Abstract
Renal cell carcinoma (RCC) is one of the three most common cancers of urinary tract cancer, accounting for 2–3% of all systemic cancers. Recent studies have found that miR-199a is lowly expressed in RCC, may act as a tumour suppressor gene to induce the occurrence of kidney cancer. In the present study, we investigated the role of miR-199a in the progression and metastasis of RCC. The results showed that miR-199a significantly downregulated in RCC and cell lines. Overexpression of miR-199a in RCC cell lines significantly inhibited cell proliferation, migration and invasion. Furthermore, the qRT-PCR and western blot results showed that miR-199a overexpression significantly downregulated ROCK-1 mRNA and protein levels. ROCK1 was identified as a target of miR-199a, and ectopic expression of miR-199a downregulated ROCK1 by direct binding to its 3′ untranslated region. Together, these findings indicate that miR-199a acts as a tumour suppressor and its downregulation in tumour tissues may contribute to the progression and metastasis of RCC through a mechanism involving ROCK1, suggesting miR-199a as a potential new diagnostic and therapeutic target for the treatment of RCC.
Introduction
Renal cancer is the seventh and eighth most common cancer and tenth most common cause of cancer death. Surgery is still the only definitive treatment for renal cell carcinoma (RCC). However, about one-third of RCC patients are presented with metastases at diagnosis and up to 50% of patients develop metastatic disease despite of surgical intervention [Citation1]. There are few options for not curative treatment of metastatic RCC including immunotherapy [Citation2] and recently developed molecular targeting drugs [Citation3,Citation4]. New therapeutic targets are urgently needed to be identified in RCC to develop more effective therapeutic approaches for the treatment of this deadly disease.
MicroRNAs (miRNAs) is a group of small and non-coding RNAs with the length of about 22 nt [Citation5]. MiRNAs can regulate genes expression by binding to their target mRNA at 3′-UTR [Citation6]. It was reported that a variety of miRNAs are located on the cancer-related regions in genome, which could act as tumour suppressors or oncogenes in cancers [Citation7]. In the previous studies, numerous miRNAs were confirmed to be correlated with initiation and development of kidney cancers, such as miR-153, miR-148a, miR-206 and so on [Citation8–10]. Among the miRNAs, miR-199 family is of great interest for cancer therapies because it is associated with various tumours including prostate cancer, breast cancer, medulloblastoma and osteosarcoma. However, the mechanistic details that are therapeutically relevant regarding the role of miR-199 in kidney cancer are not known. Additionally, a related study demonstrated that miR-199a could regulate cell behaviours, such as proliferation, migration and clonogenicity in breast cancer cell lines [Citation11]. Previous study showed that the expression of miR-199a was negatively correlated with tumour recurrence in RCC patients in T stage (p < .05). The patients with lowered miR-199a expression in the tumour tissue had a significantly shorter mean survival time than those without miR-199a down regulation (p = .017 by log-rank test). The miR-199a can serve as a promising prognostic factor of RCC [Citation12].
Rho-associated protein kinase 1 (ROCK1), which belongs to the AGC family of serine/threonine protein kinases, plays an important role in the regulation of the actin cytoskeleton through the phosphorylation of downstream substrates leading to actin filament stabilization and the modulation of actin-myosin contractility [Citation13]. Increased expression of ROCK1 has been described in several human cancers and has been correlated with poor survival in breast cancer and in renal cancer [Citation13]. Sequestration of activated ROCK1 into cancer cells prevented ROCK1 from interacting with JNK-interacting protein 3 (JIP-3) and its activation of c-Jun N-terminal kinase (JNK), a pathway triggering apoptosis, thereby protecting cells from apoptosis [Citation14].
In the present study, for the first time, we show miR-199a as a negative regulator of ROCK1-mediated renal cancer cell survival and proliferation. Moreover, we found that decreased expression of miR-199a was significantly correlated with higher tumour stage and overexpression of ROCK1 in RCCs. Our results suggest ROCK1 as a direct target of miR-199a and showed that miR-199a functions as a tumour suppressor by downregulating ROCK1, providing a potential diagnostic and therapeutic target for the treatment of renal cancer.
Materials and methods
Patient and specimens collection
This study was approved by the Ethics Committee of China-Japan Union Hospital of Jilin University, and the written informed consents were obtained from all patients or their families. Cancerous tissues and matched non-cancerous tissue specimens were collected from renal cancer patients who were all conformed by pathologists in China-Japan Union Hospital of Jilin University of Medicine between January 2007 and December 2009. None of the patients received chemotherapy or radiotherapy before specimens collection. The collected tissue samples were immediately frozen in liquid nitrogen and then stored at –80 °C until RNA extraction. After surgery, the patients were enrolled in a five-year follow-up investigation. The clinicopathological features and survival information were recorded, including age, tumour size, ER status, PR status, histological type, TNM stage and lymph node metastasis.
Analysis of miR-199a and ROCK1 expression
Fresh surgical specimens of paired malignant and normal renal tissue were immersed in RNAlater (Applied Biosystems, Tokyo, Japan) tissue storage solution and stored at –80 °C until further use. Total cellular RNA was extracted using the mirVana miRNA Isolation Kit (Applied Biosystems, Tokyo, Japan). TaqMan® MicroRNA Assays (has-miR-199a and RNU6B, Applied Biosystems, Tokyo, Japan) were performed to determine the quantity of mature miRNA. Q-PCR was performed using the 7300 Real Time PCR System (Applied Biosystems, Tokyo, Japan) with a TaqMan® Universal PCR Master Mix (Applied Biosystems, Tokyo, Japan) according to the standard protocol. The expression of miRNAs was calculated using the comparative 2–ΔΔCt method with RNU6B as endogenous control, to normalize miRNAs expression levels. Each reaction was run in triplicate and mean with SD was calculated.
Cell culture and reagents
Renal cell cancer cell lines ACHN and A498 were purchased from ATCC. The cells were cultured as described previously [Citation15]. For miR-199a reintroduction, RCC cell lines were transfected with 30 nmol/L of precursor miRNA, pre-miR-199a (Applied Biosystems Japan, Tokyo, Japan). Transfection was performed with Lipofectamine 2000 (Invitrogen Japan, Tokyo, Japan) according to the manufacture’s instruction. Samples were collected at 24, 48, 72, 96 and 120 h after the transfection and were stored at –80 °C for subsequent analysis. Cell viability was detected with the CellTiter 96® Aqueous One Solution Cell Proliferation Assay (Promega, Madison, WI), a colorimetric assay utilizing the tetrazolium compound according to the manufacturer’s protocol.
Cell proliferation assay
We performed MTT assay to evaluate the effects of miR-199a on proliferation of A498 cells. After transfection, 200 μL cell suspension were added into 96-well plates (2 × 103 cells/well) for continual three days in a 5% CO2 incubator at 37 °C. Cell number was evaluated using MTT assay every 24 h. 20 μL MTT (Sigma-Aldrich, St. Louis, MO; 0.5 mg/mL) was added to each well, and incubated for 1 h continually. Then 150 μL DMSO (Sigma-Aldrich, St. Louis, MO) was added to each well following low speed oscillation for 10 min. The absorbance values were evaluated at 570 nm with a MTT enzyme-linked immune monitor (Thermo Fisher Scientific, Waltham, MA). Every experiment was repeated at least three times.
Cell migration and invasion assays
The invasion ability of transfected A498 cells was performed by using 24-well transwell chamber and matrigel (Corning, Acton, MA). Matrigel was diluted with serum-free medium (1:3), and then added 25 μL migration into the upper well of the Transwell for 30 min at 37 °C. After digested, the transfected cells were resuspended with serum-free medium. One hundred microlitres cell suspension (5 × 105/mL) were added into upper well of the Transwell and 500 μL FBS were added into the lower chamber. Then, the assay plates were incubated at 37 °C, 5% CO2. The cells through the matrigel were erased with swab and then counted under high magnification. Every experiment selected at least three visions.
Luciferase assay
A 293 bp fragment from the 3′UTR of ROCK1 containing the miR-199a binding sites was cloned into the psiCHECK2 vector (Promega, Madison, WI). Mutant (Mut) constructs were generated by mutating the seed region of the miR-199a binding site. miR-199a expressing or control cells were cultured in 24-well plates, and transfected with 100 ng luciferase reporter plasmid and 5 ng pRL-TK vector expressing the Renilla luciferase (Promega, Madison, WI) using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA). After 48 h, cells were harvested, lysed and luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) according to the manufacturer’s protocol.
Statistical analysis
The statistical analyses were conducted using the IBM-SPSS statistical software package (version 19.0; IBM Corporation, Armonk, NY). Continuous data were compared using the unpaired t-test. The expression level of miR-199a was expressed as mean ± SD and analysed by Student’s t-test. A chi-square test was applied to evaluate the association between miR-199a expression and clinicopathological characteristics. The overall survival of each group was estimated by Kaplan–Meier’s curves. The log-rank test was used to compare survival between two groups. The effects of variables on the risk of death were modelled using the Cox proportional hazards regression. All tests were two-sided and statistical significance was defined as p < .05.
Results
The expression levels of miR-199a
One hundred and fifty patients pathologically diagnosed with kidney cancer were collected in the study, with the average age of 54 ± 11 years. The basic characteristics of the study subjects are listed in . qRT-PCR was performed to detect the miR-199a expression in kidney cancer tissues and matched non-cancerous tissues. As shown in , miR-199a expression was significantly lower in kidney cancer tissues than that in matched non-cancerous tissues (p < .05).
Figure 1. miR-199a expression levels in collected tissues specimens. miR-199a expression levels were significantly lower in kidney cancer tissues than that in the non-cancerous tissues. *p < .05.
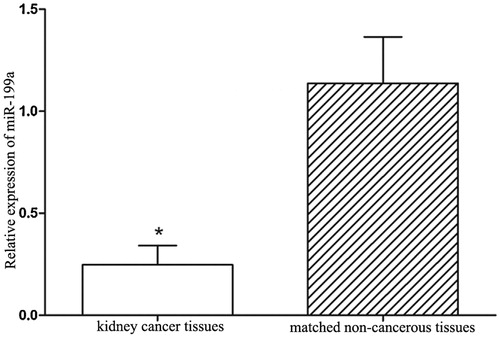
Table 1. Association between miR-199a expression and clinicopathological features in kidney cancer patients.
MiR-199a suppresses renal cancer cell proliferation, migration and invasion
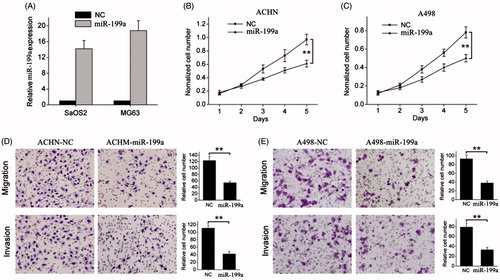
To explore the biological significance of miR-199a in renal cancer tumourigenesis, we established miR-199a stably expressing cell lines of ACHN and A498 by lentivirus infection. Increased expression of miR-199a was confirmed by qRT-PCR (). We found that overexpression of miR-199a significantly decreased the proliferation of ACHN and A498 cells compared to their corresponding controls (). Furthermore, transwell assays showed that miR-199a overexpression could suppress that migratory and invasive abilities of ACHN and A498 cell lines (). Taken together, these data demonstrate that miR-199a suppresses in vitro proliferation, migration and invasion of renal cancer cells.
Figure 2. miR-199a suppresses renal cancer cell growth and metastasis. (A) Successful overexpression of miR-199a was confirmed by qRT-PCR after infection with miR-199a-expressing or vector control lentivirus. (B, C) Cell proliferation was measured by WST-1 assay at different time points. (D, E) Transwell migration and invasion assays. Representative images are shown in the left, and the quantification of 10 randomly selected fields is shown in the right. **p < .01.
ROCK1 is a target gene of miR-199a
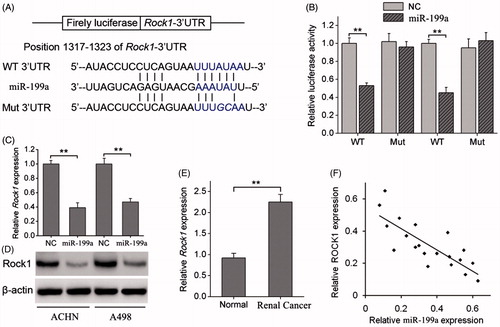
To elucidate the underlying mechanisms by which miR-199a exerts its function, we explored miR-199a targets using the TargetScan bioinformatics algorithm. Our analysis revealed that ROCK1 was a potential target of miR-199a based on putative target sequences at position 1317–1323 of the ROCK1 3′UTR (). Luciferase reporter assays showed that miR-199a significantly decreased the luciferase activity of the ROCK1 3′UTR but not that of the mutant in ACHN and A498 cells (). qRT-PCR and WB analysis showed that overexpression of miR-199a significantly downregulated the expression of ROCK1 at the mRNA and protein levels in both renal cancer cell lines (). The association between ROCK1 and renal cancer further examined by analysing the relative expression levels of ROCK1 in tissues from 20 renal patients, which showed that ROCK1 levels were more than two fold higher in renal cancer tissues than in adjacent normal tissue (). Then, we correlated ROCK1 with miR-199a expression in the same renal cancer specimens. As shown in , a significant inverse correlation was observed by Spearman’s correlation analysis between the mRNA levels of miR-199a and ROCK1 (R = –0.837, p < .001; ). Taken together, these data strongly suggest that ROCK1 is a direct target of miR-199a in renal cancer.
Figure 3. miR-199a negatively regulates ROCK1 by binding to the ROCK1 3′UTR. (A) Diagram of the ROCK1 3′UTR-containing reporter construct. Schematic representation of the miR-199a sequence, putative miR-199a targeting site in the 3′UTR of ROCK1, and the generated mutant ROCK1 3′UTR. (B) A luciferase reporter assay showed the inhibitory effect of miR-199a on ROCK1–3′UTR luciferase activity of ACHN and A498 cells. (C) ROCK1 mRNA levels were analysed by qRT-PCR in ACHN and A498 cells stably overexpressing miR-199a. (D) ROCK1 protein levels were analysed by Western blotting in ACHN and A498 stably overexpressing miR-199a. (E) Relative expression levels of ROCK1 in renal cancer tissues and adjacent normal tissues. (F) Correlation of ROCK1 expression to miR-199a expression in 20 renal cancer using simple linear regression analysis. **p < .01.
Discussion
MiRNAs have a significant role in regulating cellular activities. They can profoundly affect the expression of a large number of genes that encode proteins. Expression level of miR-199a varies among related cancers, resulting in different effects, such as miR-199a acts as a protective factor in HCC while it can induce cell proliferation in kidney cancer. MiR-199a had been reported to be upregulated in gastric cancer (GC) tissues [Citation16]. Using miRNA array, several studies demonstrated miR-199a downregulation in RCCs as compared to its normal kidney counterparts [Citation17,Citation18].
In the current study, the expression levels of miR-199a in renal cancer tissues and adjacent normal tissues were detected by using qRT-PCR. Analysis results showed that miR-199a was down-regulated in cancerous tissues. Moreover, its decreased level was significantly associated with advanced TNM stage and positive lymph node metastasis. The results above might reveal that miR-199a acted as a tumour suppressor gene and was involved in the tumour progression of renal cancer. This conclusion was consistent with the previous studies. In addition, cell experiments were performed to evaluate the effects of miR-199a on physiological processes of renal cancer cells. Results and MTT and Transwell analysis indicated that overexpression of miR-199a could inhibit renal cancer cell proliferation and invasion. Fang et al. had reported that miR-199a controlled cell behaviours in breast cancer cells via regulating HER2 expression [Citation19]. However, the mechanisms for miR-199a regulating renal cancer cell behaviours needed to be investigated.
Identification of miR-199a targets is critical for understanding its roles in tumourigenesis, and also is essential for exploring novel therapeutic targets. Previous studies indicated that several target genes of miR-199a have been identified, such as SHMT1 in lung adenocarcinoma, FUT8 in colorectal cancer [Citation20], FGFR1 in non-small-cell lung cancer [Citation21] and HGF/c-MET pathway in hepatocellular carcinoma [Citation22]. However, the detailed molecular mechanism responsible for the roles of miR-199a in RCC remains unclear. In this study, ROCK1 was identified as a novel target of miR-199a. Bioinformatics analysis revealed that the 3′UTR of ROCK1 has a complementary site for the seed region of miR-199a. In addition, we showed that a miR-199a is directly bound to the 3′UTR of ROCK1, which contains a miR-199a-binding sites using a luciferase reporter assay. Moreover, qRT-PCR and western blot analysis indicated that miR-199a over expression decreased ROCK1 expression in RCC cells.
Although the exact mechanism underlying the miRNA-mediated regulation of ROCK1 is not clear, ROCK1 has been identified as a target of several miRNAs involved in carcinogenesis and tumour progression. In GC, overexpression of miR-199a significantly inhibited GC invasion and metastasis by directly targeting the 3′UTR of ROCK1 [Citation23]. The miRNA regulation of ROCK1 expression was also shown in bladder cancer, where miR-1280 acts as a tumour suppressor by directly targeting ROCK1 and inhibiting the migration/invasion of bladder cancer cells [Citation24]. More recently, miR-148a was reported to suppress epithelial-to-mesenchymal transition by targeting ROCK1 in non-small cell lung cancer cells. These reports support the present findings that the growth and invasion of renal cancer cells may be, in part, regulated by miR-199a modulation of ROCK1 expression.
In conclusion, down-regulated miR-199a is associated with aggressive clinical characteristics in renal cancer patients and its ectopic expression inhibited cell proliferation, migration/invasion, tumour growth and metastasis. The tumour-suppressor function of miR-199a was mediated by the downregulation of its downstream target gene ROCK1. Our study provides novel insight into the mechanisms of renal cancer growth, identifies miR-199a as a negative regulator of ROCK1-mediated renal cancer cell survival and proliferation, and suggests re-expression of miR-199a as a new potential therapeutic approach in renal cancer.
Disclosure statement
No potential conflict of interest was reported by the author.
References
- YuCai B, Xu H, Xu Z, et al. Expressions of stem cell transcription factors Nanog and Oct4 in renal cell carcinoma tissues and clinical significance. Artif Cells Nanomed Biotechnol. 2016;44:1818–1823.
- Bukowski RM. Cytokine therapy for metastatic renal cell carcinoma. Semin Urol Oncol. 2001;19:148–154.
- Motzer RJ, Bukowski RM. Targeted therapy for metastatic renal cell carcinoma. J Clin Oncol. 2006;24:5601–5608.
- Motzer RJ, Molina AM. Targeting renal cell carcinoma. J Clin Oncol. 2009;27:3274–3276.
- Wang M, Xie R, Si H, et al. Integrated bioinformatics analysis of miRNA expression in osteosarcoma. Artif Cells Nanomed Biotechnol. 2017;45:936–943.
- Liu Y, Han L, Bai Y, et al. Down-regulation of microRNA-133 predicts poor overall survival and regulates the growth and invasive abilities in glioma. Artif Cells Nanomed Biotechnol. 2017 [Apr 4];[1–5]. doi: 10.1080/21691401.2017.1304551
- Palmero EI, de Campos SG, Campos M, et al. Mechanisms and role of microRNA deregulation in cancer onset and progression. Genet Mol Biol. 2011;34:363–370.
- Wang B, Teng Y, Liu Q. MicroRNA-153 regulates NRF2 expression and is associated with breast carcinogenesis. Clin Lab. 2016;62:39–47.
- Xu X, Zhang Y, Jasper J, et al. MiR-148a functions to suppress metastasis and serves as a prognostic indicator in triple-negative breast cancer. Oncotarget. 2016;7:20381–20394.
- Yin K, Yin W, Wang Y, et al. MiR-206 suppresses epithelial mesenchymal transition by targeting TGF-beta signaling in estrogen receptor positive breast cancer cells. Oncotarget. 2016;7:24537–22448.
- Andolfo I, Liguori L, De AP, et al. The micro-RNA 199b-5p regulatory circuit involves Hes1, CD15, and epigenetic modifications in medulloblastoma. Neuro Oncol. 2012;14:596–612.
- Si T, Liu C, Xu K, et al. Association of miR-199a expression with clinicopathologic characteristics and prognosis of renal cell carcinoma. J Southern Med Univ. 2012;32:1568–1571.
- Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11:9–22.
- Tsai NP, Wei LN. RhoA/ROCK1 signaling regulates stress granule formation and apoptosis. Cell Signal. 2010;22:668–675.
- BillimOugolkov V, Yuuki A, Naito K, et al. Glycogen synthase kinase-3: a new therapeutic target in renal cell carcinoma. Br J Cancer. 2009;101:2005–2014.
- Ougolkov AV, Bone ND, Fernandez-Zapico ME, et al. Inhibition of glycogen synthase kinase-3 activity leads to epigenetic silencing of nuclear factor-kappa B target genes and induction of apoptosis in chronic lymphocytic leukemia B cells. Blood. 2007;110:735–742.
- Juan D, Alexe G, Antes T, et al. Identification of a MicroRNA panel for clear-cell kidney cancer. Urology. 2010;75:835–841.
- Liu H, Brannon A, Reddy A, et al. Identifying mRNA targets of microRNA dysregulated in cancer: with application to clear cell renal cell carcinoma. BMC Syst Biol. 2010;4:51–67.
- Fang C, Zhao Y, Guo B. MiR-199b-5p targets HER2 in breast cancer cells. J Cell Biochem. 2013;114:1457–1463.
- WuZhang S, Li G, Chen P, et al. miR-198 targets SHMT1 to inhibit cell proliferation and enhance cell apoptosis in lung adenocarcinoma. Tumor Biol. 2016;4:5193–5202.
- Wang M, Wang J, Kong X, et al. MiR-198 represses tumor growth and metastasis in colorectal cancer by targeting fucosyl transferase 8. Sci Rep. 2014;4:6145.
- Tan S, Li R, Ding K, et al. miR-198 inhibits migration and invasion of hepatocellular carcinoma cells by targeting the HGF/c-MET pathway. FEBS Lett. 2011;585:2229–2234.
- Zheng B, Liang L, Wang C, et al. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res. 2011;17:7574–7583.
- Majid S, Dar AA, Saini S, et al. MicroRNA-1280 inhibits invasion and metastasis by targeting ROCK1 in bladder cancer. PLoS One. 2012;7:e46743.
