Abstract
Though long non-coding RNA LINC00858 (LINC00858) has been shown to be involved in tumours of other tissues, its involvement in colorectal cancer (CRC) is still unknown. We aimed to investigated expression and mechanism LINC00858 in human CRC. In this study, we firstly found that LINC00858 expression was significantly up-regulated in both CRC tissues and cell lines by both online data and RT-PCR assay. Then, clinical assay revealed that high LINC00858 expression was significantly associated with advanced clinical progression and poor prognosis. Multivariate analysis demonstrated that high LINC00858 expression was an independent poor prognostic factor for CRC patients. Moreover, lost-of-function assay indicated that knockdown of LINC00858 suppressed CRC cells proliferation, migration and invasion, and promoted apoptosis. Mechanistically, bioinformatics analysis, dual-luciferase reporter assays, and western blot assays showed that LINC00858 functioned as competing endogenous RNA to repress miR-22-3p, which controlled its down-stream target YWHAZ. Then, we suggested that LINC00858 exerted its function through the miR-22-3p/YWHAZ axis. To our knowledge, this is the first report which showed the role of LINC00858 in the progression of CRC. Our findings indicated that LINC00858 played an important role in CRC, and may serve as a novel prognostic factor and therapeutic target.
Introduction
Colorectal cancer (CRC) is one of the most lethal human malignancies and causes large mortalities around the world each year [Citation1]. It is projected that 412,000 individuals will be diagnosed with CRC in 2017 in China [Citation2, Citation3]. Despite great advances in chemotherapy and surgical techniques, the survival rate of CRC patients still remains unsatisfied [Citation3, Citation4]. Five-year survival rate of patients with CRC was 90.1% for patients at early stage and only 11.7% for patients with distant metastasis [Citation5]. Total survival is low, emphasizing the rationale for new preventative approaches. Thus, it is extremely necessary to identify novel and sensitive biomarkers used in the diagnosis and as therapeutic targets for human CRC.
Long non-coding RNAs (lncRNAs), initially thought to represent spurious transcriptional noise, are RNA molecules that is >200 nucleotides long and is not translated into a protein [Citation6]. Growing evidence shows that lncRNAs play functional roles in many biological processes such as cell development, proliferation, metastasis, differentiation and migration [Citation7, Citation8]. It has been confirmed that lncRNAs are dysregulated in a wide range of human diseases and disorders, including various cancers, suggesting that lncRNAs may be used as potential biomarkers for diagnosis and prognosis of cancer patients and novel therapeutic targets [Citation9–11]. Recent years, some lncRNAs have been well studied in various tumours, such as lncRNA MALAT1, lncRNA ANRIL and lncRNA HOTAIR [Citation12–14]. However, the expression pattern and potential function of most lncRNAs in CRC remain largely unclear.
Recently, growing evidence indicated that lncRNAs could competitively suppress miRNAs by acting as molecular sponges [Citation15, Citation16]. For instance, lncRNA TP73-AS1 promoted NSCLC cell proliferation, tumour growth and cycle progression by competitively sponging miR-449a [Citation17]. LncRNA FBXL19-AS1 plays oncogenic role in CRC by sponging miR-203 [Citation18]. The cross-regulation between lncRNAs and microRNAs highlights their influences on the development and progression of CRC and encouraged us to further study the potential mechanism.
LncRNA LINC00858 (LINC00858), a newly identified lncRNA, is locates in 10q23.1. Recently, the dysregulation of LINC00858 has been reported in two tumours, including osteosarcoma and lung cancer [Citation19, Citation20]. In addition, in these two tumours, LINC00858 played a positive regulator. However, to our best knowledge, the expression and function of LINC00858, as well as its potential mechanism in CRC, have not been investigated. In this study, we found that LINC00858 was highly expressed in CRC and its knockdown could suppressed CRC cells proliferation, migration and invasion by modulating miR-22-3p. This study adds to our understanding of CRC pathogenesis.
Materials and methods
Patients and tissue specimens
A total of 115 paired CRC tissues and their corresponding non-tumour normal tissue specimens were collected between January 2010 to December 2011 following surgical resection at Chongqing Yangdu Biology Institute and Chongqing No.324 hospital. None of the patients with CRC had received local or systemic therapy before surgery. The clinicopathological information of the patients was shown in . Resected CRC tissue specimens and corresponding normal tissue specimens were immediately snap frozen using liquid nitrogen and stored at −80 °C. The informed consent was obtained from all patients and the study was approved by the Ethics Committee of Chongqing Yangdu Biology Institute and Chongqing No.324 hospital.
Table 1. The Primer sequences included in this study.
Table 2. Correlation between LINC00858 expression and clinicopathological characteristics of CRC.
Cell lines and cell culture
Human CRC cell lines (HT-29, HT-15, SW837 and SW1463) and the normal colonic mucosa epithelial cell line (FHC) were purchased from the Cell Bank of the Chinese Academy of Sciences (Xuhui, Shanghai, China). Above cells were cultured using RPMI 1640 medium (BasalMedia Technologies, Shanghai, China) supplemented with 10% foetal bovine serum (FBS; Life Technologies, Carlsbad, CA) and streptomycin–penicillin antibiotic solution (Solarbio, Tongzhou, Beijing, China) at 37˚C with 5% CO2.
Cell transfection
Small interfering RNAs (siRNAs) specific targeting LINC00858 (si-LINC00858-1 and si-LINC00858-2), negative control siRNA (si-NC), miR-22-3p mimic, negative control mimic (NC mimic), miR-22-3p inhibitor and negative control inhibitor (NC inhibitor) were all purchased from Annoron Biotech Co., Ltd. (Yizhuang, Beijing, China). A plasmid vector, pcDNA3.1-LINC00858, which could consistently express LINC00858, was constructed by Vigene Biosciences Co., Ltd. (Jinang, Shandong, China) via standard molecular cloning approaches. Cell transfection of siRNA, miRNA mimic or inhibitor, plasmid vectors were all conducted using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocols.
Quantitative real-time PCR
RNA was isolated using Trizol reagents (Invitrogen, Carlsbad, CA), according to the manufacturer’s protocols. Quantitative RT-PCR was utilized to determine the levels of LINC00858, miR-22-3p and YWHAZ. All primers which were synthesized by Generay Biotech Co., Ltd (Songjiang, Shanghai, China) and the sequences were listed in . The qRT-PCR assays were all performed using a Quant Studio 6 Flex real-time PCR instrument (Thermo Fisher Scientific, Pudong, Shanghai, China). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 was used as internal reference for mRNA or miRNA, respectively. One Step qRT-PCR Kit (KangLang, Minhang, Shanghai, China) or TransScript Green miRNA Two-Step qRT-PCR SuperMix Kit (TranGen, Haidian, Beijing, China) were applied to detect mRNA or miRNA, respectively. The relative expression was analyzed using 2−△△Ct method. All experiments were performed in triplicate.
Cell proliferation assay
The cell proliferative rates of difference groups were assessed by Cell Counting Kit-8 kit (CCK-8; Solarbio, Tongzhou, Beijing, China). In brief, HT-29 or SW837 cells (at a density of 1 × 104 cells per well) were seeded into 96-well plates (ExcellBio, Taicang, Jiangsu, China) and continued to culture for 24 h. Afterwards, 10 µl CCK-8 solution were added into each well and the cells were maintained at 37 °C with 5% CO2 for additional 1–2 h. finally, A spectrophotometer was used to measure the absorbance at 450 nm.
Colony formation assay
Cells (500 cells per well) transfected with indicated siRNAs were seeded in 6-well plates (ExcellBio, Taicang, Jiangsu, China) and incubated at 37 °C with 5% CO2 for appropriate two weeks. Subsequently, the cells were fixed with ethanol and stained with 0.1% crystal violet (Sigma‐Aldrich, St Louis, MO). The number of visible colonies was counted manually using an inverted microscope (Leica MZ8, Leica Microsystems, Wetzlar, Germany).
Apoptosis assay
Flow cytometry analysis was utilized to analyse the apoptosis of HT-29 and SW837 cells using an Annexin V/propidium iodide (PI) Apoptosis Assay Kit (KeyGEN BioTech, Jiangning, Nanjing, China). Briefly, the cells were resuspended in the binding buffer and subsequently incubated with Annexin V as well as PI. Then, the cells were kept in in the dark at room temperature for 15 min. Afterwards, an FACS Calibur system (BD Biosciences, San Jose, CA) was utilized to detect the cell apoptosis. Flow Jo 7.6.1 software (Tree Star Software, San Carlos, CA) was applied to analyse the data.
Western blot analysis
Radio immunoprecipitation assay (RIPA) buffer (Beyotime, Pudong, Shanghai, China) was applied to extract the total proteins from cell lysates. The protein concentration was determined by a Bicinchoninic Acid (BCA) protein assay kit (Meilun Biotechnology, Dalian, Liaoning, China). Subsequently, protein was separated on SDS-PAGE gel (8–12%) and transferred on PVDF membranes (ThermoFisher Scientific, Waltham, MA). After blocking with 5% non-fat milk for 24 h, the membranes were incubated with primary antibodies. The primary antibodies for GAPDH, vimentin and N-Cadherin were purchased from Sinobiological.com (Yizhuang, Beijing, China). The primary antibodies for YWHAZ was purchased from Abcam Co., Ltd (Cambridge, MA). After the membranes were washed with TBST for three times, secondary antibodies were incubated with the membranes. The protein bands were visualized by ECL detection kit (Pierce, Rockford, IL) and a Gel Imaging System (GE Healthcare, Pudong, Shanghai, China). Moreover, the optical density of the protein bands was analyzed by ImageJ software (version 1.46; NIH, Bethesda, MD).
Wound healing and transwell invasion assays
The cell migratory and invasive abilities were assessed by wound-healing assays and transwell invasion assays, respectively. For wound-healing assays, the cell suspensions (70 μl, 5 × 105 cells/ml) of HT-29 or SW837 cells were seeded into the insert of a 35 mm high culture µ-dish (Ibidi, Martinsried, Germany). After culturing for 24 h at 37 °C with 5% CO2, the insert was gently removed using a tweezers and the photographs of wounded areas were taken by an inverted microscope (Leica MZ8, Leica Microsystems, Wetzlar, Germany) at 0 h and 24 h. For cell invasion assays, HT-29 or SW837 cells were re-suspended in serum-free medium and subsequently plated into the upper side of a transwell chambers (8 μm pore size; Costar, Cambridge, MA) which were pre-coated with 50 μl of Matrigel (200 mg/ml). The medium containing 20% FBS were then added into the lower chambers. After 24-h incubation, the non-invasive cells on the upper sides of the chambers were gently removed with a cotton swab. Then, 4% paraformaldehyde and 0.1% crystal violet were utilized to fix and stain the cells on the lower sides of the chambers. After washing with PBS for three times, the photographs were taken by an inverted microscope (Leica MZ8, Leica Microsystems, Wetzlar, Germany).
RNA immunoprecipitation (RIP)
Magna RIP Kit (EMD Millipore, Billerica, MA) was utilized to perform RIP assays in accordance with the manufacturer’s protocols. Briefly, cells were lysed using RIP lysis buffer and incubated with magnetic beads conjugated to human anti-Ago2 antibody (Abcam, Cambridge, MA) or isotype-matched control antibody (Millipore, Billerica, MA). Then, qRT-PCR assays were carried out to detect LINC00858 and miR-22-3p expression.
Dual luciferase reporter assays
LINC00858 wild-type (LINC00858-WT) plasmid, LINC00858 mutant (LINC00858-MUT) plasmid, YWHAZ wild-type (YWHAZ-WT) plasmid and YWHAZ mutant (YWHAZ-MUT) plasmid were all constructed by ViralTherapy Co., Ltd. (Wuhan, Hubei, China). HT-29 or SW837 cells were seeded into 24-well plates (ExcellBio, Taicang, Jiangsu, China) and incubated until the cell confluence reached appropriate 70%. Then, miRNA mimic or corresponding plasmids were co-transfected into the cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) in accordance with the manufacturer’s protocols. Thereafter, Dual-Glo Luciferase Assay Kit (Promega, Fitchburg, WI) were applied to determine the luciferase activity according to manufacturer’s instructions.
Statistical analysis
SPSS 17.0 statistics software (SPSS, Inc., Chicago, IL) and GraphPad Prism v6.0 (Graphpad Software Inc, San Diego, CA) software was used to do statistical analysis. Data in this study were expressed as mean ± the standard error of the mean (SEM). Statistical evaluation of the data was analysed using one-way ANOVA test or Student’s t-test. The differences in patient survival were analysed by the Kaplan–Meier method and log-rank test. Multivariate analyses were performed using the Cox regression model to identify independent prognostic factors. A p < .05 was considered as being statistically significant.
Results
LINC00858 was highly expressed in CRC tissues and cell lines
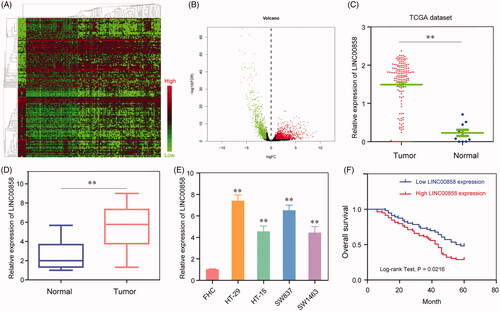
To study aberrantly expressed lncRNAs in CRC, we investigated the raw microarray data downloaded from the TCGA dataset. Hierarchical clustering identified systematic variations in the expression of lncRNAs between CRC tissues and matched adjacent non-tumorous tissues (). Moreover, we took fold change over 2 and p < .05 as the filtration standard to screen out abnormally expressed lncRNAs and found that LINC00858 may be an upregulated lncRNA in CRC (). Furthermore, the data from TCGA showed that LINC00858 expression was significantly up-regulated in CRC tissues compared normal tissues (Figurer 1(C)). In order to demonstrate online data, we further performed RT-PCR to detect the levels of LINC00858 in CRC tissues from our hospital. As shown in , the results also showed that LINC00858 expression was significantly higher expression in CRC tissues than in adjacent tissues (p < 0.01). Next, we compared the expression levels of LINC00858 between CRC cell lines and HIEC cells. Compared with FHC cells, the levels of LINC00858 in all CRC cells (HT-29, HT-15, SW837 and SW1463) were significantly increased (). Taken together, our finding indicated that LINC00858 was significantly overexpressed in CRC and may be involved in the progression of CRC.
Figure 1. Expression levels of LINC00858 in CRC tissues and its clinical significance (A) Dysregulation of lncRNAs in CRC tissues compared with normal lung tissue was analyzed by using TCGA datasets. (B) The volcano plot showed the relationship between fold change and significance of lncRNAs expression. (C) Relative expression of LINC00858 in CRC tissues compared with normal tissue was analyzed by using TCGA data. (D) LINC00858 expression level in CRC tissues (n = 115) compared with corresponding non-tumour tissues (n = 115) was examined by qPCR and normalized to GAPDH expression. (E) LINC00858 expression was assessed by qRT-PCR analysis in CRC cell lines (HT-29, HT-15, SW837 and SW1463) and the normal colonic mucosa epithelial cell line (FHC). (F) Kaplan–Meier overall survival curves according to LINC00858 expression level. * p < 0.05, **p < .01.
Correlation of LINC00858 expression with prognosis of CRC patients
Then, to explore the clinical significance of LINC00858 in CRC, Using the median LINC00858 expression in all 115 CRC patients as a cut-off, the patients were divided into a high LINC00858 expression group and a low LINC00858 expression group. As shown in , we found that high expression of LINC00858 was significantly associated with histological grade (p = .015), lymph nodes metastasis (p = .020) and TNM stage (p = .009). However, there were no significant correlations of LINC00858 expression with other clinical features such as gender, age, tumour size (all p > .05). Then, we attempted to evaluate the correlation between LINC00858 expression and clinical outcomes. As shown in , the results of Kaplan–Meier survival analysis indicated that the 5-year overall survival of high-LINC00858 group was significantly lower than that of low-LINC00858 group (p = .0216). We also performed univariate and multivariate analysis to evaluate the prognostic value of LINC00858 expression in CRC. As shown in , univariate analysis indicated that histological grade, lymph nodes metastasis and TNM stage. and LINC00858 expression were significantly correlated with the overall survival of CRC patients. More importantly, in multivariate analysis, LINC00858 was confirmed to be an independent prognostic factor for CRC patients (p = .005). Taken together, LINC00858 may have potential to be a prognostic biomarker for CRC.
Table 3. Univariate and multivariate analyses for overall survival by cox regression model.
Knockdown of LINC00858 inhibited cell proliferation and accelerated apoptosis of CRC cells
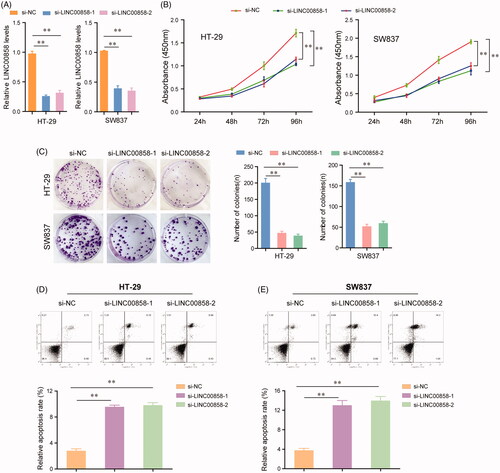
To further elucidate the role of LINC00858 in regulating the biological functions of CRC cells, loss of function assays were performed to test the role of LINC00858 on the proliferation and apoptosis of HT-29 and SW837 cells in vitro. LINC00858 expression was significantly down-regulated in HT-29 and SW837 cells transfected with LINC00858 siRNAs (si-LINC00858-1 and si-LINC00858-1) using qRT-PCR assays (). CCK-8 assays suggested that silence of LINC00858 could effectively inhibit the growth of HT-29 and SW837 cells (). Analogously, cell colony formation assays revealed that transfection of LINC00858 siRNAs remarkably suppressed the clonogenic abilities of HT-29 and SW837 cells (). In addition, flow cytometry was further utilized to analyse the apoptosis of CRC cells. The data indicated that the apoptotic rates of HT-29 and SW837 cells were significantly decreased after transfecting with LINC00858 siRNAs (). Overall, these data demonstrated that silence of LINC00858 could inhibit the proliferation and induce apoptosis of CRC cells.
Figure 2. The effects of LINC00858 on the proliferation, colony formation and apoptosis of HT-29 and SW837 cells. (A) Relative expression levels of LINC00858 in HT-29 and SW837 cells after transfecting with LINC00858 siRNAs. (B) CCK-8 assays were performed to evaluate the proliferation of HT-29 and SW837 cells. (C) Representative images of the colony formation assays and statistical analysis of cell colony numbers. (D and E) Flow cytometry analysis detected the apoptosis of HT-29 and SW837 cells transfected with LINC00858 siRNAs. *p < .05, **p < .01.
Depression of LINC00858 impaired the cells migration and invasion of CRC cells
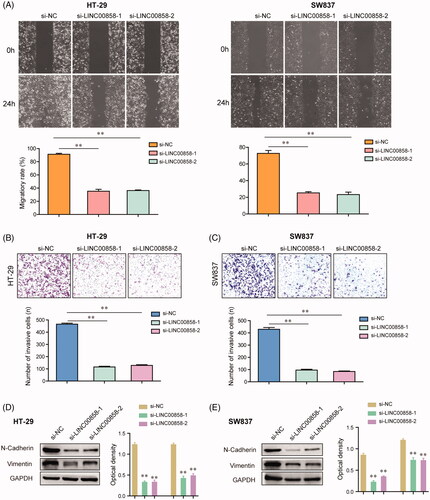
To investigate the roles of LINC00858 in CRC cells migration and invasion, wound healing and transwell invasion assays were carried out in HT-29 and SW837 cells. Wound-healing assays revealed that silence of LINC00858 resulted in a notable reduction of the migratory abilities of HT-29 and SW837 cells (). The results of transwell invasion assays suggested that the invasive capabilities of HT-29 and SW837 cells were dramatically decreased after transfection of LINC00858 siRNAs (). Besides, we also employed western blot assays to evaluate the protein expression of epithelial-mesenchymal transition (EMT)-related molecules such as N-cadherin and vimentin in CRC cells. The data confirmed that knockdown of LINC00858 remarkably reduced the protein levels of N-cadherin as well as vimentin in HT-29 and SW837 cells (). Collectively, these data demonstrated that knockdown of LINC00858 suppressed the migratory and invasive capabilities of CRC cells though orchestrating EMT-related molecules.
Figure 3. The effects of LINC00858 on the migration and invasion of HT-29 and SW837 cells. (A) The wound-healing assays showed that transfection of LINC00858 siRNAs reduced the migratory abilities of HT-29 and SW837 cells. (B and C) The invasive cell numbers of HT-29 and SW837 cells transfected with LINC00858 siRNAs were significantly decreased using transwell invasion assays. (D and E) N-cadherin and vimentin in HT-29 and SW837 were detected by western blot assays. * p < .05, **p < .01.
LINC00858 directly targeted miR-22-3p in CRC cells
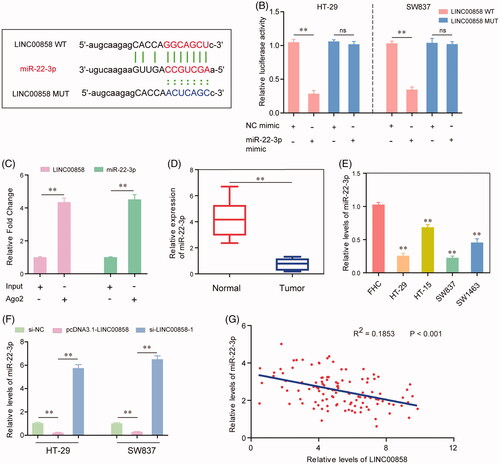
Given that LINC00858 played essential roles in modulating the biological functions of CRC cells, we next aimed to investigate the underlying molecular mechanisms. The results of bioinformatics analysis using “starBase V2.0” (http://starbase.sysu.edu.cn/) discovered that miR-22-3p might be a potential target of LINC00858 (). To further validate that, we next conducted dual luciferase reporter assays in CRC cells. The results demonstrated that ectopic miR-22-3p significantly reduced the relative luciferase activities in HT-29 and SW837 cells transfected with LINC00858 wild type plasmid vector (LINC00858 WT), while co-transfection of miR-22-3p mimic and LINC00858 mutant plasmid vector (LINC00858 MUT) had no effects on the relative luciferase activities of HT-29 and SW837 cells (). Furthermore, the results of RIP assays confirmed that LINC00858 and miR-22-3p were dramatically enriched in Ago2-containing beads compared with the input group (). In addition, qRT-PCR assays proved that the expression levels of miR-22-3p in CRC tissues as well as cell lines were significantly lower than that in the adjacent normal tissues and normal cells ( and ). Moreover, based on the data of qRT-PCR assays, overexpression of LINC00858 remarkably inhibited the expression of miR-22-3p, whereas the relative levels of miR-22-3p were notably increased after silence of LINC00858 (). Besides, Pearson’s correlation analysis suggested that there was a remarkable inverse correlation between LINC00858 and miR-22-3p (). In summary, our data clearly demonstrated that LINC00858 directly interacted with miR-22-3p and indicated that LINC00858 acted as a ceRNA by sponging miR-22-3p.
Figure 4. MiR-22-3p was a direct target of LINC00858. (A) Binding region between miR-22-3p and LINC00858 which was predicted by “starBase V2.0”. (B) Dual luciferase reporter assays showed that LINC00858 directly targeting miR-22-3p. (C) RIP assay determined the enrichment of LINC00858 and miR-22-3p. (D and E) The expression levels of miR-22-3p in CRC tissues and cell lines were evaluated by qRT-PCR assays. (F) The expression levels of miR-22-3p in HT-29 and SW837 cells after transfecting pcDNA3.1-LINC00858 overexpressing plasmid or LINC00858 siRNAs. (G) Pearson’s correlation analysis was applied to analyse the expressing relationship between LINC00858 and miR-22-3p. *p < .05, **p < .01.
LINC00858 modulated YWHAZ via competitively binding miR-22-3p
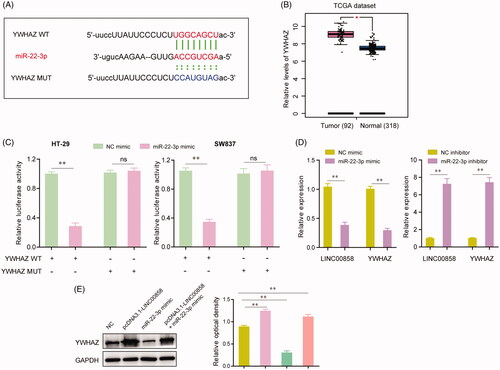
We next performed bioinformatics analysis to identify the downstream target gene of miR-22-3p in CRC cells. Based on the prediction of “starBase V2.0” (http://starbase.sysu.edu.cn/), YWHAZ was selected as a potential target of miR-22-3p and the binding region between miR-22-3p and YWHAZ was exhibited in . Besides, in silico analysis using TCGA dataset suggested that YWHAZ was highly expressed in CRC tissues compared with corresponding normal tissues (). Thereafter, we performed dual luciferase reporter assays to verify the relationship between miR-22-3p and YWHAZ in CRC cells. The data revealed that enhancing expression of miR-22-3p led to a marked decrease in the luciferase activity of the YWHAZ wild-type plasmid (YWHAZ WT); meanwhile, no significant changes of luciferase activity were observed in the cells transfected with YWHAZ mutant plasmid (YWHAZ MUT) and miR-22-3p mimic (). Additionally, qRT-PCR assays indicated that both the expression levels of LINC00858 and YWHAZ were notably impeded by enhancing the expression of miR-22-3p, while knockdown of miR-22-3p remarkably abrogated the inhibitory effects of miR-22-3p on the expression of LINC00858 and YWHAZ (). Besides, we further conducted western blot assays to assess the alteration of YWHAZ protein levels under various conditions. The data showed that ectopic expression of LINC00858 resulted in a significant increase of the YWHAZ expression, and re-introduction of LINC00858 reversed the inhibitory effects of miR-22-3p on the expression of YWHAZ (). To sum up, our results demonstrated that LINC00858 exerted tis biological functions on CRC cells through miR-22-3p/YWHAZ axis.
Figure 5. YWHAZ was a target gene of miR-22-3p. (A) The putative binding sites of miR-22-3p within the 3′-UTR of YWHAZ as predicted by “starBase V2.0”. (B) The relative expression of YWHAZ were analysed by “GEPIA” using TCGA dataset. (C) Dual-luciferase reporter assays showed the luciferase activities of the combination of miR-22-3p and YWHAZ wild-type (WT) or mutant (MUT). (D) The relative expression levels of LINC00858 or YWHAZ in HT-29 cells after transfecting with miR-22-3p mimic or inhibitor. (E) The protein levels of YWHAZ in HT-29 cells after transfection of pcDNA3.1-LINC00858, or miR-22-3p mimic, or co-transfection of pcDNA3.1-LINC00858 as well as miR-22-3p mimic. *p <.05, **p < .01.
Discussion
The incidence and mortality rate of CRC in China have increased rapidly over the past several decades [Citation21]. In order to improve clinical prognosis of CRC patients, it is necessary to screen useful diagnostic and prognostic biomarkers of CRC, which can contribute to early diagnosis and improve the management of individualized treatment [Citation22, Citation23]. Recently, lncRNAs as potential biomarkers for prognosis of CRC patients become a hotspot due its important role in progression of CRC and frequent dysregulation in CRC tissues [Citation24]. In this study, using TCGA dataset, we screened a dysregulated lncRNA LINC00858, which was significantly highly expressed in CRC tissues. Then, the results of RT-PCR demonstrated the above results. In clinical assay, it was found that high expression of LINC00858 was associated with histological grade, lymph nodes metastasis and advanced TNM stage, indicating a positive role of LINC00858 in clinical progression of CRC. Then, we further explored the prognostic value of LINC00858 in CRC patients by Kaplan-Meier assay, finding that LINC00858 expression were significantly correlated with poor overall survival of CRC patients. Moreover, multivariate analyses confirmed that LINC00858 was an independent prognostic parameter of patient outcomes. Taken together, our miRNAs LINC00858 potential use as novel biomarkers of CRC and may be useful in clinical management for CRC patients.
Recently, several studies reported that LINC00858 was implicated in the regulation of several tumor progression. For instance, Zhu et al. reported that LINC00858 was highly expressed in non-small cell lung cancer and its forced expression promoted cell proliferation and induced cell migration and invasion by acting a molecular sponge for miR-422a [Citation20]. Gu et al. found that LINC00858 expression was up-regulated in osteosarcoma and its knockdown inhibits the proliferation and invasion of osteosarcoma cells through inhibiting CDK14 by up-regulating miR-139, suggesting the effect of LINC00858/miR-139/CDK14 axis on osteosarcoma tumorigenesis [Citation19]. In addition, recent study by Yamada et al. firstly reported that LINC00858 was a upregulated lncRNA in CRC [Citation25]. However, the express pattern, clinical significance and biological function of LINC00858 in CRC remain largely unclear. In this study, in order to explore the potential function of LINC00858 in CRC, we use siRNA to down-regulated the levels of LINC00858 and performed a series of cells experiments. We found that knockdown of LINC00858 significantly suppressed cells proliferation, migration and invasion of CRC. Furthermore, we confirmed that LINC00858 displayed its oncogenic role by promoting apoptosis and EMT pathway. Overall, our findings indicated that LINC00858 may function as a tumour promoter in CRC.
Growing findings indicated the existence of a widespread interaction network involving ceRNAs, where ncRNAs could regulate modulatory RNA by binding and titrating them off their binding sites on protein coding messengers [Citation26]. Recently, several lncRNAs were reported to be able to competitively inhibit miRNAs by acting as a molecular sponge in various tumors, including CRC [Citation27, Citation28]. In this study, in order to explore the potential mechanism by which LINC00858 promoted CRC cells proliferation, migration and invasion and influenced the clinical outcome of CRC patients, we wonder whether LINC00858 could regulate some important miRNAs. Then, we performed bioinformatics analysis and found that LINC00858 contained binding sites for several miRNAs. We focused on miR-22-3p, which has been shown to be involved in the regulation of CRC cell proliferation and invasion in previous study. By luciferase reporter assays, we validated that LINC00858 directly bond to miR-22-3p. In addition, we also found that overexpression of LINC00858 suppressed the expression of miR-22-3p, while knockdown of LINC00858 display the opposite effect. Thus, our findings revealed that LINC00858 may exhibited its oncogenic role by suppressing miR-22-3p expression.
YWHAZ, also known as 14–3-3zeta, has been reported to be up-regulated in several tumors and serves as a positive regulator [Citation29, Citation30]. Overexpression of YWHAZ promotes hepatocellular carcinoma venous metastasis by modulating hypoxia-inducible factor-1α [Citation31]. In lung cancer, forced YWHAZ expression promotes lung cancer cell invasion by increasing the Snail protein expression [Citation32]. Recent study by Chen et al. reported that miR-22 suppressed metastasis of hepatocellular carcinoma by targeting YWHAZ [Citation33]. However, whether miR-22-3p could target YWHAZ in CRC remains unknown. In this study, our results confirmed that YWHAZ was a direct target of miR-22-3p by using luciferase reporter assay. In addition, we also found that YWHAZ expression was significantly increased by up-regulating LINC00858 expression, whereas miR-22-3p mimics reversed the suppressed role of LINC00858 up-regulation in the expression levels of YWHAZ. Thus, our findings indicated that LINC00858 promoted the tumorigenesis and progression of CRTC through miR-22-3p-YWHAZ axis. However, more in vitro and in vivo experiments were needed to further confirm our results.
Conclusion
In summary, we firstly showed that increased LINC00858 expression is a common event and an independent prognostic biomarker in CRC. LINC00858 acted as a tumor-promoting gene, which promoted the malignant behaviour of CRC cells through miR-22-3p-YWHAZ axis. This novel finding triggers a new perspective for clinical treatment of glioma and might provide a therapeutic strategy.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Hashim D, Boffetta P, La Vecchia C, et al. The global decrease in cancer mortality: trends and disparities. Ann Oncol. 2016;27:926–933.
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132.
- Binefa G, Rodriguez-Moranta F, Teule A, et al. Colorectal cancer: from prevention to personalized medicine. Wjg. 2014;20:6786–6808. PubMed PMID: 24944469; PubMed Central PMCID: PMCPMC4051918.
- Xiang B, Snook AE, Magee MS, et al. Colorectal cancer immunotherapy. Discov Med. 2013; 15:301–308. PubMed PMID: 23725603; PubMed Central PMCID: PMCPMC4042089.
- Sridharan M, Hubbard JM, Grothey A. Colorectal cancer: how emerging molecular understanding affects treatment decisions. Oncology (Williston Park). 2014;28:110–118.
- St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–251. PubMed PMID: 25869999; PubMed Central PMCID: PMCPMC4417002.
- Meller VH, Joshi SS, Deshpande N. Modulation of chromatin by noncoding RNA. Annu Rev Genet. 2015;49:673–695.
- Booton R, Lindsay MA. Emerging role of MicroRNAs and long noncoding RNAs in respiratory disease. Chest 2014;146:193–204.
- Luo D, Deng B, Weng M, et al. A prognostic 4-lncRNA expression signature for lung squamous cell carcinoma. Artif Cells Nanomed Biotechnol. 2018;46:1207–1214.PubMed PMID: 28835135.
- Cheetham SW, Gruhl F, Mattick JS, et al. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419. PubMed PMID: 23660942; PubMed Central PMCID: PMCPMC3694235.
- Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–1109.
- Gutschner T, Hammerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med. 2013;91:791–801.
- Zhu H, Li X, Song Y, et al. Long non-coding RNA ANRIL is up-regulated in bladder cancer and regulates bladder cancer cell proliferation and apoptosis through the intrinsic pathway. Biochem Biophys Res Commun. 2015;467:223–228.
- Svoboda M, Slyskova J, Schneiderova M, et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis. 2014;35:1510–1515.
- Qi X, Zhang DH, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52:710–718.
- Ergun S, Oztuzcu S. Oncocers: ceRNA-mediated cross-talk by sponging miRNAs in oncogenic pathways. Tumor Biol. 2015;36:3129–3136.
- Zhang L, Fang F, He X. Long noncoding RNA TP73-AS1 promotes non-small cell lung cancer progression by competitively sponging miR-449a/EZH2. Biomed Pharmacother. 2018;104:705–711.
- Shen B, Yuan Y, Zhang Y, et al. Long non-coding RNA FBXL19-AS1 plays oncogenic role in colorectal cancer by sponging miR-203. Biochem Biophys Res Commun. 2017;488:67–73.
- Gu Z, Hou Z, Zheng L, et al. Long noncoding RNA LINC00858 promotes osteosarcoma through regulating miR-139-CDK14 axis. Biochem Biophys Res Commun. 2018;503:1134–1140.
- Zhu SP, Wang JY, Wang XG, et al. Long intergenic non-protein coding RNA 00858 functions as a competing endogenous RNA for miR-422a to facilitate the cell growth in non-small cell lung cancer. Aging (Albany NY). 2017;9:475–486. PubMed PMID: 28177876; PubMed Central PMCID: PMCPMC5361675.
- Li X, Hu F, Wang Y, et al. CpG island methylator phenotype and prognosis of colorectal cancer in Northeast China. Biomed Res Int. 2014;2014:1. PubMed PMID: 25243122; PubMed Central PMCID: PMCPMC4163374.
- Shah R, Jones E, Vidart V, et al. Biomarkers for early detection of colorectal cancer and polyps: systematic review. Cancer. Epidemiol Biomarkers Prev. 2014;23:1712–1728.
- Fakih MG. Metastatic colorectal cancer: current state and future directions. JCO. 2015;33:1809–1824.
- Smolle M, Uranitsch S, Gerger A, et al. Current status of long non-coding RNAs in human cancer with specific focus on colorectal cancer. IJMS.. 2014;15:13993–14013. PubMed PMID: 25119862; PubMed Central PMCID: PMCPMC4159835.
- Yamada A, Yu P, Lin W, et al. A RNA-Sequencing approach for the identification of novel long non-coding RNA biomarkers in colorectal cancer. Sci Rep. 2018;8:575. PubMed PMID: 29330370; PubMed Central PMCID: PMCPMC5766599.
- An Y, Furber KL, Ji S. Pseudogenes regulate parental gene expression via ceRNA network. J Cell Mol Med. 2017;21:185–192. PubMed PMID: 27561207; PubMed Central PMCID: PMCPMC5192809.
- Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. PubMed PMID: 26068968; PubMed Central PMCID: PMCPMC4673179.
- Christensen LL, True K, Hamilton MP, et al. SNHG16 is regulated by the Wnt pathway in colorectal cancer and affects genes involved in lipid metabolism. Mol Oncol. 2016;10:1266–1282. PubMed PMID: 27396952; PubMed Central PMCID: PMCPMC5423192.
- Ruenauver K, Menon R, Svensson MA, et al. Prognostic significance of YWHAZ expression in localized prostate cancer. Prostate Cancer Prostatic Dis. 2014;17:310–314.
- Moraru E, Vilcea ID, Mirea CS, et al. c-abl and YWHAZ gene expression in gastric cancer. Rom J Morphol Embryol. 2015;56:717–723.
- Tang Y, Liu S, Li N, et al. 14-3-3zeta promotes hepatocellular carcinoma venous metastasis by modulating hypoxia-inducible factor-1alpha. Oncotarget. 2016; 7:15854–15867. PubMed PMID: 26910835; PubMed Central PMCID: PMCPMC4941282.
- Tong S, Xia T, Fan K, et al. 14-3-3zeta promotes lung cancer cell invasion by increasing the Snail protein expression through atypical protein kinase C (aPKC)/NF-kappaB signaling. Exp Cell Res. 2016;348:1–9.
- Chen M, Hu W, Xiong CL, et al. miR-22 targets YWHAZ to inhibit metastasis of hepatocellular carcinoma and its down-regulation predicts a poor survival. Oncotarget. 2016;7:80751–80764. PubMed PMID: 27811373; PubMed Central PMCID: PMCPMC5348352.
