?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Aim
The aim of the present study was to produce chrysin–curcumin-loaded PCL-PEG nanofibres by an electrospinning technique and to evaluate the biological activity of the chrysin–curcumin-loaded PCL-PEG fibres for wound healing and its related genes using in vivo methods.
Materials and methods
The electrospinning method was carried out for the preparation of the chrysin, curcumin and chrysin–curcumin-loaded PCL-PEG nanofibres with different concentrations. FTIR and SEM were performed to characterize the chemical structures and morphology of the nanofibres. In vitro drug release, as well as in vivo wound-healing studies were investigated in male rats. The expressions of genes related to the wound-healing process were also evaluated by real-time PCR.
Results
Our study showed that the chrysin–curcumin-loaded nanofibres have anti-inflammatory properties in several stages of the wound-healing process by affecting the IL-6, MMP-2, TIMP-1, TIMP-2 and iNOS gene expression. Our results demonstrated that the effect of the chrysin-loaded nanofibre, the curcumin-loaded nanofibre and the chrysin–curcumin-loaded nanofibre in the wound-healing process is dose dependent and in accordance with the obtained results in that it might affect the inflammation phase more than the other stages of the wound-healing process.
Conclusion
We have introduced chrysin–curcumin-loaded PCL-PEG nanofibres as a novel compound for shortening the duration of the wound-healing process.
Keywords:
Introduction
Chronic wound healing can result in infection and eventually cancer. One of the aims of modern medicine is shortening wound-healing time, increasing bioavailability and reducing the side effects of drugs [Citation1–3]. Among the many proposed wound-healing protocols, physical methods such as infrared waves [Citation4], high-voltage electric waves [Citation5], and drug delivery involving biomolecules such as endostatin [Citation6], vitamin A [Citation7], ascorbic acid [Citation8] and growth factors [Citation9] can be considered. A myriad of studies has also introduced natural compounds and “therapeutic agents” for decreasing wound “or ulcer” healing time [Citation10–13] for decreasing wound-healing time. However, none of these approaches has been widely accepted to be effective enough since wound healing is a very complicated biological process that involves intracellular and extracellular pathways [Citation14]. Different types of immune cells, endothelial cells, keratinocytes and fibroblasts play a pivotal role in wound healing. Moreover, the proliferation, differentiation and cellular migration of the aforementioned cells can play a vital role in the wound healing process [Citation15–17].
Using a high dose of antibiotics also appears to not have a strong effect on the wound-healing process since sometimes a low level of bacteria may aid in the wound-healing process through the recruitment of immune cells [Citation18]. Thus, a balance of the normal flora should be maintained in the wound area [Citation19]. In this regard, several studies showed that natural compounds such as herbal and mushroom extracts possess biomedical potential [Citation20–24]. It has been demonstrated that honey can remove harmful bacteria such as E.coli, salmonella and Staphylococcus aureus [Citation25,Citation26] and also plays a role in maintaining wound moisture which is necessary for healing [Citation27,Citation28]. It has also been declared that honey can promote angiogenesis in the wound area and epithelial tissue regeneration [Citation27].
Chrysin (5, 7-dihydroxyflavone) is a natural and biologically active flavonoid in many extracts, such as plants gum, honey, and propolis. Previous studies have reported that Chrysin possesses potent anti-inflammatory and anti-oxidant properties [Citation29,Citation30]. On the other hand, curcumin is a naturally occurring polyphenolic compound with a broad range of favourable biological functions, including anti-cancer, anti-oxidant and anti-inflammatory activities [Citation31]. The low bioavailability and in vivo stability of curcumin, however, require the development of suitable carrier vehicles to deliver the molecule in a sustained manner at therapeutic levels.
Despite the aforementioned functions, the poor water solubility of chrysin and curcumin hinders its application as a proper drug for wound healing [Citation32–34]. This limitation calls for the creation of an appropriate conveyor to increase the in vivo stability of the bioactive compound, bioavailability and eliminate their poor water solubility.
The wound-healing process involves a series of genes such as IL-6 [Citation35,Citation36], tissue inhibitor of metalloproteinase (TIMPs) [Citation37,Citation38], inducible nitric oxide synthase (iNOS) [Citation39,Citation40] and matrix metalloproteinases (MMPs) [Citation41,Citation42]. There are several studies that suggest the role of IL-6 in the early phase of wound healing [Citation35,Citation43]. MMPs are involved in extracellular matrix (ECM) degradation which makes for a poor wound healing process. On the other hand, TIMPs are essential for wound healing in smooth muscle cells, keratinocytes, fibroblasts and also endothelial cells by targeting MMPs [Citation44]. Furthermore, iNOS plays a significant role in wound healing when free radical levels are elevated [Citation45]. In recent years, the effect of reactive oxygen species (ROS) is under focus in different tissues because of its great influence on healing pathways [Citation46–49].
The aim of the present study is to fabricate chrysin–curcumin-loaded PCL-PEG nanofibres using the electrospinning technique [Citation50,Citation51] and then evaluate the biological activity of the chrysin–curcumin-loaded PCL-PEG fibres on the wound healing process and its related genes using an in vivo rat method.
Materials and methods
Materials
Polyethylene glycol (PEG) with a molecular weight (Mw) of 4000, dichloromethane, ε-caprolactone (ε-CL), dimethyl sulfoxide (DMSO), diethyl ether and methanol were purchased from Merck (Darmstadt, Germany). Tween20, chrysin and curcumin were provided by Sigma-Aldrich (St Louis, MO, USA) and Stannous octoate (Sn(Oct)2) was supplied by Alfa Aesar. Healthy male rats (2 and 3 weeks of age) were purchased from Pasteur Institute of Iran. TRIzol reagent, the QuantiTect reverse transcription kit, and the QuantiTect SYBR Green real-time PCR (RT-PCR) kit were purchased from Roche (Mannheim, Germany). A cDNA synthesis kit and a Gene JET RNA purification kit were purchased from Thermo Fisher Scientific (Waltham, USA).
Synthesis of PCL/PEG/PCL triblock copolymer
The most methods that follow goes on, described before in our previous study [Citation1]. As we showed in our previous study, PCL/PEG/PCL copolymers were prepared by the ring-opening polymerization (ROP) of ε-CL in the presence of PEG, using stannous octoate as a catalyst. In summary, 1 g of polyethylene glycol (PEG4000), 10 ml of ε-CL, and Sn(Oct)2 (0.5 wt.% of monomers) were added to a 25-ml round-bottom flask supported by a nitrogen atmosphere [Citation1]. The solution was heated to 130 °C for 6 h while stirring. dichloromethane was used for dissolving the resulting polymer and precipitated in cold diethyl ether three times and after that dried at room temperature for 72 h.
Preparation of chrysin- and curcumin-loaded nanofibre with utilization of electrospinning method
Like as this method shown before [Citation1], PCL/PEG/PCL (10% w/v) was dissolved in acetone, chloroform, methanol (at a 2: 1.5: 1.5 ratio) and DMSO (minimal amount for dissolving chrysin) to prepare the PCL/PEG/PCL solution. For the preparation of chrysin-loaded nanofibers, 5, 10 and 15% (w/w) chrysin (relative to the ratio of PCL-PEG-PCL) were added to the solvent. In the next step, 5, 10 and 15% (w/w) curcumin (relative to the ratio of PCL-PEG-PCL) were added to the solvent. To prepare the chrysin–curcumin-loaded nanofibre, three different conditions were followed as the addition of: (1) 5% (w/w) chrysin and 10% (w/w) curcumin; (2) 10% (w/w) chrysin and 5% (w/w) curcumin; (3) 7.5% (w/w) chrysin and 7.5% (w/w) curcumin (relative to PCL-PEG-PCL w/w) to the solvent.
Finally, a PCL-PEG-PCL triblock copolymer with a ratio of 10% (w/v) was added to the above-described solutions and stirred for 24 h at room temperature. A 5-ml plastic syringe with a blunt-ended metal needle tip (gauge 16) was used for each solution. The spinning solution was delivered at a controlled feeding rate of 2 ml/h, and the voltage between the ground collector on the electrospinning device and the needle tip (Fanavaran Nano-Meghyas, Tehran, Iran) was adjusted to a range of 28–30 kV. The distance between the nozzle and the collector and the rotating speed of the drum were fixed at 10 cm and 200 rpm, respectively. The metal collector was covered with an aluminium foil. All electrospinning experiments were carried out at room temperature [Citation52,Citation53]. To remove the excess DMSO after electrospinning, all nanofibres were washed with deionized water and dried in vacuum at 40 °C.
Scanning electron microscopy
By using scanning electron microscopy (SEM; model 6400; JEOL, Boston, MA, USA) the morphology and diameter of the PCL-PEG nanofibres with different concentrations of chrysin and curcumin-loaded PCL-PEG nanofibres were determined. At an accelerating voltage of 26 kV, samples were imaged.
Fourier transform infrared spectroscopy (FTIR)
The chemical structure of the PCL-PEG nanofibres, as well as the chrysin- and curcumin-loaded PCL-PEG nanofibres were performed by FTIR. The FTIR spectra of the nanofibres were obtained by using a Shimadzu 8400 s infrared spectrophotometer (Kyoto, Japan).
In vitro chrysin and curcumin release assay
To investigate the release kinetics of chrysin and curcumin from the nanofibres mats, a piece of fibre mat (20–30 mg, 2525
0.1 mm3) was placed into a 20 ml vial of phosphate-buffered saline (PBS, pH 7.4) containing Tween 20 (0.5%) and DMSO (5%) following by incubation at 37 °C with a shaking rate of 150 rpm for 15 days. In specific intervals, 1 ml of the media solution was taken to evaluate the absorbance of the chrysin and curcumin nanofibres by a UV spectrophotometer at a wavelength of 425 nm, and then, 1 ml fresh buffer was replaced in the solution. The buffer solution was used for the medium to determine the calibration curve. The release percentages of the chrysin and curcumin nanofibres were calculated according to the following equation:
In vivo wound-healing studies
In the Faculty of Advanced Sciences at Tabriz University of Medical Sciences this experiment was conducted. We purchased ninety male rats (weight: 250–300 grams) from the Tabriz University of Medical Sciences central animal house, and they caged in pathogen-free and in addition to food and water ad libitum, facilities on a 12-h12-h lightdark cycle. University of Tabriz animal ethics committee have approved all experiments. The rats were allocated to 10 groups and each group included nine rats. The most steps of wound creation and the other manners method is similar to our previous study [Citation1]. After anaesthesia, we created an open excision-type wound (22 cm2) on the thoracic-lumbar region of the rats to the depth of the panniculus carnosus region of rats. The wound was not dressed or covered. After recovery of the animals from anaesthesia, appropriate disinfected individual cages were prepared for caging them. All nanofibres were UV sterilized before use and each of them was placed on the wound site and slightly around the wound. We considered 10 groups; The first group was control group which received the PCL-PEG nanofibres for treatment while the 2nd, 3th and 4th groups were treated with 5% (w/w), 10% (w/w) and 15% (w/w) chrysin-loaded nanofibres and the 5th, 6th and 7th groups received 5% (w/w), 10% (w/w) and 15% (w/w) curcumin-loaded nanofibres, respectively. The 8th, 9th and 10th groups were treated with the chrysin 5%-curcumin 10% (w/w), chrysin 10%-curcumin 5% (w/w) and chrysin 7.5%-curcumin 7.5% (w/w) loaded nanofibres, respectively. Differences in the wound area were measured at 5, 10 and 15 days after treatment with nanofibres. The extent of wound healing was reported as the percentage of wound area that remained exposed. Each group, as well as the control, were tested on three rats and the mean value was recorded.
where W5, W10 and W15 are the wound areas after 5, 10 and 15 days treatment, and W0 is the initial wound area. Also wound sampling was done at 5, 10 and 15 days after treatment. At the end of each treatment time point, full-thickness sections of the wound were dissected and stored in −70 °C until RNA extraction.
RNA extraction and cDNA synthetic
As described before [Citation1], the sections which included full thickness of the wound were stored in −70 °C until RNA extraction. For RNA recovery from tissue biopsies by disruption, 500 µl TRIzol solution (Invitrogen, Carlsbad, CA, USA) was used, and then, for homogenization, we used a pestle mixer (Scientific Specialties, Randallstown, MD, USA), and then, RNA was extracted in 50 µl chloroform that prepared form Sigma-Aldrich. After that, isopropanol (Sigma, Castle Hill, NSW, Australia) used for precipitating RNA from the aqueous phase, and ice-cold 75% (w/w) ethanol used for washing the pellet. Then, in 100 μl Milli-Q water the pellet was dried and resuspended. 1 μl of Oligo (dT) primers, 1 μg of total RNA and 11 μl of diethylpyrocarbonate (DEPC) treated water were mixed in dry ice followed by incubation at 65 °C for 5 min and then stored on dry ice for 1 min to synthesize the complementary DNA (cDNA). 4 μl of 5X reaction buffer, 1 μl of RiboLockTM RNase Inhibitor (20 U/μl) and 2 μl of (10 mM) dNTP Mix, mixed as the reaction mixture of the components, was added before incubation at 25 °C for 5 min. 2 μl of M-MuL V reverse transcriptase (20 U/μl) was used for RNA reverse-transcription and then incubated at 25 °C for 5 min and further incubated at 42 °C for 60 min. The reaction was terminated at 70 °C for 5 min.
Real-time quantitative PCR
Based on the same previous method [Citation1], for all tissue samples, reverse transcription was performed in a single batch and in the same PCR run were analysed for a given primer set. Each reaction volume (20 µl total) in the PCR contained 3 μl of cDNA and a SYBR Green PCR Master Mix that purchased from Applied Biosystems was used as the amplification reagent supplement. The program for real-time PCR included the following: initial denaturation at 95 °C for 10 s, followed by cycles of denaturation at 94 °C for 15 s, annealing at 58 °C for 30 s, and 72 °C for 30 s. After that, the melting curve analysis of 70 °C to 95 °C used for assessing the amplicons. reports the primers sequences for the RT-PCR. The 2−ΔΔCT method was used to calculate ΔCT values in relation to β-actin CT values, in which ΔCT used for representing the difference between the CT value of the target genes and the CT value of β-actin.
Table 1. The primer sequences used in the present study.
Results
Morphology of the nanofibrous scaffolds
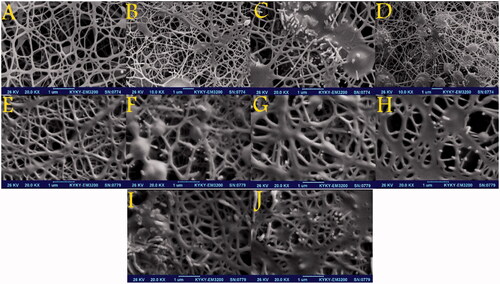
To investigate the physicochemical characterization of the nanofibres, the scaffolds were characterized by SEM. The average diameter of the PCL-PEG nanofibres and chrysin-loaded PCL-PEG nanofibres, curcumin-loaded PCL-PEG nanofibres and the chrysin–curcumin-loaded PCL-PEG nanofibres were in the range of 50–300 nm. shows the drug-free nanofibre of PCL-PEG with a nearly smooth surface and net-like structure. Likewise, the chrysin- and curcumin-loaded PCL-PEG nanofibre at 5% (w/w) showed a net-like structure with beads (). Increasing the chrysin concentration to 10% (w/w) significantly changed the size of beads and granular beads (). According to , although the chrysin-loaded PCL-PEG nanofibres 15% (w/w) were almost the same as the 10% (w/w), the granular beads were more populated. As shown in , the diameter of the nanofibres increased in the curcumin-loaded PCL-PEG nanofibre 10% (w/w) and according to the diameter increased even more for the curcumin-loaded PCL-PEG nanofibre 15% (w/w). reveals that the diameter of nanofibres increased for the 5% chrysin 10% curcumin (w/w). According to the which is related to 7.5% chrysin–7.5% curcumin (w/w) and 10% chrysin-5% curcumin (w/w), the beads transformed to the granular surface and their diameter increased. Due to the precipitation of curcumin at higher concentrations, the maximum concentration of curcumin that could be loaded in the PCL-PEG nanofibres was 15% (w/w).
Figure 1. SEM micrographs showing the morphology of the PCL-PEG nanofibres: (A) free PCL-PEG; (B) Chrysin 5% loaded PCL-PEG nanofibre; (C) Chrysin 10% loaded PCL-PEG nanofibre; (D) Chrysin 15% loaded PCL-PEG nanofibre; (E) Curcumin 5% loaded PCL-PEG nanofibre; (F) Curcumin 10% loaded PCL-PEG nanofibre; (G) Curcumin 15% loaded PCL-PEG nanofibre; (H) Chrysin 5%-Curcumin 10% loaded PCL-PEG; (I) Chrysin 7.5%-Curcumin 7.5% loaded PCL-PEG and (J) Chrysin 10%-Curcumin 5% loaded PCL-PEG.
FTIR results
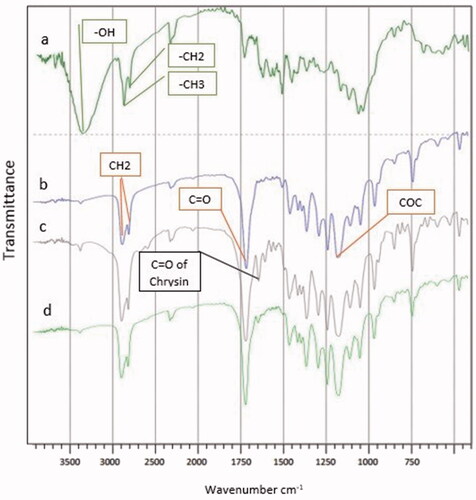
shows the FT-IR spectra of the PCL-PEG-PCL triblock copolymers. The peak at 1107 cm−1 is attributed to the C–O–C stretching vibration of the repeated -OCH2CH2 units present in PEG. Stretching vibrations of the ester carbonyl group appeared at 1726 cm−1. The peak at 1630 cm−1 is attributed to the C–C stretching. Characteristic bonds appeared at 3442 cm−1 (O–H, stretching) for the phenolic hydroxyl group. Signals at 2945 and 2866 cm−1 correspond to the characteristic absorption of the C–H stretching bonds of –CH2CH2, which are similar to those of ε-CL. These signals demonstrate the formation of the PCL-PEG-PCL triblock copolymer. are related to the curcumin PCL-PEG nanofibre, the chrysin PCL-PEG nanofibre and the curcumin–chrysin PCL-PEG nanofibre, respectively.
In vitro release study
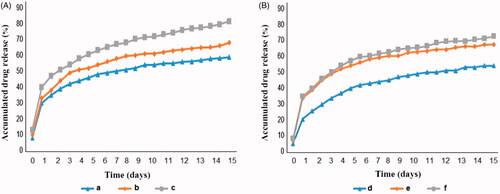
shows the release behaviour of the chrysin and curcumin in PBS from the nanofibre mat (A: chrysin and B: curcumin). The maximum amount of chrysin and curcumin were released from the nanofibres for day 1 (). As shown, the maximum release of chrysin and curcumin from the nanofibres is related to the amount of chrysin and curcumin. For example, the maximum release of chrysin from the nanofibre mat with 5, 10 and 15% chrysin was about 59, 68 and 81.5%, respectively. Based on the , the in vitro release of chrysin from the nanofibre mat was greater and faster than the curcumin.
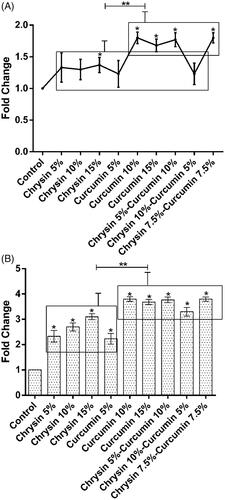
In vivo wound closure study
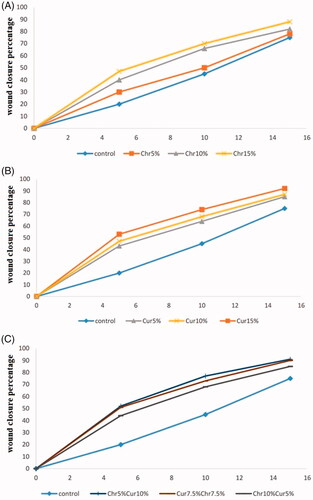
shows the picture of one of the rats treated with curcumin nanofibres (using chrysin 7.5%–curcumin 7.5% nanofibre, 5 days after treatment). The wound closure percentage in all rats was measured on 5, 10, and 15 days post-injury and the effect of chrysin, curcumin and chrysin–curcumin-loaded nanofibres on wound closure is shown in –C), respectively. According to these graphs, in all groups, the percentage of wound closure was higher than controls. In higher doses, the chrysin-loaded nanofibre significantly promoted the wound-healing process, in a way that the percentage of wound closure on day 15 was 78, 82 and 88%, for 5, 10 and the 15% chrysin-loaded nanofibres, respectively (). On day 15, the percentage of wound closure for 5, 10, and 15% curcumin nanofibre was around 85, 87, and 92%, respectively (). In general, although the wound closure efficacy of the nanofibres containing curcumin was higher than that of chrysin, there was no significant difference between the different percentage of curcumin compared to chrysin. also demonstrates that there is no synergistic effect between chrysin and curcumin for wound closure.
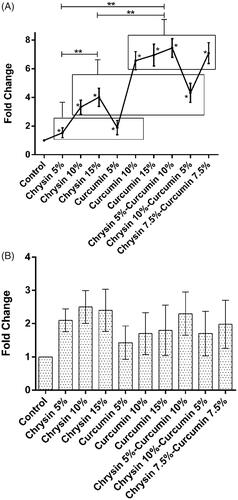
The effect of chrysin and curcumin nanofibres on IL-6 expression
On day 5, the results revealed that the chrysin-loaded PCL-PEG nanofibre 15% (w/w) significantly elevated the expression of IL-6 compared to 5, 10% (w/w) and the curcumin-loaded PCL-PEG 10% (w/w) more significantly elevated the expression of IL-6 than 5, 15% (w/w) in comparison with the control group (p values <.05). Likewise, the chrysin–curcumin-loaded PCL-PEG 7.5% (w/w) more significantly elevated IL-6 expression than the other chrysin–curcumin-loaded PCL-PEG nanofibres compared to the control group (p values <.05) ().
Figure 6. The expression of IL-6 after (A) 5 days treatment (B) 10 days treatment. X-axis represents the study groups and Y-axis shows fold change of the genes. Statistical analysis was performed by ANOVA. Each point represents the mean ± SEM. *p values <.05 vs. control. **p < .05.
On day 10, the chrysin-loaded PCL-PEG nanofibre 15% (w/w) significantly elevated the expression of IL-6 than the 5, 10% (w/w) and the curcumin-loaded PCL-PEG 10% (w/w) even more significantly elevated the expression of IL-6 than the nanofibres 5, 15% (w/w) in comparison with the control group (p values <.05). The chrysin–curcumin-loaded PCL-PEG 7.5% (w/w) and chrysin 5%-curcumin 10% were more effective for promoting the expression of IL-6 than the chrysin 10%-curcumin 5% in comparison with the control group (p values <.05) ().
The effect of chrysin and curcumin nanofibres on MMP-2 expression
On day 5, the chrysin loaded PCL-PEG 15% (w/w) more significantly elevated the expression of MMP-2 than the nanofibre 5, 10% (w/w) and compared to control group. Additionally, the curcumin-loaded PCL-PEG 15% (w/w) showed the same trend in comparison with nanofibers 5, 10% (w/w) (p values <.05). Compared to the control group, the chrysin 5%-curcumin 10% and the chrysin–curcumin-loaded PCL-PEG 7.5% (w/w) elevated the expression of MMP-2 more than the chrysin 10%-curcumin5% loaded PCL-PEG (w/w) (p values <.05) (). On day 10 post-treatment, the chrysin-loaded PCL-PEG 10% (w/w) remarkably elevated the expression of MMP-2 than the nanofibres 5, 15% (w/w) and the control group; and the same trend was observed for the curcumin-loaded PCL-PEG 10% (w/w) compared to the nanofibres 5, 15% and the control group (w/w) (p values <.05). The chrysin–curcumin-loaded PCL-PEG 7.5% (w/w) were more effective in promoting the expression of MMP-2 than the chrysin 5%-curcumin 10% and chrysin 10%-curcumin 5% (w/w) as well as the control group (p values <.05) ().
Figure 7. The expression of MMP-2 after (A) 5 days of treatment and (B) 10 days of treatment. The X-axis represents the study groups and the Y-axis shows the fold change of the genes. Statistical analysis was done by ANOVA. Each point represents the mean ± SEM. *p values < .05 vs. control. **p < .05.
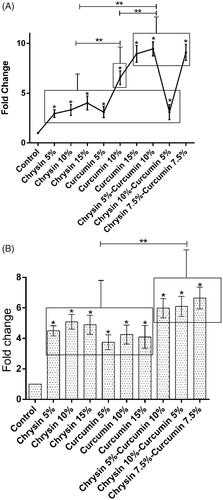
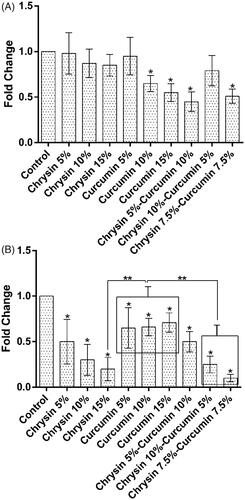
The effect of chrysin and curcumin nanofibres on TIMP-1 expression
On day 5, in comparison with the control group, the chrysin-loaded PCL-PEG 15% (w/w) significantly elevated the expression of TIMP-1 compared to the nanofibres 5, 10% (w/w); and the same trend was observed in the effect of curcumin-loaded PCL-PEG 15% (w/w) on the expression of TIMP-1 than the nanofibres 5, 10% (w/w) (p values <.05). Likewise, the chrysin 5%-curcumin 10% remarkably elevated the expression of TIMP-1 more than the chrysin 10%-curcumin 5% loaded PCL-PEG and the chrysin–curcumin-loaded PCL-PEG 7.5% (w/w) (p values <.05) (). On day 10, the chrysin-loaded PCL-PEG 10% (w/w) promoted the expression of TIMP-1 more than the nanofibres 5, 15% (w/w) and the control group; and a similar trend was noticed for the curcumin-loaded PCL-PEG 15% (w/w) in the expression of TIMP-1 compared to the nanofibres 5, 10% (w/w) and the control group, but these levels were not significantly different. In comparison with control group, the chrysin 5%–curcumin 10% elevated the expression of TIMP-1 more than the chrysin 10%-curcumin5% loaded PCL-PEG and the chrysin–curcumin-loaded PCL-PEG 7.5% (w/w), but these levels were not significant ().
The effect of chrysin and curcumin nanofibres on TIMP-2 expression
On day 5, in comparison with the control group, the chrysin-loaded PCL-PEG 15% (w/w) significantly elevated the expression of TIMP-2 more than the nanofibres 5, 10% (w/w) and the curcumin-loaded PCL-PEG 15% (w/w) more significantly elevated the expression of TIMP-2 than the nanofibres 5, 10% (w/w) (p values <.05). Furthermore, in comparison with control group, the chrysin 5%–curcumin 10% elevated the expression of TIMP-2 more than the chrysin 10%–curcumin 5% loaded PCL-PEG and the chrysin–curcumin-loaded PCL-PEG 7.5% (w/w) (p values <.05) (). On day 10, in comparison with the control group, the chrysin-loaded PCL-PEG 15% (w/w) significantly elevated the expression of TIMP-2 more than the nanofibres 5, 10% (w/w); and the curcumin-loaded PCL-PEG 15% (w/w) more significantly elevated the expression of TIMP-2 than the nanofibres 5, 10% (w/w) (p values <.05). Likewise, the same trend was observed for the chrysin 10%–curcumin 5% in terms of promoting the expression of TIMP-2 compared to the chrysin 5%–curcumin 10% loaded PCL-PEG and the chrysin–curcumin-loaded PCL-PEG 7.5% (w/w) (p values <.05) ().
Figure 9. The expression of TIMP-2 after (A) 5 days of treatment and (B) 10 days of treatment. The X-axis represents the study groups and the Y-axis shows the fold change of the genes. Statistical analysis was done by ANOVA. Each point represents the mean ± SEM. *p values < .05 vs. control. **p < .05.
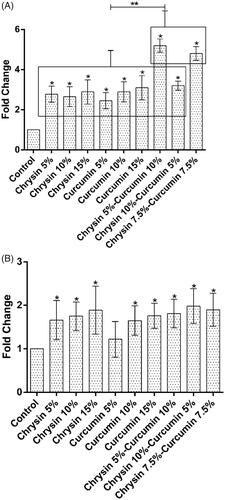
The effect of chrysin and curcumin nanofibres on iNOS expression
On day 5, in comparison with the control group, the chrysin-loaded PCL-PEG 15% (w/w) reduced more the expression of iNOS than the nanofibres 5, 10% (w/w) and the curcumin-loaded PCL-PEG 15% (w/w) more significantly reduced the expression of iNOS than the nanofibres 5, 10% (w/w) (p values <.05). The chrysin 5%–curcumin 10% reduced the expression of iNOS more than the chrysin 10%-curcumin 5% loaded PCL-PEG and the chrysin–curcumin-loaded PCL-PEG 7.5% (w/w) as well as the control group (p values <.05) (). On day 10, the chrysin-loaded PCL-PEG 15% (w/w) more significantly reduced the expression of iNOS compared to the nanofibers 5, 10% (w/w) and the curcumin-loaded PCL-PEG 5% (w/w) even more reduced the expression of iNOS in comparison with the nanofibres 10, 15% (w/w) (p values <.05). The chrysin–curcumin-loaded PCL-PEG 7.5% reduced the expression of iNOS more than the chrysin 10%–curcumin 5% loaded PCL-PEG and chrysin 5%-curcumin 10% (w/w) in comparison with the control group (p values <.05) ().
Discussion
The purpose of using the nanofibres in this study was to solve the problem associated with the insolubility of chrysin and curcumin-based on a simple principle that increasing the surface area of the drug and carrier can enhance the dissolution rate of the drug and provide a helpful pathway for the delivery of water-insoluble drugs. Therefore, the chrysin–curcumin-loaded nanofibre can be a more effective and promising candidate in wound healing applications [Citation54].
This interesting phenomenon for the chrysin-curcumin-loaded nanofibre has not been reported elsewhere. In this study, chrysin and curcumin presented a positive effect on an open wound-healing model in rats and also its effect on the expression of the different type of genes that are related to wound healing such as IL-6, TIMPs, iNOS and MMPs were systematically investigated.
IL-6 plays a critical role in inflammation and is produced by inflammatory cells [Citation43,Citation55]. Based on previous studies, IL-6 has a regulatory role in leukocyte recruitment in inflammatory conditions [Citation56,Citation57]. Also, it has been shown that IL‐6 deficient mice have a low level of macrophages and neutrophils [Citation36]. As the results show, chrysin and curcumin nanofibres elevated IL-6 gene expression during the wound-healing process after 10 days, and on day 10, the maximum expression was elevated by chrysin 7.5%–curcumin 7.5% loaded PCL-PEG. Based on these results, high doses of curcumin play a crucial role in inducing IL-6 expression.
Previous studies have been shown that MMP-2 gene is expressed at wound sites during the healing process and also MMPs have a crucial role in keratinocyte detachment from the basement membrane and the other healing process [Citation58–62]. Based on these results, MMP-2 gene expression was variable between groups for day 5 and day 10, so that in chrysin and curcumin groups, the 15% concentration elevated more MMP-2 expression and chrysin 10%–curcumin 5% loaded PCL-PEG reduced MMP-2 expression. For day 10, MMP-2 gene expression reduced in comparison with day 5 and also the curcumin group, especially curcumin 5% more greatly reduced MMP-2 expression.
TIMP genes are related to the cell proliferation stage in the process of wound healing [Citation23], and any disturbance in the expression of these genes may lead to a delayed wound healing [Citation36]. As our results showed, the expression of TIMP-1 reduced from day 1 to day 10. Also, it has been shown that for day 5, a higher level of TIMP-1 expression was related to the chrysin 5%-curcumin 10% loaded PCL-PEG, and for day 10, it was related to the chrysin 10% loaded PCL-PEG. On day 5, TIMP-2 expression higher level was related to chrysin 5%–curcumin 10% loaded PCL-PEG and for day 10, it was related to the chrysin 10%–curcumin 5% loaded PCL-PEG, but generally, TIMP-2 gene expression levels for day 10 were lower than day 5.
Wound-healing processes have an interconnected relationship with many components and one of these components is iNOS which plays a key role in wound healing and closure [Citation63]. As the present results showed, the down-regulation of iNOS gene for day 5 was related to the chrysin 5%-curcumin 10% loaded PCL-PEG and for day 10, the chrysin group in comparison with curcumin group more greatly reduced iNOS gene expression especially chrysin 15%. On the other hand, chrysin 7.5%–curcumin 7.5% loaded PCL-PEG reduced more iNOS gene expression than all of the other groups.
Conclusion
In conclusion, we have introduced here chrysin–curcumin-loaded PCL-PEG nanofibres as a novel compound to shorten the duration of the wound-healing process. The electrospinning of a PCL-PEG-chrysin–curcumin solution was performed and ended by the production of a yellow colour electrospun mat without any significant accumulation of chrysin on the surface. Our results showed that the chrysin–curcumin-loaded nanofibres have anti-inflammatory properties in several stages of the wound-healing process by affecting the IL-6, MMP-2, TIMP-1, TIMP-2 and iNOS gene expression. These results demonstrated that the effect of the chrysin-loaded nanofibre, the curcumin-loaded nanofibre and the chrysin–curcumin-loaded nanofibre in the wound-healing process is somewhat dose dependent and in accordance with the obtained results, it might affect the inflammation phase more than the other stages of wound-healing process. In this study, we focused on gene expression related to the inflammation phase and the evaluation of the expression of genes related to other phases, the proliferation and wound closure properties need to be investigated further.
Acknowledgements
This research was supported by the Medical Biotechnology Department of the Tabriz University of Medical Sciences, Tabriz, Iran.
Disclosure statement
The authors report that they have no conflicts of interest.
Additional information
Funding
References
- Mohammadi Z, Sharif Zak M, Majdi H, et al. The effect of chrysin-loaded nanofiber on wound healing process in male rat. Chem Biol Drug Des. 2017;90:1106–1114.
- Tan L, Hou Z, Gao Y. Efficacy of combined treatment with vacuum sealing drainage and recombinant human epidermal growth factor for refractory wounds in the extremities and its effect on serum levels of IL-6, TNF-α and IL-2. Exp Ther Med. 2018;15:288–294.
- Victor-Vega C, Desai A, Montesinos MC, et al. Adenosine A2A receptor agonists promote more rapid wound healing than recombinant human platelet-derived growth factor (Becaplermin gel). Inflammation. 2002;26:19–24.
- Braverman B, McCarthy RJ, Ivankovich AD, et al. Effect of helium-neon and infrared laser irradiation on wound healing in rabbits. Lasers Surg Med. 1989;9:50–58.
- Kloth LC. Electrical stimulation for wound healing: A review of evidence from in vitro studies, animal experiments, and clinical trials. Int J Low Extrem Wounds. 2005;4:23–44.
- Bloch W, Huggel K, Sasaki T, et al. The angiogenesis inhibitor endostatin impairs blood vessel maturation during wound healing. FASEB J. 2000;14:2373–2376.
- Hunt TK. Vitamin A and wound healing. J Am Acad Dermatol. 1986;15:817–821.
- Kaplan B, Gonul B, Dincer S, Dincer Kaya FN, et al. Relationships between tensile strength, ascorbic acid, hydroxyproline, and zinc levels of rabbit full-thickness incision wound healing. Surg Today. 2004;34:747–751.
- Cross KJ, Mustoe TA. Growth factors in wound healing. Surg Clin North Am. 2003;83:531–545.
- Tejada S, Manayi A, Daglia M, et al. Wound healing effect of curcumin: a review. Curr Pharm Biotechnol 2016; 17: 1002-1007.
- Firooz A, Bouzari N, Mojtahed F, et al. Topical immunotherapy with diphencyprone in the treatment of extensive and/or long-lasting alopecia areata. J Eur Acad Dermatol Venereol. 2005;19:393–394.
- Pazoki-Toroudi H, Nilforoushzadeh MA, Ajami M, et al. Combination of azelaic acid 5% and clindamycin 2% for the treatment of acne vulgaris. Cutan Ocul Toxicol. 2011;30:286–291.
- Toroudi HP, Rahgozar M, Bakhtiarian A, et al. Potassium channel modulators and indomethacin-induced gastric ulceration in rats. Scand J Gastroenterol. 1999;34:962–966.
- Lee DS, Sinno S, Khachemoune A. Honey and wound healing: An overview. Am J Clin Dermatol. 2011;12:181–190.
- Broughton G, Janis JE, Attinger CE. The basic science of wound healing. Plast Reconstr Surg. 2006;117:12S–34S.
- Martin P. Wound healing-aiming for perfect skin regeneration. Science. 1997;276:75–81.
- Reinders Y, Felthaus O, Brockhoff G, et al. Impact of platelet-rich plasma on viability and proliferation in wound healing processes after external radiation. IJMS. 2017;18:1819.
- Edwards R, Harding KG. Bacteria and wound healing. Curr Opin Infect Dis. 2004;17:91–96.
- Browne AC, Vearncombe M, Sibbald RG. High bacterial load in asymptomatic diabetic patients with neurotrophic ulcers retards wound healing after application of Dermagraft. Ostomy Wound Manage. 2001;47:44–49.
- Shivakumar M, Ramalingam S, Natarajan D. Larvicidal potential of some India medicinal plant extracts against Aedes aegypti (L). Asian J Pharm Clin Res. 2013;6: 77-80.
- Veer V, Gopalakrishnan R. Herbal insecticides, repellents and biomedicines: effectiveness and commercialization. New Delhi, India: Springer; 2016.
- Mukhtar MD, Sani A, Yakasai AA. Cytotoxicity of fractions of Pistia stratiotes l. On larvae of Culex mosquito and A. salina. Anim Res Int. 2004;1; 95-99.
- Karou SD, Savadogo A, Canini A, et al. Antibacterial activity of alkaloids from Sida acuta. Afr J Biotechnol. 2006;5; 195-200.
- Chelela B, Chacha M, Matemu A. Larvicidal potential of wild mushroom extracts against Culex quinquefasciatus Say, Aedes aegypti and Anopheles gambiae Giles S.S. Am J Res Commun. 2014;2:2325–4076.
- Mandal S, DebMandal M, Pal NK, et al. Antibacterial activity of honey against clinical isolates of Escherichia coli, Pseudomonas aeruginosa and Salmonella enterica serovar Typhi. Asian Pacific J Trop Med. 2010;3:961–964.
- Willix DJ, Molan PC, Harfoot CG. A comparison of the sensitivity of wound-infecting species of bacteria to the antibacterial activity of manuka honey and other honey. J Appl Bacteriol. 1992;73:388–394.
- Al-Waili N, Salom K, Al-Ghamdi AA. Honey for wound healing, ulcers, and burns; data supporting its use in clinical practice. ScientificWorldJ. 2011;11:766–787.
- Mostafavi, E. Soltantabar, P. Webster, TJ. Nanotechnology and picotechnology: A new arena for translational medicine. In: Biomaterials in translational medicine. Academic Press; 2019. p. 191–212.
- Cho H, Yun CW, Park WK, et al. Modulation of the activity of pro-inflammatory enzymes, COX-2 and iNOS, by chrysin derivatives. Pharmacol Res. 2004;49:37–43.
- Fonseca SF, Padilha NB, Thurow S, et al. Ultrasound-promoted copper-catalyzed synthesis of bis-arylselanyl chrysin derivatives with boosted antioxidant and anticancer activities. Ultrason Sonochem. 2017;39:827–836.
- Asadi N, Annabi N, Mostafavi E, et al. Synthesis, characterization and in vitro evaluation of magnetic nanoparticles modified with PCL-PEG-PCL for controlled delivery of 5FU. Artif Cells Nanomed Biotechnol. 2018;46:938–945.
- Munin A, Edwards-Lévy F. Encapsulation of natural polyphenolic compounds; a review. Pharmaceutics. 2011;3:793–829.
- Mat Nor MS, Manan Z, Mustaffa A, et al. Solubility prediction of flavonoids using new developed UNIFAC-based model. Chem Eng Trans. 2017;56: 799-804.
- Rahmani Del Bakhshayesh A, Mostafavi E, Alizadeh E, et al. Fabrication of three-dimensional scaffolds based on nano-biomimetic collagen hybrid constructs for skin tissue engineering. ACS Omega. 2018;3:8605–8611.
- Hafezi F, Gatzioufas Z, Angunawela R, et al. Absence of IL-6 prevents corneal wound healing after deep excimer laser ablation in vivo. Eye (Lond). 2018;32:156–157.
- Lin ZQ, Kondo T, Ishida Y, et al. Essential involvement of IL-6 in the skin wound-healing process as evidenced by delayed wound healing in IL-6-deficient mice. J Leukoc Biol. 2003;73:713–721.
- Kunkemoeller B, Kyriakides TR. Redox signaling in diabetic wound healing regulates extracellular matrix deposition. Antioxid Redox Signal. 2017;27:823–838.
- Vaalamo M, Leivo T, Saarialho-Kere U. Differential expression of tissue inhibitors of metalloproteinases (TIMP-1, -2, -3, and -4) in normal and aberrant wound healing. Hum Pathol. 1999;30:795–802.
- A Abd El-Aleem S, Muftah AA, B Jude E. Immunohistochemical characterization of the inflammatory responses in wound healing and the use of the subcutaneous polyvinyl alcohol (PVA) sponge implantation for evaluation of the healing process. J Cytol Histol. 2018;09: 519.
- Shi HP, Most D, Efron DT, et al. The role of iNOS in wound healing. Surgery 2001;130:225–229.
- Ellies LG, Hingorani DV, Lippert CN, et al. Impact of MMP-2 and MMP-9 activation on wound healing. Tumor Growth and RACPP Cleavage. BioRxiv . 2018: 327791.
- Wysocki AB, Staiano-Coico L, Grinnell F. Wound fluid from chronic leg ulcers contains elevated levels of metalloproteinases MMP-2 and MMP-9. J Invest Dermatol. 1993;101:64–68.
- Akira S, Kishimoto T. IL-6 and NF-IL6 in acute-phase response and viral infection. Immunol Rev. 1992;127:25–50.
- Xue M, Jackson CJ. Extracellular matrix reorganization during wound healing and its impact on abnormal scarring. Adv Wound Care (New Rochelle). 2015;4:119–136.
- Badr G, Hozzein WN, Badr BM, et al. Bee venom accelerates wound healing in diabetic mice by suppressing activating transcription factor-3 (ATF-3) and inducible nitric oxide synthase (iNOS)-mediated oxidative stress and recruiting bone marrow-derived endothelial progenitor cells. J Cell Physiol. 2016;231:2159–2171.
- Javedan G, Shidfar F, Davoodi SH, et al. Conjugated linoleic acid rat pretreatment reduces renal damage in ischemia/reperfusion injury: unraveling antiapoptotic mechanisms and regulation of phosphorylated mammalian target of rapamycin. Mol Nutr Food Res. 2016;60:2665–2677.
- Mehrjerdi FZ, Aboutaleb N, Pazoki-Toroudi H, et al. The protective effect of remote renal preconditioning against hippocampal ischemia reperfusion injury: role of KATP channels. J Mol Neurosci. 2015;57:554–560.
- Habibey R, Ajami M, Ebrahimi SA, et al. Nitric oxide and renal protection in morphine-dependent rats. Free Radic Biol Med. 2010;49:1109–1118.
- Arabian M, Aboutaleb N, Soleimani M, et al. Role of morphine preconditioning and nitric oxide following brain ischemia reperfusion injury in mice. Iran J Basic Med Sci. 2015;18:14–21.
- Mellor LF, Huebner P, Cai S, et al. Fabrication and evaluation of electrospun, 3D-bioplotted, and combination of electrospun/3D-bioplotted scaffolds for tissue engineering applications. Biomed Res Int. 2017;2017:6956794.
- Saghati, S, Akbarzadeh, A, Del Bakhshayesh, A, et al. Electrospinning and 3D Printing: Prospects for Market Opportunity. Electrospinning 2018;136–155.
- Mei L, Wang Y, Tong A, et al. Facile electrospinning of an efficient drug delivery system. Expert Opin Drug Deliv. 2016;13:741–753.
- Mei L, Fan R, Li X, et al. Nanofibers for improving the wound repair process: the combination of a grafted chitosan and an antioxidant agent. Polym Chem. 2017;8:1664–1671.
- Kamble P, Sadarani B, Majumdar A, et al. Nanofiber based drug delivery systems for skin: A promising therapeutic approach. J Drug Deliv Sci Technol. 2017;41:124–133.
- Kishimoto T, Akira S, Taga T. Interleukin-6 and its receptor: A paradigm for cytokines. Science. 1992;258:593–597.
- Natsume M, Tsuji H, Harada A, et al. Attenuated liver fibrosis and depressed serum albumin levels in carbon tetrachloride-treated IL-6-deficient mice. J Leukoc Biol. 1999;66:601–608.
- Romano M, Sironi M, Toniatti C, et al. Role of IL-6 and its soluble receptor in induction of chemokines and leukocyte recruitment. Immunity. 1997;6:315–325.
- Salo T, Mäkelä M, Kylmäniemi M, et al. Expression of matrix metalloproteinase-2 and -9 during early human wound healing. Lab Invest. 1994;70:176–182.
- Caley MP, Martins VLC, O'Toole EA. Metalloproteinases and wound healing. Adv Wound Care (New Rochelle). 2015;4:225–234.
- Ravanti L, Toriseva M, Penttinen R, et al. Expression of human collagenase-3 (MMP-13) by fetal skin fibroblasts is induced by transforming growth factor beta via p38 mitogen-activated protein kinase. FASEB J. 2001;15: 1098-1100.
- Nwomeh BC, Liang HX, Diegelmann RF, et al. Dynamics of the matrix metalloproteinases MMP-1 and MMP-8 in acute open human dermal wounds. Wound Repair Regen. 1998;6:127–134.
- Hartenstein B, Dittrich BT, Stickens D, et al. Epidermal development and wound healing in matrix metalloproteinase 13-deficient mice. J Invest Dermatol. 2006;126:486–496.
- Park JE, Barbul A. Understanding the role of immune regulation in wound healing. The Am J Surg. 2004;187:S11–S16.