Abstract
Long non-coding RNAs (lncRNAs) have been identified as critical players in tumorigenesis. Previous studies revealed that lncRNA SBF2-AS1 was involved in tumor progression. However, the role and underlying mechanism of SBF2-AS1 in cervical cancer (CC) remain unknown. In the present study, our data showed that SBF2-AS1 expression was significantly increased in CC. High SBF2-AS1 expression was associated with advanced FIGO stage and lymph node metastasis of CC patients. Function assays showed that SBF2-AS1 inhibition significantly reduced CC cells proliferation both in vitro and in vivo. Mechanistically, we showed that SBF2-AS1 upregulation restrained the activity of miR-361-5p and led to overexpression of FOXM1 in CC cells. Furthermore, we found that miR-361-5p inhibitors could rescue the effects of SBF2-AS1 inhibition on CC cells proliferation. Taken together, we demonstrated that the SBF2-AS1/miR-361-5p/FOXM1 axis might play an important role in CC progression. SBF2-AS1 might serve as a potential therapeutic target for CC treatment.
Introduction
Cervical cancer (CC) is the second most common malignant cancer, and accounts for a large proportion of cancer-associated mortalities occurring in females [Citation1,Citation2]. Although improvements and advancements have been made in current treatments of CC by operative treatment, chemotherapy and radiotherapy, the long-term survival rate of CC patients is still unsatisfactory because of metastasis and recurrence [Citation3,Citation4]. Thus, it is urgent to explore the underlying molecular mechanism of CC tumorigenesis.
Long non-coding RNAs (lncRNAs) are a type of noncoding RNAs with more than 200 nucleotides (nt) in length and no or limited coding potential [Citation5]. Recently, increasing evidence showed that lncRNAs could serve as oncogenes or tumor suppressors through regulating cell proliferation, invasion and survival [Citation6,Citation7]. Recent studies revealed that lncRNAs might act as crucial regulators in CC development. For example, Zhang et al. found that lncRNA ANRIL promoted CC carcinogenesis via PI3K/Akt pathways [Citation8]. Zhang et al. found that lncRNA PVT1 facilitated CC progression through negative modulation of miR-128-3p [Citation9]. Guo et al. revealed that lncRNA SNHG20 promoted CC cells proliferation and invasion via miR-140-5p-ADAM10 axis [Citation10]. However, the functions of lncRNAs in CC progression remain largely unknown.
SBF2 antisense RNA 1 (SBF2-AS1), which is located at the 11p15.1 locus. Previous studies showed that lncRNA SBF2-AS1 could act an important regular in tumor progression. For example, Zhao et al. showed that high SBF2-AS1 expression was associated with advanced tumor progression and poor prognosis in patients with non-small cell lung cancer [Citation11]. Lv et al. revealed that SBF2-AS1 could promote proliferation of NSCLC cells both in vitro and in vivo [Citation12]. Zhang et al. found that SBF2-AS1 promoted hepatocellular carcinoma metastasis by regulating EMT and predicted unfavorable prognosis [Citation13]. Chen et al. revealed that SBF2-AS1 was significantly upregulated in esophageal squamous cell carcinoma (ESCC), and silencing of SBF2-AS1 inhibited the proliferative and invasive ability of ESCC cells [Citation14]. However, the function and underlying mechanism of SBF2-AS1 in CC remain unclear.
Materials and methods
Patients and tissue samples
A total of 66 paired CC tissues and adjacent non-tumor tissues (ANT) were obtained from patients who underwent surgery at Cangzhou Central Hospital between January 2015 and March 2018. All samples were processed and stored in liquid nitrogen immediately until use. None of the patients had received treatment. The study was approved by the Ethics Committee of Cangzhou Central Hospital Written informed consent was obtained from all patients.
Cell culture and transfection
Human CC cell lines (SiHa, HeLa, C33a, Me180 and Ms751) and human normal cervical cell lines (Ect1/E6E7) were purchased from American Type Culture Collection (ATCC, Manassas, VA). All cells were cultured with Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS, Hyclone, Rockville, Md) in humidified air at 37 °C with 5% CO2.
Si-SBF2-AS1, miR-361-5p mimics, miR-361-5p inhibitor (miR-361-5p-in) or negative control were bought from GenePharma (Shanghai, China). Cell transfection was using Lipofectamine 2000 reagents (Invitrogen, CA, Carlsbad, CA) according to manufacturer’s instructions. The siRNA sequences for SBF2-AS1 were SBF2-AS1-1: 5′-CAGAAGGAGUCUACUGCUAAG-3′ (Sense) and 5′-UAGCAGUAGACUCCUUCUGGG-3′ (Antisense) and SBF2-AS1-2: 5′-GCAAGCCUGCAUGGUACAUTT-3′ (Sense) and 5′-AUGUACCAUGCAGGCUUGCTT-3′ (Antisense).
RNA extraction and quantitative real-time PCR
Total RNAs were isolated from tissues or cell lines using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RNAs were inversely transcribed into cDNA using the PrimeScript RT reagent Kit (Takara, Dalian China). qRT-PCR experiments were completed using the SYBR Green Kit (Takara). The qRT-PCR results were normalized to U6 or GAPDH and expression fold change was calculated according to the 2−ΔΔCt method.
Cell proliferation assay
Cell proliferation was determined by using the Cell Counting Kit-8 (CCK-8, Beyotime, China) Kit. In brief, 2 × 103 cells were seeded into the 96-well plates and cultured for different times. Then 10 μL of the CCK-8 solution was added into the wells and incubated for 2 h at 37 °C. The absorbance at 450 nm was determined by a microplate reader (Bio-Rad, Carlsbad, CA).
Colony formation assay
Cells counted and seeded in six-well plates at 1000 cells/well. Cells were incubated for 14 days to form colonies. Then cells were washed by PBS, fixed by paraformaldehyde for 1 h, stained with Giemsa for 20 min, washed three times by ddH2O and then photographed with a digital camera. The number of colonies (>50 cells/colony) was counted under microscopy (Nikon, Japan)483.
Cell cycle assay
After the cells were treated for 24 h, cells were collected and fixed in 70% ethanol overnight. Then, cells were added with RNase (50 μg/mL) and PI (50 μg/mL) (BD Biosciences, Franklin Lakes, NJ) for incubation for 30 min, then the samples were subjected to flow cytometer (FACScan, BD Biosciences) for analysis. Data were analyzed using CELL Quest 3.0 software.
Cell apoptosis assay
After the cells were treated for 24 h, the cells were collected and washed with cold PBS buffer. The cells were then stained with Annexin V-FICT/PI followed the instruction of Annexin V-FITC/PI Apoptosis Detection Kit (eBioscience, San Diego, CA).
Luciferase reporter assay
The potential binding sites for miR-361-5p in SBF2-AS1 were predicted using DIANA tools. The sequences containing these wild-type (WT) or mutant (MUT) binding sites were inserted into pmirGLO luciferase vector (Promega, Madison, WI) to generate reporter plasmids. For luciferase reporter assay, the reporter plasmid and miR-361-5p mimics were co-transfected into HeLa cells and cultured for 24 h. Then the luciferase activity was measured using the dual-luciferase reporter assay system according to the manufacturer’s instructions.
RNA immunoprecipitation assay
RNA immunoprecipitation (RIP) assay was conducted using EZMagna RIP RNA-binding protein immunoprecipitation kit (Millipore, Billerica, MA) according to the manufacturer’s instructions. Cells in different groups were lysed using RNA lysis buffer containing protease inhibitor and RNase inhibitor. Cells were incubated with the RIP buffer containing magnetic beads coated with Ago2 antibodies (Millipore). IgG acted as a negative control. After incubation for 2 h at 4 °C, co-precipitated RNAs were isolated by TRIzol reagent and measured by PCR analysis.
Animal experiments
Four-week-old female BALB/C nude mice were randomly divided into two groups. HeLa cells transfected with si-NC or si-SBF2-AS1 were subcutaneously injected into the right flank of the mice. Tumor volumes and weights were measured at indicated time points. The animal experiment was approved by the Ethics Committee of Cangzhou Central Hospital.
Statistical analysis
All data were shown as mean ± SD. Statistical analysis was carried out using the SPSS software version 17.0(IBM, Armonk, NY). One-way analysis of variance (ANOVA) or two-tailed Student’s t-test was applied to determine the group differences. p < .05 was considered as significant difference.
Results
LncRNA SBF2-AS1 expression was increased in CC
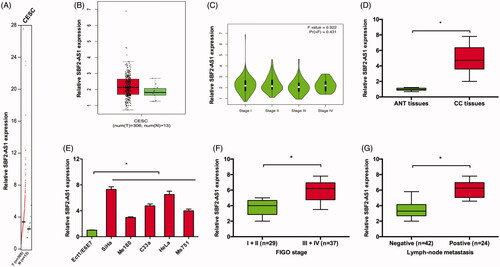
Firstly, we used GEPIA database to explore lncRNA SBF2-AS1 expression in CC, results showed that SBF2-AS1 expression was significantly increased in CC tissues compared to ANT tissues (), indicating SBF2-AS1 might be involved in CC development. Then, we explored SBF2-AS1 expression in CC tissues and cell lines. qRT-PCR showed that SBF2-AS1 expression was significantly increased in CC tissues and cell lines (). Notably, we found that SBF2-AS1 expression was positively correlated with advanced FIGO stage and lymph node metastasis in CC patients (). These data suggested that SBF2-AS1 might play critical roles in CC progression.
Figure 1. Increased SBF2-AS1 expression in CC. (A–C) SBF2-AS1 expression in CC tissues and ANT tissues was analyzed according to GEPIA database. (D,E) SBF2-AS1 expression in CC tissues and cell lines was determined by qRT-PCR. (F,G) High SBF2-AS1 expression was associated with advanced FIGO stage and lymph node metastasis of CC patients. *p < .05. CESC: cervical squamous cell carcinoma and endocervical adenocarcinoma.
LncRNA SBF2-AS1 inhibition decreased CC cells progression
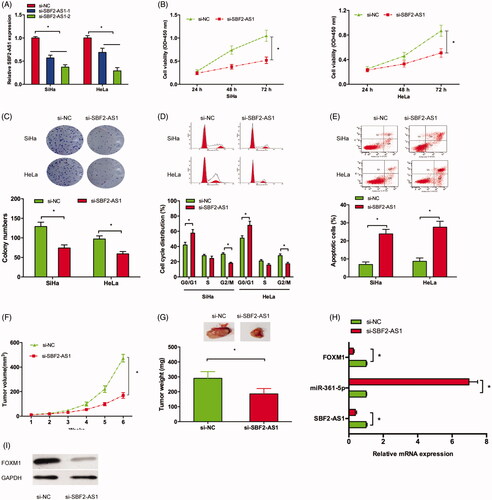
To explore the roles of SBF2-AS1 on CC progression, we transfected si-SBF2-AS1 into CC cells (). CCK-8 and colony formation assays showed that SBF2-AS1 inhibition significantly suppressed SiHa and HeLa cells viability in vitro (). Next, flow cytometry assay showed that SBF2-AS1 silencing arrested SiHa and HeLa cells in G0/G1 phase and induced cell apoptosis (). Then, we evaluate the effects of SBF2-AS1 on CC growth in vivo. Results showed that SBF2-AS1 suppression significantly inhibited the tumor volumes and weights in vivo (). In addition, qRT-PCR showed that SBF2-AS1 suppression decreased SBF2-AS1, FOXM1 expression, while increased miR-361-5p expression in xenograft tumor (). Moreover, Western blot assay showed that SBF2-AS1 suppression decreased FOXM1 protein expression in xenograft tumor (). Thus, these data suggested that SBF2-AS1 might act as a tumor oncogenic lncRNA in CC progression.
Figure 2. SBF2-AS1 inhibition reduced CC growth both in vitro and in vivo. (A) si-SBF2-AS1 transfection led to SBF2-AS1 inhibition in SiHa and HeLa cells. (B,C) CCK-8 and colony formation assays showed that SBF2-AS1 suppression inhibited CC cells viability. (D,E) Flow cytometry assay showed that SBF2-AS1 silencing arrested CC cells in G0/G1 phase and induced cell apoptosis. (F,G) The xenograft tumor volume and weight in the SBF2-AS1 inhibition group were significantly suppressed. (H) qRT-PCR was used to determine SBF2-AS1, miR-361-5p and FOXM1 expression in xenograft tumor. (I) Western blot showed SBF2-AS1 inhibition decreased FOXM1 protein expression in xenograft tumor. *p < .05.
LncRNA SBF2-AS1 acted as a sponge for miR-361-5p
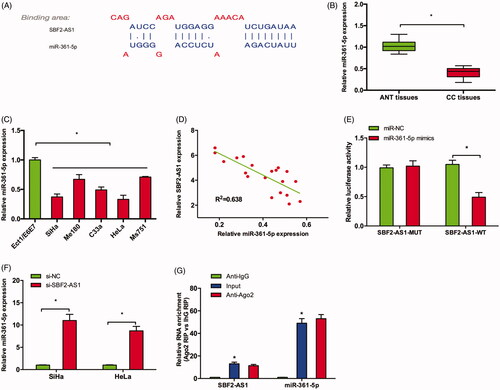
Recently, increasing studies showed that lncRNAs are involved in tumor progression through acting as competing endogenous RNAs (ceRNAs) for miRNAs [Citation15]. Thus, we explore the interaction between SBF2-AS1 and miRNA in CC. Bioinformatics analysis showed that SBF2-AS1 might interact with miR-361-5p (). qRT-PCR showed that miR-361-5p expression was downregulated in CC tissues and cell lines (), which was significantly negatively correlated with SBF2-AS1 expression in CC tissues (). Luciferase reporter assay revealed that miR-361-5p mimics significantly reduced the luciferase activity of SBF2-AS1-Wt reporter in CC cells (). qRT-PCR revealed that SBF2-AS1 inhibition increased miR-361-5p expression in SiHa and HeLa cells (). Moreover, RIP assay showed that SBF2-AS1 and miR-361-5p expression were significantly enriched in Ago2 pellet compared to IgG control ().
Figure 3. SBF2-AS1 acted as a sponge for miR-361-5p. (A) Potential binding site for miR-361-5p in SBF2-AS1. (B,C) miR-361-5p expression in CC tissues and cell lines was determined by qRT-PCR. (D) Correlation between SBF2-AS1 expression and miR-361-5p expression in CC tissues. (E) Luciferase reporter assay revealed that miR-361-5p mimics significantly reduced the luciferase activity of SBF2-AS1-Wt reporter in CC cells. (F) SBF2-AS1 inhibition increased miR-361-5p expression in SiHa and HeLa cells (G) RIP assay showed that SBF2-AS1 and miR-361-5p expression was significantly enriched in Ago2 pellet compared to IgG control. *p < .05.
SBF2-AS1 regulated FOXM1 expression via miR-361-5p
Previous studies revealed that FOXM1 could act as a target of miR-361-5p in tumor progression [Citation16,Citation17]. In the present study, GEPIA database showed that FOXM1 was significantly increased in CC tissues compared to ANT tissues (), which was further confirmed in CC tissues by qRT-PCR and IHC in our studies (). Kaplan-Meier Plotter analysis showed that high FOXM1 expression was associated with poor overall survival of CC patients (). Correlation analysis demonstrated that there was a negative correlation between miR-361-5p expression and FOXM1 expression in CC tissues ().
Figure 4. SBF2-AS1 regulated FOXM1 expression via miR-361-5p. (A,B) FOXM1 expression in CC tissues and ANT tissues was analyzed by GEPIA. (C,D) FOXM1 expression in CC tissues was determined by qRT-PCR and IHC. (E) Kaplan–Meier plotter analysis showed that high FOXM1 expression was associated with poor overall survival of CC patients. (F) Correlation between miR-361-5p expression and FOXM1 expression in CC tissues. (G) miR-361-5p inhibitors reversed the effects of SBF2-AS1 silencing on FOXM1 expression in protein levels. (H,I) Correlation between SBF2-AS1 expression and FOXM1 expression in CC tissues. (J) CCK-8 assay showed that miR-361-5p inhibitors significantly rescued the proliferation of SBF2-AS1 silenced CC cells. *p < .05.
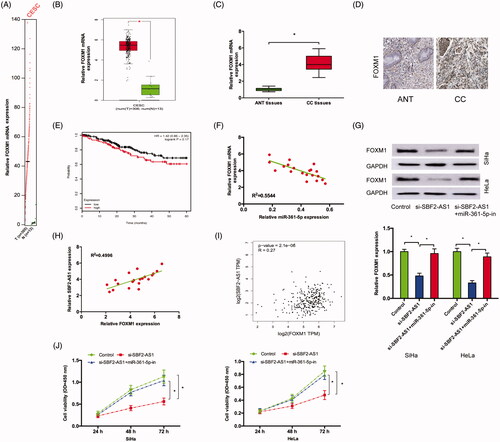
Next, we explored whether SBF2-AS1 can regulate FOXM1 via miR-361-5p. Western blot assay showed that SBF2-AS1 suppression significantly reduced FOXM1 protein expression, while miR-361-5p inhibitors reversed the effects (). Next, correlation analysis revealed that FOXM1 expression was positively correlated with SBF2-AS1 expression in CC tissues both in our data and GEPIA database (). Furthermore, CCK-8 assay showed that miR-361-5p inhibitors significantly rescued the proliferation of SBF2-AS1 silenced CC cells (). Thus, these data suggested that SBF2-AS1 promoted CC progression via regulating miR-361-5p/FOXM1axis.
Discussion
Recently, increasing studies showed that lncRNAs might play critical roles in the initiation and progression of human cancers. For example, Zhang et al. showed that upregulation of lncRNA MALAT1 correlated with tumor progression and poor prognosis in clear cell renal cell carcinoma [Citation18]. Gao et al. revealed that lncRNA ZFAS1 could act as an unfavourable prognostic factor and promoted glioma cell progression by activation of the Notch signaling pathway [Citation19]. Wang et al. found that lncRNA ADAMTS9-AS2 regulated the progression of ovarian cancer through targeting miR-182-5p/FOXF2 signaling pathway [Citation20]. However, the roles and underlying mechanisms of lncRNAs in tumor progression remain largely unknown. In the present study, we found that SBF2-AS1 was significantly increased in CC tissues and cell lines, high SBF2-AS1 expression was significantly correlated with advanced FIGO stage and lymph node metastasis in CC patients. In vitro function assays, CCK-8 and colony formation assays showed that SBF2-AS1 inhibition significantly suppressed CC cells viability. Flow cytometry assay revealed that SBF2-AS1 silencing arrested CC cells in G0/G1 phase and induced cell apoptosis. Moreover, we showed that SBF2-AS1 inhibition could reduce tumor growth in vivo. Thus, those data indicated that SBF2-AS1 might play important roles in CC progression.
Recently, increasing studies showed that lncRNAs regulated tumorigenesis through working as ceRNA [Citation15]. For example, Zhang et al. showed that lncRNA UCA1 promoted cell progression by acting as a ceRNA of ATF2 in prostate cancer [Citation21]. Chen et al. showed that lncRNA CCAT1 acted as a ceRNA to regulate cell growth and differentiation in acute myeloid leukemia via regulating miR-155/c-Myc axis [Citation22]. In the present study, miR-361-5p was confirmed as a novel interactive miRNA of SBF2-AS1. The function of miR-361-5p has not been discovered before. In the present study, we showed that miR-361-5p expression was significantly decreased and negatively correlated with SBF2-AS1 expression in CC. In addition, luciferase reporter and RIP assays showed that SBF2-AS1 might act as a molecular sponge for miR-361-5p in CC progression.
Forkhead box (FOX) superfamily of proteins play important roles in regulating a wide range of transcriptional activities ranging from cell homeostasis and development [Citation23]. FOXM1 is pro-oncogene transcription factor recognized as a master regulator of physiological and pathological processes, including cancer [Citation24]. For example, Luo et al. showed that FOXM1 promoted cell proliferation, invasion, and stem cell properties in nasopharyngeal carcinoma [Citation25]. Wang et al. showed that miR 876 5p inhibited the progression of glioblastoma multiforme by directly targeting FOXM1 [Citation26]. Recently, Hou et al. [Citation17] and Tian et al. [Citation17] revealed that FOXM1 could act as a target of miR-361-5p in tumor progression. In the present study, we found that FOXM1 was significantly increased and negatively correlated with miR-361-5p expression in CC tissues. High FOXM1 expression was associated with poor overall survival of CC patients. Western blot assay showed that SBF2-AS1 suppression significantly reduced FOXM1 expression, while miR-361-5p inhibitors reversed the effects. Moreover, the function assays revealed that miR-361-5p inhibitors significantly rescued the proliferation of SBF2-AS1 silenced CC cells.
In conclusion, we revealed that lncRNA SBF2-AS1 might work as a ceRNA for miR-361-5p to upregulate FOXM1 expression, consequently promoting CC progression. Thus, these findings indicated that SBF2-AS1 might act as a therapeutic target for CC treatment.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29.
- Chen W, Sun K, Zheng R, et al. Cancer incidence and mortality in China, 2014. Chin J Cancer Res. 2018;30:1.
- Franco EL, Duarte-Franco E, Ferenczy A. Cervical cancer: epidemiology, prevention and the role of human papillomavirus infection. Canadian Med Assoc J. 2001;164:1017–1025.
- Reynoso-Noverón N, Peña-Nieves A, Rodríguez MO, et al. Cervical cancer epidemiology: cervical cancer. Cham: Springer; 2017. p. 19–33.
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155.
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7.
- Shi X, Sun M, Liu H, et al. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166.
- Zhang D, Sun G, Zhang H, et al. Long non-coding RNA ANRIL indicates a poor prognosis of cervical cancer and promotes carcinogenesis via PI3K/Akt pathways. Biomed Pharmacother. 2017;85:511–516.
- Zhang D, Sun G, Zhang H, et al. Long non-coding RNA PVT1 facilitates cervical cancer progression through negative modulation of miR-128-3p. Int J Clin Exp Pathol. 2017;10:4522–4529.
- Guo H, Yang S, Li S, et al. LncRNA SNHG20 promotes cell proliferation and invasion via miR-140-5p-ADAM10 axis in cervical cancer. Biomed Pharmacother. 2018;102:749–757.
- Zhao QS, Li L, Zhang L, et al. Over-expression of lncRNA SBF2-AS1 is associated with advanced tumor progression and poor prognosis in patients with non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2016;20:3031–3034.
- Lv J, Qiu M, Xia W, et al. High expression of long non-coding RNA SBF2-AS1 promotes proliferation in non-small cell lung cancer. J Exp Clin Cancer Res. 2016;35:75.
- Zhang YT, Li BP, Zhang B, et al. LncRNA SBF2-AS1 promotes hepatocellular carcinoma metastasis by regulating EMT and predicts unfavorable prognosis. Eur Rev Med Pharmacol Sci. 2018;22:6333–6341.
- Chen R, Xia W, Wang X, et al. Upregulated long non-coding RNA SBF2-AS1 promotes proliferation in esophageal squamous cell carcinoma. Oncol Lett. 2018;15:5071–5080.
- Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859:169–176.
- Hou XW, Sun X, Yu Y, et al. miR-361-5p suppresses lung cancer cell lines progression by targeting FOXM1. Neoplasma 2017;64:526–534.
- Tian L, Zhao Z, Xie L, et al. MiR-361-5p suppresses chemoresistance of gastric cancer cells by targeting FOXM1 via the PI3K/Akt/mTOR pathway. Oncotarget. 2018;9:4886.
- Zhang H, Yang F, Chen SJ, et al. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol. 2015;36:2947–2955.
- Gao K, Ji Z, She K, et al. Long non-coding RNA ZFAS1 is an unfavourable prognostic factor and promotes glioma cell progression by activation of the Notch signaling pathway. Biomed Pharmacother. 2017;87:555–560.
- Wang A, Jin C, Li H, et al. LncRNA ADAMTS9-AS2 regulates ovarian cancer progression by targeting miR-182-5p/FOXF2 signaling pathway. Int J Biol Macromol. 2018;120:1705–1713.
- Zhang S, Dong X, Ji T, et al. Long non-coding RNA UCA1 promotes cell progression by acting as a competing endogenous RNA of ATF2 in prostate cancer. Am J Transl Res. 2017;9:366.
- Chen L, Wang W, Cao L, et al. Long non-coding RNA CCAT1 acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol Cells. 2016;39:330.
- Myatt SS, Lam EWF. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847.
- Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102.
- Luo W, Gao F, Li S, et al. FoxM1 promotes cell proliferation, invasion, and stem cell properties in nasopharyngeal carcinoma. Front Oncol. 2018;8:483.
- Wang L, Lu J, Zhang H, et al. MicroRNA-876-5p inhibits the progression of glioblastoma multiforme by directly targeting Forkhead box M1. Oncol Rep. 2019;41:702–710.
