Abstract
More and more circular RNAs (circRNAs) revealed to play a critical role in the initiation and progression of cancer, however, the effects of circRNAs on non-small cell lung cancer (NSCLC) remain largely undetermined. In the present study, we screened the dysregulated circRNAs in paired NSCLC and normal samples from GEO database and identified circ_0043278 was to be significantly up-regulated in NSCLC and demonstrated it promotes NSCLC progression in vitro and in vivo. Then, we revealed the expression of miR-520f, a downstream factor of circ_0043278, was significantly down-regulated in NSCLC and acted as a tumor inhibitor. In addition, we revealed that circ_0043278 sponged miR-520f, which was demonstrated to target ROCK1, CDKN1B, and AKT3 in NSCLC cells. In conclusion, circ_0043278 promoted NSCLC cell proliferation, invasion, and migration by increasing ROCK1, CDKN1B, and AKT3 expressions through direct inhibition of miR-520f.
Introduction
Non-small cell lung cancer (NSCLC) is the major subtype of lung cancer, accounting for more than 80% of lung cancer incidence [Citation1,Citation2]. It is frequently diagnosed at an advanced stage due to the lack of effective diagnostic methods and biomarkers, resulting in a poor prognosis, despite recent improvements in NSCLC treatments [Citation3]. The combination of chemotherapy and surgical resection currently becomes the standard therapeutic strategy for NSCLC patients, however, its effects are seriously limited by drug resistance and tumor recurrence, even along with several unexpected side effects [Citation4,Citation5]. Detection of novel accurate diagnostic biomarkers might predict tumor correctly and timely with an aim to improve the prognosis of NSCLC, however, this action was still on the road.
Non-coding RNAs (ncRNAs), characterized by the lack of protein-encoding capacity, are important RNA molecules that function in multiple biological processes, such as cell proliferation, migration, invasion, and differentiation [Citation6,Citation7]. It was well documented that the members of ncRNAs including miRNAs and lncRNAs play critical roles in the tumorigenesis of multiple human tumors, including NSCLC [Citation8]. Expression profiles of miRNAs and lncRNAs in NSCLC were reported by numerous studies during the past decades [Citation9,Citation10]. Unfortunately, whether these specific ncRNAs could act as diagnostic biomarkers of NSCLC remain controversial. Circular RNAs (circRNAs), marked by its covalently closed loop, is a novel subtype of ncRNAs without 5′ caps and 3′ tails [Citation11,Citation12]. It was reported that circRNAs exist in mammal cells widely and play an important role in various aspects of tumor progression, including tumor cell invasion, proliferation, and migration [Citation13]. Recently, multiple circRNAs were found to be involved in the tumorigenesis of NSCLC [Citation14], however, the exact roles of circRNAs in NSCLC remain largely undetermined.
In order to better characterize the effects of circRNAs on NSCLC, we analyzed the data of a microarray dataset (GSE101586) from the Gene Expression Omnibus database (GEO, http://www.ncbi.nlm.nih.gov/geo) and identified as novel NSCLC-related circRNA (circ_0043278). In the present study, we aimed to investigate the effects and mechanisms of circ_0043278 on NSCLC progression. We designed functional assays to show the oncogenic role of circ_0043278 in NSCLC in vitro and in vivo. In mechanism, we explored circ_0043278 could regulate the expression of ROCK1, CDKN1B, and AKT3 through inhibiting miR-520f in NSCLC cells. Our findings circ_0043278 may serve as a novel potential biomarker and therapeutic target of NSCLC.
Materials and methods
NSCLC tissue samples and cell lines
Forty-four pairs of NSCLC tissues and corresponding normal tissues were collected from patients who were diagnosed at Suzhou Municipal Hospital as NSCLC during 2015–2018. Written informed consents were provided by all subjects and the present study was approved by the ethics committee of Suzhou Municipal Hospital. The human airway epithelial cell line 16HBE and human NSCLC cell lines H1975, A549, SPC-A1, and H1299 were all obtained from the type Culture Collection of Chinese Academy of Sciences (Shanghai, China). All types of cell lines were maintained in the RPMI-1640 medium (HyClone Laboratories Inc., Logan, Utah,USA ) containing 10% fetal bovine serum (FBS), 1% penicillin and streptomycin at 37 °C in a cell incubator with 5% CO2 and 95% air.
circRNAs expression profile analysis
The gene expression profile GSE101586 of NSCLC was downloaded from the Gene Expression Omnibus database (GEO, http://www.ncbi.nlm.nih.gov/geo). The differentially expressed circRNAs of GSE101586 was analyzed by an online tool GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/).
RNA extraction and quantitative real-time PCR (RT-PCR) assay
Total RNAs of NSCLC tissues and cell lines, as well as normal tissues and cells, were extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA quality was examined by NanoDro2000c (Thermo Scientific, Waltham, MA, USA) following the instructions provided by the manufacturers. Then, 5 μg of total RNAs were reverse transcribed into cDNA through BestarTM qPCR RT kit (#2220, DBI Bioscience, Shanghai, China, China), and the quantitative polymerase chain reaction (RT-PCR) reaction was carried out via BestarTM qPCR MasterMix (#2043, DBI Bioscience, China) on an ABI7500 system. Primers used in this study were all synthesized in RiboBio (Guangzhou, China). Sequence of all primers were showed as follow: GAPDH, F: 5′-TGT TCG TCA TGG GTG TGA AC-3′, R: 5′-ATG GCA TGG ACT GTG GTC AT-3′; U6, F: 5′-CTC GCT TCG GCA GCA CA-3′, R: 5′-AAC GCT TCA CGA ATT TGC GT-3′; circ_0043278, F: 5′-AAT GTG CAC CAA GAC CAA GG-3′, R: 5′-CCA AAG CCA CAG TCC ATC AC-3′; miR-520f, F: 5′-ACA CTC CAG CTG GGA AGT GCT TCC TTT TAG-3′, R: 5′-CTC AAC TGG TGT CGT GGA GTC GGC AAT TCA GTT GAG AAC CCT CT-3′; ROCK1, F: 5′-AAG AGA GTG ATA TTG AGC AGT TGC G-3′, R: 5′-TTC CTC TAT TTG GTA CAG AA AGC CA-3′; CDKN1B, F: 5′- CTG CAA CCG ACG ATT CTT CTA CT-3′, R: 5′-GGG CGT CTG CTC CAC AGA-3′; AKT3, F: 5′-TGA AGT GGC ACA CAC TCT AAC T-3′, R: 5′-CCG CTC TCT CGA CAA ATG GA-3′. The expression of circ_0043278, ROCK1, CDKN1B, and AKT3 were normalized to GAPDH, miR-520f expression was normalized to U6, gene expression was quantified via 2−ΔΔCt method.
RNA interference and transfection assay
Two siRNAs against circ_0043278 (si-Circ#1 and si-Circ#2) were constructed to block the expression of circ_0043278 in H1299 cells, negative control siRNAs (si-NC) was utilized as a control. Target sequence was: si-Circ#1: 5′-ATTCCATTTCACTACTTCAGA-3′; si-Circ#2: 5′-TCCATTTCACTACTTCAGATT-3′. To overexpress circ_0043278, the cDNA of circ_0043278 was inserted into pcDNA vector to construct pcDNA-circ_0043278 (Circ OE), empty vector was used as a control. Inhibitor and mimics of miR-520f were used to silence and enhance the expression of miR-520f, respectively. For transfection, H1299 and A549 cells were cultured in RPMI-1640 medium in six-well plates. After growth to 80% confluence, H1299 and A549 cells were transfected with 50 nM si-Circ#1 or si-Circ#2, 100 nM miR-520f inhibitors or mimics, and 50 ng pcDNA-circ_0043278 by using Lipofectamine 2000 (Invitrogen, Karlsruhe, Germany).
Cell viability analysis (MTT assay)
Effects of circ_0043278 and miR-520f on NSCLC cell viability were assessed through MTT assay. In brief, treated H1299 and A549 cells were seeded into 96-well plates and cultured at 37 °C for 24 h, followed by the incubation of 20 μl dye solution for another 4 h. After stopping the reaction with 200 μl stop solution, the absorbance was determined with Infinite® 200 PRO (FPRO-T; Tecan, Seestrasse, Switzerland) at 570 nm.
Cell proliferation analysis (Colony formation assay)
Effects of circ_0043278 and miR-520f on NSCLC cell proliferation were evaluated using colony formation assay. Briefly, treated H1299 and A549 cells were seeded into six-well plates at a density of 2000 cells/well. After incubation at 37 °C under 5% CO2 and 95% air for at least 14 days, the colonies were fixed and stained with Giemsa solution and the number of colonies was calculated under a microscope.
Tumor sphere formation assay
Treated H1299 and A549 cells were seeded onto six-well plates at a concentration of 100 cells/well and incubated in serum-free RPMI-1640 medium containing B27 (1:50, Invitrogen, Karlsruhe, Germany), 20 ng/ml EGF (BD Biosciences,San Jose, USA), and insulin (4 mg/ml, Sigma, St. Louis, MO, USA) for two weeks. Subsequently, the number of H1299 and A549 cell sphere were imaged and counted.
Cell invasi analysis (Transwell assay)
Transwell assay was performed to examine the effects of circ_0043278 and miR-520 on NSCLC cell invasion using the chamber (8-µm pores, Corning Incorporated, USA) coated with Matrigel matrix (BD Biosciences, USA). Treated H1299 or A549 cells were harvested and re-suspended in culture medium without FBS, making the final concentration 3 × 105 cells/ml. Subsequently, 200 µl PC NSCLC cell suspension was added into the upper chamber, while 500 µl culture medium containing 20% FBS was added into the lower chamber. After incubated at 37 °C for 24 h, the invaded NSCLC cells were fixed and stained with 0.5% crystal violet (Beyotime Institute of Biotechnology, Haimen, China).
Cell migration analysis (Wound-healing assay)
Wound-healing assay was utilized to detect the influences of circ_0043278 and miR-520f on NSCLC cell migration. Treated H1299 and A549 cells were plated into 6-well plates and incubated at 37 °C under 5% CO2 and 95% air until confluence. A straight wound was made in the cell layer by a sterile plastic pipette, followed by PBS washing twice. After maintained for 24 h, the migratory rate was calculated.
In vivo tumor growth assay
Male BALB/c mice (8 weeks) were applied for the in vivo tumor growth assay to evaluate the effects of circ_0043278 and miR-520f on tumor growth. Animals were provided by Suzhou University, animal manipulates in the present study were approved by the Institutional Animal Care and Use Committee of the Suzhou University. H1299 cells stably transfected with si-Circ#1+#2 were harvested and re-suspended in RPMI-1640, making the final concentration 2 × 105 cells/ml. Then, 100 μL of circ_0043278 blocked H1299 cell suspensions were injected into the left flank of nude mice. Tumor growth was examined after injection, volumes of tumors were measured as the length × width2×0.5.
Plasmid construction and dual luciferase activity assay
For the establishment of recombinant luciferase reporter plasmids, the wild type (WT) and mutant (Mut) fragments of circ_0043278 containing putative miR-520f binding sites were inserted into the pGL3 (Promega, Madison, WI, USA) vector. The recombinant luciferase reporter plasmids were named as pGL3-circ_0043278 WT and pGL3-circ_0043278 Mut, respectively. For the dual-luciferase reporter assay, H1299 cells (1 × 104 cells/well) were plated into 96-well plates and cultured for 16 h at 37 °C, followed by the co-transfection with pGL3-circ_0043278 WT and miR-520f inhibitor; A549 cells (1 × 104 cells/well) were plated into 96-well plates and cultured for 16 h at 37 °C, followed by the co-transfection with pGL3-circ_0043278 WT and miR-520f mimics. The Dual-Luciferase Assay System (Promega) was utilized to examine the firefly and renilla luciferase activities. Renilla luciferase activity was normalized to firefly luciferase activity. Dual-luciferase reporter assay between circ_0043278 and miR-1229, miR-203, miR-450b-3p, miR-769-3p, miR-526b, or miR-638, as well as between miR-520f and ROCK1, CDKN1B or AKT3, were performed as same as miR-520f.
Annotation for circRNA/miRNA interaction and KEGG analysis
Putative target miRNAs of circ_0043278 were predicted by using Targetscan (http://www.targetscan.org/vert_71/). Biological pathways of miRNAs were defined by Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/).
CircRNA-miRNA-mRNA co-expression network analysis
The circRNA-miRNA-mRNA co-expression network of circ_0043278 was constructed according to the prediction of target miRNAs and their target genes using Cytoscape software (Version 3.4.0).
RNA-pull down assay
RNA-pull down assay was performed to confirm the interaction between circ_0043278 and miR-520f in H1299 and A549 cells using the specific biotin-labeled miR-520f probe, which was obtained from Sangon Biotech (Shanghai, China). After lysed in RIP lysis buffer, cell lysates of H1299 and A549 were incubated overnight at 4 °C with magnetic beads that recognized biotin. The enrichment of circ_0043278 was detected using qRT-PCR.
Statistical analysis
Data in the present study were presented as mean ± SEM. One-way analysis of variance, conducted with Graphpad (Ver. Prism 7, GraphPad Prism Software, La Jolla, CA, USA), was utilized to analyze the difference between groups and p values less than .05 was considered significant.
Results
Circ_0043278 was identified as a NSCLC-related circRNA
In order to identify unique gene involved in NSCLC, circRNAs microarray analysis was performed using NSCLC and normal samples of GSE101586 (). Hsa_102049 was identified to be one of the top 30 dysregulated circRNAs in GSE101586 (). Data from UCSC genome browser showed that hsa_102049 derived from TADA2A gene and named as circ_0043278 (). In the GSE101586 dataset, circ_0043278 expression was significantly higher in the lung tumor samples than control samples (), implying that circ_0043278 might play a role in the tumorigenesis of NSCLC. To further confirm the oncogenic role of circ_0043278 in NSCLC, we measured the expression of circ_0043278 in 44 paired NSCLC and corresponding normal tissues (our cohort in Suzhou) by qRT-PCR assay. Results showed that circ_0043278 expression was significantly up-regulated in NSCLC tissues compared with normal tissues (, p < .05). Moreover, circ_0043278 expression was found to be remarkably higher in metastasis tissues than non-metastasis tissues (, p < .05). In addition, the 44 NSCLC patients were divided into high circ_0043278 group and low circ_0043278 group and the statistical results indicated that circ_0043278 expression was closely correlated with differentiation grade (p = .040), distal metastasis (p = .012), and tumor, node, metastasis (TNM) stage (p = .021), not relevant to age (p = .588), gender (p = .263), and tumor size (p = .422, ). These results suggested that increased circ_0043278 might involve in the tumorigenesis of NSCLC.
Figure 1. Circ_0043278 was identified as a NSCLC-related circRNA. (a) Hierarchical clustering analysis of circRNAs that were differentially expressed in NSCLC tissues and corresponding normal tissues in GSE101586. (b) Top 30 dysregulated circRNAs of GSE101586, the wireframe marked has_circ_102049 has the most fold change level (upregulated). (c) Chromosomal map showing the location of circ_0043278 using UCSC genome browser. (d) Expression of circ_0043287 in the control and tumor samples of GSE101586. (e and f) Expression of circ_0043287 in 44 paired normal and corresponding NSCLC samples was evaluated by qRT-PCR (*p < .05 vs. normal or non-metastasis group).
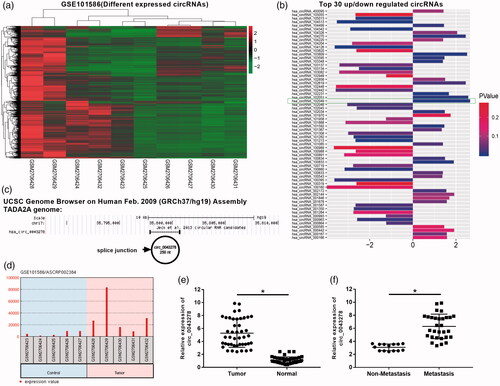
Table 1. Correlations between has_circ_0043278 expression and clinicopathologic characteristics in NSCLC.
Circ_0043278 acts as an oncogenic molecule in NSCLC
Results from qRT-PCR showed that circ_0043278 expression was markedly increased in four NSCLC cell lines (H1975, A549, SPC-A1, and H1299) compared to 16HBE (human bronchial epithelial cells as normal control) cells (, p < .05). To further explore the functions of circ_0043278 in NSCLC, we established a circ_0043278 blocked cell model in H1299 cells and a circ_0043278 overexpressed cell model in A549 cells by treating cells with circ_0043278 siRNAs (si-Circ#1, si-Circ#2) and Circ OE (Circ_0043278 over-expression), respectively. Results showed that circ_0043278 was significantly decreased in si-Circ#1 and si-Circ#2 treated H1299 cells compared to those cells treated with si-NC (, p < .05) and it was significantly increased in Circ OE treated A549 cells compared to those cells treated with vector (, p < .05). MTT assay showed that cell viability was significantly reduced in circ_0043278 knockdown H1299 cells (, p < .05 vs. si-NC), while it was significantly increased in circ_0043278 overexpressed A549 cells (, p < .05). Tumor sphere formation assay showed that sphere number was remarkably lower in si-Circ#1 and si-Circ#2 treated H1299 cells compared with those cells treated with si-NC (, p < .05), while the sphere number of Circ OE treated A549 cells was obviously higher than vector treated A549 cells (, p < .05). In the cell proliferation analysis, colony formation assay indicated that si-Circ#1 and si-Circ#2 treatment significantly decreased the colony number of H1299 cells compared to si-NC group (, p < .05), while Circ OE treatment remarkably increased the colony number of A549 cells compared to the vector group (, p < .05). To better characterize the effects of circ_0043278 on tumor growth, we performed in vivo tumor growth in nude mice by injecting transfected H1229 cells. Results indicated that tumor weight and volume were significantly reduced in si-circ#1+#2 treated group compared to si-NC group (, p < .05). The knockdown efficiency of si-circ#1+#2 treated H1299 cells was examined by qRT-PCR (, p < .05).
Figure 2. Circ_0043278 acts as an oncogenic molecule in NSCLC. (a) Expression of circ_0043278 in 16HBE cell line and four NSCLC cell lines: H1975, A549, SPC-A1, and H1299 (*p < .05 vs. 16HBE cells). (b) Circ_0043278 expression of H1299 cells treated with si-NC, si-Circ#1 and si-Circ#2 was detected by qRT-PCR (*p < .05 vs. si-NC group). (c) Circ_0043278 expression of A549 cells transfected with empty vector and Circ OE was measured via qRT-PCR (*p < .05 vs. vector group). MTT assay was performed to determine cell viability of (d) H1299 cells treated with si-NC, si-Circ#1, and si-Circ#2, or (e) A549 cells treated with empty vector and Circ OE at 0, 24, 48, and 72 h (*p < .05 vs. si-NC or vector group). Tumor sphere formation assay was performed to evaluate cell self-renewal capability in (f) circ_0043278 blocked H1299 cells and (g) circ_0043278 overexpressed A549 cells (*p < .05 vs. si-NC or vector group). Colony formation assay was carried out in (h) si-NC, si-Circ#1 and si-Circ#2 treated H1299 cells or (i) empty vector and Circ OE treated A549 cells to assess the effects of circ_0043278 knockdown and overexpression on cell proliferation (*p < .05 vs. si-NC or vector group). (j–l) Hypodermic injection of H1229 cells transfected with si-circ#1+#2 or si-NC into BALB/c nude mice established xenograft model, tumor weight, and volume were then assessed (*p < .05 vs. si-NC group). (m) Expression of circ_0043278 in H1299 cells treated with si-NC and si-circ#1+#2 was examined by qRT-PCR (*p < .05 vs. si-NC group).
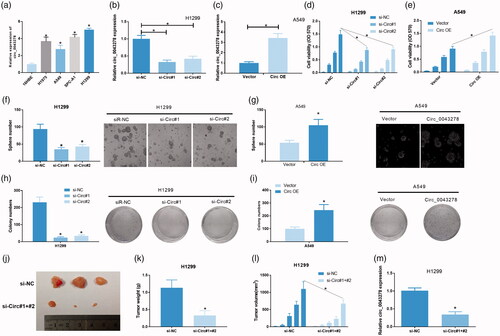
Circ_0043278 promotes NSCLC cell invasion and migration
Furthermore, we evaluated the effects of circ_0043278 knockdown and overexpression on NSCLC cell invasion and migration by transwell and wound-healing assays. Compared with si-NC treated H1299 cells, si-Circ#1 and si-Circ#2 treated H1299 cells showed a significant decrease in invasive cell number (, p < .05) and migration rate (, p < .05C). Similarly, Circ OE treated A549 cells showed a significant increase of invasive cell number ( < .05) and migration rate (, p < .05) compared with vector treated A549 cells.
Figure 3. Circ_0043278 promotes NSCLC cell invasion and migration. (a) H1299 cells treated with si-NC, si-Circ#1 and si-Circ#2 were subjected to transwell assay to assess the effects of circ_0043278 knockdown on cell invasion (*p < .05 vs. si-NC group). (b) Effects of circ_0043278 knockdown on cell migration was evaluated by wound-healing assay in H1299 cells (*p < .05 vs. si-NC group). (c) A549 cells treated with vector and Circ OE were subjected to transwell assay to examine the effects of circ_0043278 overexpression on cell invasion (*p < .05 vs. vector group). (d) Effects of circ_0043278 overexpression on cell migration was assessed using wound-healing assay in A549 cells (*p < .05 vs. vector group).
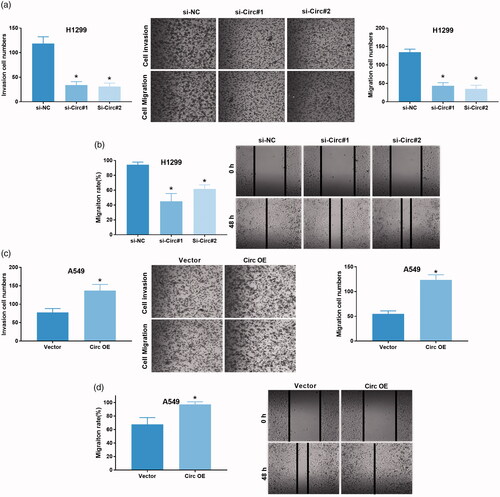
KEGG analysis of circ_0043278 target miRNAs
In order to investigate the underlying molecular mechanisms of circ_0043278 in NSCLC progression, we screened its target miRNAs through Targetscan ( http://www.targetscan.org/vert_71/), results showed that circRNAs possessed 7 miRNAs binding sites (Supplementary Excel 1). Dual-luciferase reporter assay was then performed to verify the interaction between circ_0043278 and miR-1299, miR-203, miR-520f, miR-450b-3p, miR-769-3p, miR-526b, or miR-638 in H1299 and A549 cells. Results showed that mimics of miR-1299, miR-203, miR-520f, miR-450b-3p, and miR-526b, but not of miR-769-3p and miR-638, could significantly attenuate the luciferase activity of H1299 cells driven by circ_0043278 (, p < .05), moreover, inhibitors of miR-1299, miR-203, miR-520f, miR-450b-3p, and miR-526b, but not of miR-769-3p and miR638, were found to enhance the luciferase activity of A549 cells driven by circ_0043278 (, p < .05). We subsequently predicted the target genes of miR-1299, miR-203, miR-520f, miR-450b-3p, and miR-526b using Targetscan7.2 and analyze their target genes via KEGG (). By using the data from KEGG analysis of miR-1299, miR-203, miR-520f, miR-450b-3p, and miR-526b, we established the ceRNA network of circ_0043278 ().
Figure 4. KEGG analysis of circ_0043278 target miRNAs. (a and b) Dual-luciferase reporter assay was performed in H1299 and A549 cells to verify the interaction between circ_0043278 and miR-1229, miR-203, miR-520f, miR-450b-3p, miR-769-3p, miR-526b, or miR-638. (c) Schematic diagram of the establishment of circ_0043278 ceRNA network. (d) KEGG analysis of miR-1299, miR-203, miR-450-3p, miR-520f and miR-526b. (e) The ceRNA network of circ_0043278 within miR-1299, miR-203, miR-450-3p, miR-520f and miR-526b.
MiR-520f expression was significantly decreased in NSCLC
MiR-520f shares the most changed miRNA that is induced by circ_0043278 in NSCLC cells, so it was selected for further investigation. The interaction between circ_0043278 and miR-520f was further confirmed by dual-luciferase reporter assay and RNA pull down assay in H1299 and A549 cells. In the dual-luciferase reporter assay, we cloned the wild type and mutant circ_0043278 fragment containing miR-520f binding site () into pGL3 vector. Results showed that miR-520f mimics could significantly reduce the luciferase activity of H1299 cells driven by pGL3/circ_0043278 WT compared to miR-NC group (, p < .05), and miR-520f inhibitors could remarkably increase the luciferase activity of A549 cells driven by pGL3/circ_0043278 WT (, *p < .05 vs. miR-NC). Luciferase activities in H1299 and A549 cells driven by pGL3/circ_0043278 Mut were not affected by miR-520f mimics or inhibitors (). RNA pull down assay followed by the detection of qRT-PCR were carried out in H1299 and A549 cells to further confirm the interaction between circ_0043278 and miR-520f using the biotin-labeled miR-520f probe. Results showed a specific enrichment of circ_0043278 in Bio-miR-520f specific probe group compared to Bio-NC and Bio-miR-520f Mut groups in both H1299 and A549 cells (, p < .05 vs. Bio-NC or Bio-miR-520f Mut). Overall, survival analysis showed that NSCLC patients with low miR-520f expression exhibited a worse prognosis than those patients with high miR-520f expression (). qRT-PCR analysis of miR-520f expression in 44 paired NSCLC and normal tissues showed that miR-520f expression was significantly down-regulated in NSCLC samples compared to normal samples (, p < .05), and its expression was significantly lower in the metastasis group than non-metastasis group (, p < .05). The 44 NSCLC patients were divided into high miR-520f group and low miR-520f group and the statistical results indicated that miR-520f expression was closely correlated with differentiation grade (p = .009), distal metastasis (p = .009), and TNM stage (p = .016), not relevant to age (p = .368), gender (p = .500), and tumor size (p = .146, ). Moreover, miR-520f expression in NSCLC was found to be a negative correlation with circ_0043278 expression (). These data claimed miR-520f acts as a downstream factor of circ_0043278 in NSCLC.
Figure 5. miR-520f expression was significantly decreased in NSCLC. (a) Wild type and mutant sequence of putative binding site between circ_0043278 and miR-520f. (b and c) Interaction between circ_0043278 and miR-520f was verified by dual-luciferase reporter assay in H1299 and A549 cells. (d and e) RNA pull down assay was used to further confirm the interaction between circ_0043278 and miR-520f using biotin-labeled miR-520f. (f) Kaplan–Meier survival analysis of NSCLC patients with low or high miR-520f expression. (g and h) Expression of miR-520f was detected by qRT-PCR in normal and NSCLC tissues. (i) Correlation between circ_0043278 and miR-520f expression.
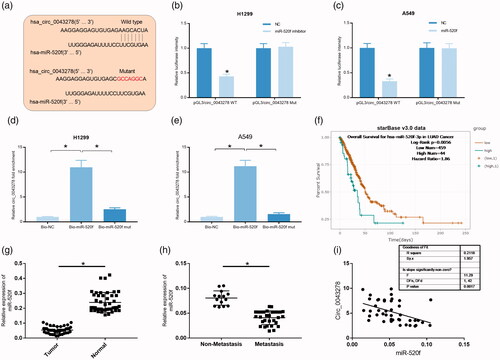
Table 2. Correlations between miR-520f expression and clinicopathologic characteristics in NSCLC.
MiR-520f acts as a tumor inhibitor in NSCLC
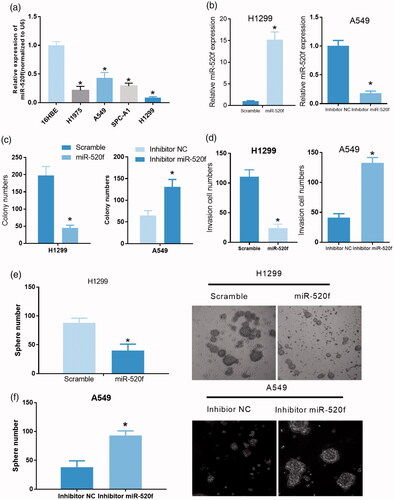
To further confirm the role of miR-520f in NSCLC, we detected the expression of miR-520f in 16HBE, H1975, A549, SPC-A1, and H1299 cells by qRT-PCR assay. The results showed a significant down-regulation of miR-520f in H1975, A549, SPC-A1, and H1299 cells compared with 16HBE cells (, p < .05). In order to investigate the functions of miR-520f in NSCLC, we constructed a miR-520f overexpressed cell model in H1299 cells and a miR-520f knockdown cell model in A549 cells by treating cells with miR-520f mimics and inhibitors, respectively. Efficiency of miR-520f overexpression and knockdown in H1299 and A549 cells were evaluated by qRT-PCR (, p < .05). Subsequently, the effects of miR-520f overexpression and knockdown on cell proliferation, invasion, and tumor sphere formation capability of NSCLC cells were measured, respectively. Results from colony formation assay showed a significant down-regulation of colony number in miR-520f mimics treated H1299 cells compared to those cells treated with scramble control, while the colony number in miR-520f inhibitor-treated A549 cells was significantly increased compared to inhibitor NC treated A549 cells (, p < .05). In transwell assay, overexpression of miR-520f in H1299 cells significantly reduced the invasive cell number, while knockdown of miR-520f in A549 cells remarkably increased the invasive cell number (, p < .05). Moreover, sphere number was found to be obviously reduced by treating H1299 cells with miR-520f mimics, while it was significantly increased by treating A549 cells with miR-520f inhibitor (, p < .05). These findings suggested that miR-520f played as a tumor inhibitor of NSCLC in vitro.
Figure 6. miR-520f acts as a tumor inhibitor in NSCLC. (a) Relative expression of miR-520f was detected in 16HBE, H1975, A549, SPC-A1 and H1299 cells via qRT-PCR (*p < .05 vs. 16HBE cells). (b) Relative expression of miR-520f in miR-520f mimics treated H1299 cells and miR-520f inhibitor treated A549 cells (*p < .05 vs. scramble or inhibitor NC group). (c) Effects of miR-520f overexpression and knockdown were evaluated by colony formation assay in H1299 and A549 cells, respectively (*p < .05 vs. scramble or inhibitor NC group). (d) Transwell assay was carried out in miR-520f mimics treated H1299 cells and miR-520f inhibitor treated A549 cells to assess the effects of miR-520f on cell invasion (*p < .05 vs. scramble or inhibitor NC group). (e and f) Tumor sphere formation assay was performed to assess the effects of miR-520f overexpression and knockdown on cell self-renewal capability in H1299 and A549 cells, respectively.
Circ_0043278 regulates ROCK1, CDKN1B and AKT3 expression in NSCLC cells by sponging miR-520f
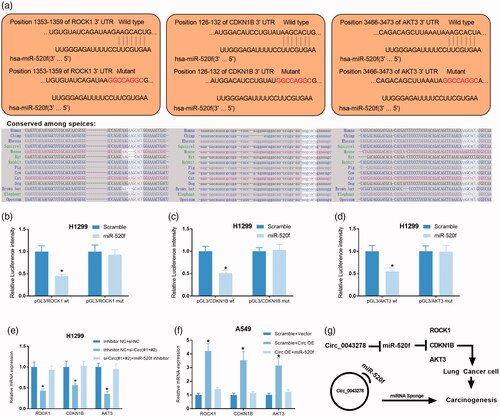
In the ceRNA network of circ_0043278 within miR-520f, we found that miR-520f could target to multiple key oncogenes (). Bioinformatics analysis showed there are putative binding sites between miR-520f and ROCK1, CDKN1B, or AKT3 and all of them are conserved among species (). So, we chose these genes for further validation. Dual-luciferase reporter assay showed that miR-520f mimics significantly attenuated the luciferase activity of H1299 cells driven by pGL3ROCK1 WT, pGL3CDKN1B WT, and pGL3AKT3 WT, but not that driven by pGL3ROCK1 Mut, pGL3CDKN1B Mut, and pGL3AKT3 Mut, compared to scramble control group (, p < .05). In addition, to investigate the roles of circ_0043278 in miR-520f/ROCK1-CDKN1B-AKT3 axis, relative mRNA expression of ROCK1, CDKN1B, and AKT3 were examined in H1299 cells treated with si-Circ#1+#2 and miR-520f inhibitor, as well as in A549 cells treated with Circ OE and miR-520f mimics, by qRT-PCR. Results indicated that circ_0043278 knockdown significantly decreased the mRNA production of ROCK1, CDKN1B, and AKT3 of H1299 cells, while circ_0043278 overexpression remarkably up-regulated the mRNA expression level of ROCK1, CDKN1B, and AKT3 of A549 cells (). Also, all of the effects could be reversed by the application of miR-520f or miR-520f mimics, respectively (). Altogether, we concluded circ_0043278 promoted the tumorigenesis of NSCLC by increasing the expression of ROCK1, CDKN1B, and AKT3 through miR-520f ().
Figure 7. Circ_0043278 regulates ROCK1, CDKN1B, and AKT3 expression in NSCLC cells by sponging miR-520f. (a) Wild type and mutant sequence of putative binding site between miR-520f and ROCK1, CDKN1B or AKT3. (b–d) Interaction between miR-520f and ROCK1, CDKN1B, or AKT3 was verified by dual-luciferase reporter assay in H1299 cells. (e) Relative mRNA expression of ROCK1, CDKN1B, and AKT3 were examined by qRT-PCR in H1299 cells treated with si-Circ#1+#2 + miR-520f inhibitor. (f) Relative mRNA expression of ROCK1, CDKN1B, and AKT3 in A549 cells treated with Circ OE + miR-520f were measured by qRT-PCR assay. (g) The diagram of the mechanisms underlying the circ_0043278/miR-520f axis in NSCLC.
Discussion
Although the tremendous advances have been made in medical technologies during the past decades, the incidence and prognosis of NSCLC remain unsatisfying [Citation15]. It is widely accepted that exploring novel diagnostic biomarkers and therapeutic targets are essential for improving the overall survival rate of NSCLC patients [Citation16]. Therefore, it is urgent to better understand the molecular pathways associated with NSCLC initiation and progression. CircRNAs are a novel class of ncRNAs with unclear biological functions in mammal cells and often exhibit tissue or time-specific expression [Citation17]. Highly conserved and remarkably stable are the two key features of circRNAs [Citation18,Citation19]. With these proprieties, circRNAs were considered to be promising biomarkers of tumor in precision medicine [Citation11]. Numerous studies have revealed a strong association of circRNAs with the pathogenesis of multiple human diseases, including Alzheimer’s disease, type 2 diabetes, and a variety of cancers [Citation17,Citation20,Citation21], implying a potential role of circRNA serving as molecular diagnostic biomarkers. Moreover, circRNAs were found to be involved in the tumorigenesis of NSCLC, recently [Citation22]. For example, a total of 185 circRNAs were detected to be dysregulated by an RNA sequencing performed by Hongbing Shen et al. in 10 pairs NSCLC and normal samples, among which circFARSA expression was found to be increased in NSCLC tissues and patients’ plasma, suggesting that circFARSA might act as a potential non-invasive biomarker for NSCLC [Citation23]. In this study, we focus on a circRNA microarray to detect the circRNA expression profile of NSCLC samples and paired normal samples to better characterize the relationship between circRNAs expression and NSCLC and to offer a preliminary theoretical basis for a search of biomarkers for the early diagnosis of NSCLC. We identified that circ_0043278 was increased in NSCLC compared to normal samples, implying a role of circ_0043278 in the pathogenesis of NSCLC. After validating the up-regulation of circ_0043278 in NSCLC tissues and cell lines, we investigated its role in NSCLC tumor progression in vitro and in vivo. Our findings indicating circ_0043278 might be a promising diagnostic and prognostic biomarker for NSCLC patients. However, more clinical samples need to validate the potential diagnoses value of circ_0043278 in NSCLC. In addition, a single circRNA may not reach a high-diagnostic accuracy. We think we should analyze more mircroarray data to fetch a panel of circRNAs in NSCLC biomarker in further study.
In the present study, we also investigated the underlying regulatory mechanism of circ_0043278 in NSCLC cells. Circ_0043278 is located at chr17: 35800000–35810000 with 250 length in gene TADA2A. As a newly identified circRNA, circ_043278 has not been reported by any studies to our best knowledge. However, circRNAs were demonstrated by increasing evidences to play a role in NSCLC progression through regulation of corresponding oncogene or inhibitory cancer genes by sponging target miRNAs [Citation24]. For instance, Yang W et al. reported that the promotive effects of circABCB10 on NSCLC progression in vitro and in vivo were demonstrated to be mediated by miR-1252/FOXR2 axis [Citation25]. In our study, we predicted the target miRNAs of circ_0043278 by bioinformatics analysis and verified circ_0043278/miR-520f axis could regulate ROCK1, CDKN1B, and AKT3 expression in NSCLC that gives strength as a evidence for the ceRNA regulation theory. In addition, we also investigated the role of miR-520f in NSCLC, which has been reported to be a famous tumor suppressor in gastric cancer, breast cancer, and hepatocellular carcinoma [Citation26–28].
In conclusion, our findings suggest that circ_0043278 promoted NSCLC progression by sponging miR-520f to release ROCK1, CDKN1B, and AKT3, providing multiple therapeutic targets and diagnostic biomarkers for NSCLC patients.
Supplementary_Excel_1.xlsx
Download ()Disclosure statement
The authors have no commercial or other associations that might pose a conflict of interest.
Additional information
Funding
References
- Qin H, Wang F, Liu H, et al. New advances in immunotherapy for non-small cell lung cancer. Am J Transl Res. 2018;10:2234–2245.
- Szejniuk WM, Robles AI, McCulloch T, et al. Epigenetic predictive biomarkers for response or outcome to platinum-based chemotherapy in non-small cell lung cancer, current state-of-art. Pharmacogenomics J. 2019;19(1):5–14.
- Sui H, Ma N, Wang Y, et al. Anti-PD-1/PD-L1 therapy for non-small-cell lung cancer: toward personalized medicine and combination strategies. J Immunol Res. 2018;2018:1.
- Reck M. Pembrolizumab as first-line therapy for metastatic non-small-cell lung cancer. Immunotherapy. 2018;10:93–105.
- Nagano T, Tachihara M, Nishimura Y. Mechanism of resistance to epidermal growth factor receptor-tyrosine kinase inhibitors and a potential treatment strategy. Cells. 2018;7:212–227.
- Beermann J, Piccoli MT, Viereck J, et al. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–1325.
- de Almeida RA, Fraczek MG, Parker S, et al. Non-coding RNAs and disease: the classical ncRNAs make a comeback. Biochem Soc Trans. 2016;44:1073–1078.
- Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J. 2017;474:4219–4251.
- Wei MM, Zhou GB. Long non-coding RNAs and their roles in non-small-cell lung cancer. Genomics Proteomics Bioinformatics. 2016;14:280–288.
- Chen Y, Lu L, Feng B, et al. Non-coding RNAs as emerging regulators of epithelial to mesenchymal transition in non-small cell lung cancer. Oncotarget. 2017;8:36787–36799.
- Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–148.
- Wang PL, Bao Y, Yee MC, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859.
- Wang X, Fang L. Advances in circular RNAs and their roles in breast cancer. J Exp Clin Cancer Res. 2018;37:206.
- Xu N, Chen S, Liu Y, et al. Profiles and bioinformatics analysis of differentially expressed circrnas in taxol-resistant non-small cell lung cancer cells. Cell Physiol Biochem. 2018;48:2046–2060.
- Osmani L, Askin F, Gabrielson E, et al. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): Moving from targeted therapy to immunotherapy. Semin Cancer Biol. 2018;52:103–109.
- Zhou Q, Huang SX, Zhang F, et al. MicroRNAs: a novel potential biomarker for diagnosis and therapy in patients with non-small cell lung cancer. Cell Prolif 2017;50(6):e12394.
- Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94.
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. CircRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66.
- Shao Y, Li J, Lu R, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6:1173–1180.
- Haque S, Harries LW. Circular RNAs (circRNAs) in health and disease. Genes (Basel). 2017;8(12):353.
- Zhao Z, Li X, Jian D, et al. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017;54:237–245.
- Yan B, Zhang W, Mao XW, et al. Circular RNA ciRS-7 correlates with advance disease and poor prognosis, and its down-regulation inhibits cells proliferation while induces cells apoptosis in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2018;22:8712–8721.
- Hang D, Zhou J, Qin N, et al. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018;7:2783–2791.
- Qu D, Yan B, Xin R, et al. A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am J Cancer Res. 2018;8:1387–1402.
- Tian X, Zhang L, Jiao Y, et al. CircABCB10 promotes nonsmall cell lung cancer cell proliferation and migration by regulating the miR-1252/FOXR2 axis. J Cell Biochem. 2019:120(3):3765–3772.
- Du X, Fan W, Chen Y. microRNA-520f inhibits hepatocellular carcinoma cell proliferation and invasion by targeting TM4SF1. Gene 2018;657:30–38.
- Meshkat M, Mesrian Tanha H, Ghaedi K, et al. Association of a potential functional mir-520f rs75598818 G > A polymorphism with breast cancer. J Genet. 2018;97:1307–1313.
- Hong S, Bi M, Chen S, et al. MicroRNA-520f suppresses growth of gastric carcinoma cells by target ATPase family AAA domain-containing protein 2 (ATAD2). Neoplasma. 2016;63:873–879.
