are effective in only a portion of patients or during a limited period of the disease[Citation60]. Thus, the comprehensive study of the pathogenesis of neuropathic pain and the development of effective drugs are crucial. Our results showed that chemokines and their targeting genes may be promising targets for the treatment of neuropathic pain. Maraviroc, a Ccr5 antagonist, is the first chemokine receptor-targeted drug approved by the Food and Drug Administration and is currently being used to treat human immunodeficiency virus-1 infections. It has also been shown to reduce tumor growth[Citation61] and neuropathic pain[Citation62]. Plerixafor, known as AMD3100, is in clinical use for hematopoietic stem cell mobilization[Citation63] and can also can alleviate neuropathic pain[Citation64]. Aside from drugs, neutralizing antibodies can also be used for the treatment of neuropathic pain. Mogamulizumab-kpkc, a Ccr4 antagonist, is approved by the FDA for the treatment of adults with mycosis fungoides and Sézary syndrome[Citation65] but may be applied to pain in the future.
5. Conclusion
We found that the ACC was involved not only in emotion regulation but also in connection with inflammatory and immune responses in the development of neuropathic pain. Moreover, chemokines and their targeting genes in the ACC may be differentially involved in the initiation and maintenance of neuropathic pain affection. These genes may be a target for not only nociception but also pain affection following nerve injury.
Highlights
1. Chemokines show different temporal expression kinetics after nerve injury.
2. Chemokines in anterior cingulate cortex are involved in pain affection.
3. Expressional files of chemokines may contribute to strategies of pain treatment.
Consent for publication
No individual person’s data in any form were involved in this study.
Ethics approval and consent to participate
This study was approved by the Institutional Ethics Committee of Central South University
Data sharing statement
The datasets of GSE172133 and GSE175760 can be found in online repositories.
Author contributions
YZ, ZH and XY carried out the experiments. SJ, LF, XH and YZ analyzed data and presentation. YZ and ZH wrote the manuscript. QG participated in conception of the study. CH conducted the study design. All authors read and approved the final version of the manuscript.
Supplemental Material
Download Zip (1.4 MB)Supplementary material
Supplemental data for this article can be accessed here
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Scholz J, Finnerup NB, Attal N, et al. The IASP classification of chronic pain for ICD-11: chronic neuropathic pain. Pain. 2019;160:53–59.
- Taylor AM, Harris AD, Varnava A, et al. Neural responses to a modified Stroop paradigm in patients with complex chronic musculoskeletal pain compared to matched controls: an experimental functional magnetic resonance imaging study. BMC Psychol. 2016;4:5.
- Buffington AL, Hanlon CA, McKeown MJ. Acute and persistent pain modulation of attention-related anterior cingulate fMRI activations. Pain. 2005;113:172–184.
- Fomberstein K, Qadri S, Ramani R. Functional MRI and pain. Curr Opin Anaesthesiol. 2013;26:588–593.
- Zhuo M. Cortical excitation and chronic pain. Trends Neurosci. 2008;31:199–207.
- Luo C, Kuner T, Kuner R. Synaptic plasticity in pathological pain. Trends Neurosci. 2014;37:343–355.
- Zhao MG, Ko SW, Wu LJ, et al. Enhanced presynaptic neurotransmitter release in the anterior cingulate cortex of mice with chronic pain. J Neurosci. 2006;26:8923–8930.
- Xu H, Wu LJ, Wang H, et al. Presynaptic and postsynaptic amplifications of neuropathic pain in the anterior cingulate cortex. J Neurosci. 2008;28:7445–7453.
- Ko HG, Choi JH, Park DI, et al. Rapid turnover of cortical NCAM1 regulates synaptic reorganization after peripheral nerve injury. Cell Rep. 2018;22:748–759.
- Li XY, Ko HG, Chen T, et al. Alleviating neuropathic pain hypersensitivity by inhibiting PKMzeta in the anterior cingulate cortex. Science. 2010;330:1400–1404.
- Wang H, Xu H, Wu LJ, et al. Identification of an adenylyl cyclase inhibitor for treating neuropathic and inflammatory pain. Sci Transl Med. 2011;3:65ra3.
- Zhou Y, Lutz PE, Wang YC, et al. Global long non-coding RNA expression in the rostral anterior cingulate cortex of depressed suicides. Transl Psychiatry. 2018;8:224.
- Akula N, Marenco S, Johnson K, et al. Deep transcriptome sequencing of subgenual anterior cingulate cortex reveals cross-diagnostic and diagnosis-specific RNA expression changes in major psychiatric disorders. Neuropsychopharmacol. 2021;46:1364–1372.
- Kao CY, He Z, Zannas AS, et al. Fluoxetine treatment prevents the inflammatory response in a mouse model of posttraumatic stress disorder. J Psychiatr Res. 2016;76:74–83.
- Logan RW, Xue X, Ketchesin KD, et al. Sex differences in molecular rhythms in the human cortex. Biol Psychiatry. 2022;91(1): 152–162.
- Li M, Su S, Cai W, et al. Differentially expressed genes in the brain of aging mice with cognitive alteration and depression- and anxiety-like behaviors. Front Cell Dev Biol. 2020;8:814.
- Su S, Li M, Wu D, et al. Gene transcript alterations in the spinal cord, anterior cingulate cortex, and amygdala in mice following peripheral nerve injury. Front Cell Dev Biol. 2021;9:634810.
- Bennett GJ, Xie YK. A peripheral mononeuropathy in rat that produces disorders of pain sensation like those seen in man. Pain. 1988;33:87–107.
- Shen Y, Ding Z, Ma S, et al. SETD7 mediates spinal microgliosis and neuropathic pain in a rat model of peripheral nerve injury. Brain Behav Immun. 2019;82:382–395.
- Shen Y, Ding Z, Ma S, et al. Targeting Aurora kinase B alleviates spinal microgliosis and neuropathic pain in a rat model of peripheral nerve injury. J Neurochem. 2020;152:72–91.
- Wang D, An SC, Zhang X. Prevention of chronic stress-induced depression-like behavior by inducible nitric oxide inhibitor. Neurosci Lett. 2008;433:59–64.
- Han M, Xiao X, Yang Y, et al. SIP30 is required for neuropathic pain-evoked aversion in rats. J Neurosci. 2014;34:346–355.
- Pertea M, Kim D, Pertea GM, et al. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–1667.
- Mortazavi A, Williams BA, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628.
- Liu P, Yang S, Wang Z, et al. Feasibility and mechanism analysis of shenfu injection in the treatment of idiopathic pulmonary fibrosis. Front Pharmacol. 2021;12:670146.
- Huang J, Ma J, Wang J, et al. Whole-transcriptome analysis of rat cavernosum and identification of circRNA-miRNA-mRNA networks to investigate nerve injury erectile dysfunction pathogenesis. Bioengineered. 2021;12:6516–6528.
- Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191.
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–d8.
- Liu K, Fu Q, Liu Y, et al. An integrative bioinformatics analysis of microarray data for identifying hub genes as diagnostic biomarkers of preeclampsia. Biosci Rep. 2019;39. DOI:10.1042/BSR20190187
- Wu XB, He LN, Jiang BC, et al. Increased CXCL13 and CXCR5 in anterior cingulate cortex contributes to neuropathic pain-related conditioned place aversion. Neurosci Bull. 2019;35:613–623.
- Harrison NA, Brydon L, Walker C, et al. Neural origins of human sickness in interoceptive responses to inflammation. Biol Psychiatry. 2009;66:415–422.
- Slavich GM, Way BM, Eisenberger NI, et al. Neural sensitivity to social rejection is associated with inflammatory responses to social stress. Proc Natl Acad Sci U S A. 2010;107:14817–14822.
- Holmes SE, Hinz R, Conen S, et al. Elevated translocator protein in anterior cingulate in major depression and a role for inflammation in suicidal thinking: a positron emission tomography study. Biol Psychiatry. 2018;83:61–69.
- Wang Z, Song K, Zhao W, et al. Dendritic cells in tumor microenvironment promoted the neuropathic pain via paracrine inflammatory and growth factors. Bioengineered. 2020;11:661–678.
- Miu J, Mitchell AJ, Müller M, et al. Chemokine gene expression during fatal murine cerebral malaria and protection due to CXCR3 deficiency. J Immunol. 2008;180:1217–1230.
- Kwiatkowski K, Piotrowska A, Rojewska E, et al. Beneficial properties of maraviroc on neuropathic pain development and opioid effectiveness in rats. Prog Neuropsychopharmacol Biol Psychiatry. 2016;64:68–78.
- Liou JT, Yuan HB, Mao CC, et al. Absence of C-C motif chemokine ligand 5 in mice leads to decreased local macrophage recruitment and behavioral hypersensitivity in a murine neuropathic pain model. Pain. 2012;153:1283–1291.
- Liou JT, Mao CC, Ching-Wah Sum D, et al. Peritoneal administration of Met-RANTES attenuates inflammatory and nociceptive responses in a murine neuropathic pain model. J Pain. 2013;14:24–35.
- Wu XB, He LN, Jiang BC, et al. Spinal CXCL9 and CXCL11 are not involved in neuropathic pain despite an upregulation in the spinal cord following spinal nerve injury. Mol Pain. 2018;14:1744806918777401.
- Raghu H, Lepus CM, Wang Q, et al. CCL2/CCR2, but not CCL5/CCR5, mediates monocyte recruitment, inflammation and cartilage destruction in osteoarthritis. Ann Rheum Dis. 2017;76:914–922.
- Tacke F, Alvarez D, Kaplan TJ, et al. Monocyte subsets differentially employ CCR2, CCR5, and CX3CR1 to accumulate within atherosclerotic plaques. J Clin Invest. 2007;117:185–194.
- Pawlik K, Piotrowska A, Kwiatkowski K, et al. The blockade of CC chemokine receptor type 1 influences the level of nociceptive factors and enhances opioid analgesic potency in a rat model of neuropathic pain. Immunology. 2020;159:413–428.
- Wennerberg K, Der CJ. Rho-family GTPases: it’s not only Rac and Rho (and I like it). J Cell Sci. 2004;117:1301–1312.
- Yang YK, Lu XB, Wang YH, et al. Identification crucial genes in peripheral neuropathic pain induced by spared nerve injury. Eur Rev Med Pharmacol Sci. 2014;18:2152–2159.
- Wang J, Ma SH, Tao R, et al. Gene expression profile changes in rat dorsal horn after sciatic nerve injury. Neurol Res. 2017;39:176–182.
- Diebold BA, Bokoch GM. Molecular basis for Rac2 regulation of phagocyte NADPH oxidase. Nat Immunol. 2001;2:211–215.
- Dooley JL, Abdel-Latif D, St Laurent CD, et al. Regulation of inflammation by Rac2 in immune complex-mediated acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2009;297:L1091–102.
- Zotova E, Holmes C, Johnston D, et al. Microglial alterations in human Alzheimer’s disease following Aβ42 immunization. Neuropathol Appl Neurobiol. 2011;37:513–524.
- Haruwaka K, Ikegami A, Tachibana Y, et al. Dual microglia effects on blood brain barrier permeability induced by systemic inflammation. Nat Commun. 2019;10:5816.
- Bui TM, Wiesolek HL, Sumagin R. ICAM-1: a master regulator of cellular responses in inflammation, injury resolution, and tumorigenesis. J Leukoc Biol. 2020;108:787–799.
- Jean-Toussaint R, Tian Y, Chaudhuri AD, et al. Proteome characterization of small extracellular vesicles from spared nerve injury model of neuropathic pain. J Proteomics. 2020;211:103540.
- Machelska H, Mousa SA, Brack A, et al. Opioid control of inflammatory pain regulated by intercellular adhesion molecule-1. J Neurosci. 2002;22:5588–5596.
- Chen J, Zhang B, Pan C, et al. Effects of monocyte chemotactic protein-3 on ICAM-1, VCAM-1, TF, and TFPI expression and apoptosis in human umbilical vein endothelial cells. Nan Fang Yi Ke Da Xue Xue Bao. 2013;33:86–92.
- Gu W, Yao L, Li L, et al. ICAM-1 regulates macrophage polarization by suppressing MCP-1 expression via miR-124 upregulation. Oncotarget. 2017;8:111882–111901.
- Sousa C, Golebiewska A, Poovathingal SK, et al. Single-cell transcriptomics reveals distinct inflammation-induced microglia signatures. EMBO Rep. 2018;19. DOI:10.15252/embr.201846171
- Gris D, Marsh DR, Oatway MA, et al. Transient blockade of the CD11d/CD18 integrin reduces secondary damage after spinal cord injury, improving sensory, autonomic, and motor function. J Neurosci. 2004;24:4043–4051.
- Tsuda M, Koga K, Chen T, et al. Neuronal and microglial mechanisms for neuropathic pain in the spinal dorsal horn and anterior cingulate cortex. J Neurochem. 2017;141:486–498.
- Chen T, Koga K, Descalzi G, et al. Postsynaptic potentiation of corticospinal projecting neurons in the anterior cingulate cortex after nerve injury. Mol Pain. 2014;10:33.
- Chen T, Wang W, Dong YL, et al. Postsynaptic insertion of AMPA receptor onto cortical pyramidal neurons in the anterior cingulate cortex after peripheral nerve injury. Mol Brain. 2014;7:76.
- Cohen SP, Mao J. Neuropathic pain: mechanisms and their clinical implications. BMJ. 2014;348:f7656.
- Nie Y, Huang H, Guo M, et al. Breast phyllodes tumors recruit and repolarize tumor-associated macrophages via secreting CCL5 to promote malignant progression, which can be inhibited by CCR5 inhibition therapy. Clin Cancer Res off J Am Assoc Cancer Res. 2019;25:3873–3886.
- Piotrowska A, Kwiatkowski K, Rojewska E, et al. Maraviroc reduces neuropathic pain through polarization of microglia and astroglia - Evidence from in vivo and in vitro studies. Neuropharmacology. 2016;108:207–219.
- Hoggatt J, Singh P, Tate TA, et al. Rapid mobilization reveals a highly engraftable hematopoietic stem cell. Cell. 2018;172:191–204.e10.
- Liu ZY, Song ZW, Guo SW, et al. CXCL12/CXCR4 signaling contributes to neuropathic pain via central sensitization mechanisms in a rat spinal nerve ligation model. CNS Neurosci Ther. 2019;25:922–936.
- Kasamon YL, Chen H, de Claro RA, et al. FDA approval summary: mogamulizumab-kpkc for mycosis fungoides and sézary syndrome. Clin Cancer Res off J Am Assoc Cancer Res. 2019;25:7275–7280.