Abstract
Accumulating studies showed that microRNAs are maintaining a variety of important biological processes but the underlying mechanism in proliferation and tumourigenicity is unclear. In this study we show that miR-342 expression in bone marrow and patients’ sera of childhood acute myeloid leukemia (AML) was both significantly higher than those in the corresponding normal controls. Functional assays demonstrated that forced expression of miR-342 significantly suppresses AML cell proliferation and G1/S transition of leukemia cells. Mechanistically, bioinformatics prediction and luciferase reporter assay identified N-a-acetyltransferase 10 protein (Naa10p) as a direct molecular target of miR-342, Naa10p siRNA significantly repressed cell proliferation and increased cell apoptosis. In conclusion, our study confirmed that miR-342/Naa10p plays key roles in AML progression, providing insights into underlying mechanisms of AML pathogenesis and also a potential therapeutic target for this malignancy.
Keywords:
Introduction
Acute myeloid leukemia (AML) is one of the most common and fatal forms of hematopoietic malignancies and arises from the differentiation arrest of myeloid precursor and malignant proliferation in bone marrow and blood [Citation1,Citation2]. During the last decade, much progress has been made in the clinical treatment of AML including chemotherapies [Citation3,Citation4]. However, the majority of patients with AML fail to survive longer than 5 years [Citation5]. Thus, the molecular mechanisms underlying the pathogenesis of AML is crucial to explore novel therapeutic strategies and improve the clinical outcome of AML.
The biological processes of AML development are regulated by several lineage-specific regulators, such as microRNAs (miRNAs), which demonstrates key roles in hematopoiesis.
MicroRNAs (miRNAs) are a class of small, non-coding RNAs that regulate gene expression by directly targeting the 3’UTR of their target genes. Accumulating studies showed that miRNAs may have links to tumourigenesis in various cancers [Citation6]. Recent studies showed that the biological processes of AML lineages are regulated by several lineage-specific regulators including microRNAs [Citation7]. For example, Krowiorz et al. showed that MiR-139-5p is a potent tumour suppressor in adult AML [Citation8]. Zhang et al. detected the expression levels of miRNAs in 112 de novo AML patients and 28 controls and found that miR-186 is down-regulated and predicts poor prognosis in AML patients [Citation9]. miR-99a was reported as an oncogene and is associated with poor prognosis of AML [Citation10]. Recently, aberrant expression of miR-342 has been frequently observed and may be associated with the development of a variety of prevalent cancers including hepatocellular carcinoma, cervical cancer, lung cancer and so on [Citation11–14]. However, the expression and the molecular mechanisms of miR-342 underlying the pathogenesis of AML remain unclear.
N-a-acetyltransferase 10 protein (Naa10p). also known as the paralog of the yeast gene arrest-defective-1 (ARD1) was first identified as the catalytic subunit of N-acetyltransferase A (NatA), one of the major N-terminal acetyltransferase complexes in eukaryotes. Studies showed that Naa10p plays an important role in several biological progression including cell proliferation [Citation15,Citation16], apoptosis [Citation17], metastasis [Citation18], neuronal development [Citation19] and autophagy [Citation20]. Recently, dysregulated Naa10p had been detected in several types of cancer [Citation16,Citation21,Citation22] associated with tumourigenesis [Citation17,Citation23], However, the correlation between Naa10p and AML remain unclear.
Here, we investigated the expression of miR-342 in AML progression and Naa10p. serve as a potential target of miR-342 in AML, implying potential therapeutic applications in AML patients.
Methods
AML samples and cell lines
The AML patient samples were obtained at the time of diagnosis with informed consent from the participants and were approved by the Affiliated Hospital of Jining Medical University. Samples were stored in liquid nitrogen until used. The MNC normal control samples were isolated from normal BM cells purchased from AllCells, LLC (Emeryville, CA) by use of NycoPrep 1.077 A (Axis-Shield, Oslo, Norway) according to the manufacturer’s manual. All patients were treated according to the protocols of the corresponding institutes/hospitals.
Cell culture and transfection
Human macrophage promyeloblast (Kg1a and HL60) and HEK-293T cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Kg1a and HL60 cells were maintained in RPMI1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS). The HEK-293T cells were cultured in DMEM medium (Gibco, Grand Island, NY, USA), supplemented with 10% fetal bovine serum (HyClone, Logan, Utah, USA), 100 U/ml penicillin, and 100 mg/ml streptomycin (Sigma-Aldrich, St. Louis, MO, USA) in 5% CO2 at 37 °C. The cells used for all experiments were in the log phase of growth. The synthetic miR-342 mimic, miR-342 inhibitor, and negative controls were purchased from GenePharma (Shanghai, China). Short hairpin RNA targeting human Naa10p was synthesized from Ribobio (Guangzhou, China). Transfection of miRNAs and shRNAs was performed with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer's protocol.
RNA extraction and qRT-PCR
Total RNA was extracted from the gastric cancer tissues or cells with Trizol (Invitrogen, CA, USA) according to RNA extraction protocol. And then, we used 2 µg total RNA to synthesis cDNA using the Taqman™ microRNA reverse transcription kit. Finally, TaqMan™ MicroRNA Assay kit (Applied Biosystems, USA) was used to detect the expression levels of miRNA and then normalized to U6 snRNA.
Cell proliferation assay
Cell proliferation assays of SGC-7901 cells transfected with indicated siRNA were performed 5 days after the transfection by incubating the cells with MTT (3–(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide).
Cell cycle assay
Cell cycle phases of leukemia cells were analyzed by measuring the DNA fragment staining with propidium iodide (PI, Sigma-Aldrich Co.) The cells were harvested, washed with PBS twice, fixed with 75% ethanol for 1 h at −20 °C. After washing with PBS, cells were incubated with incubation with PI dye solution containing RNaseA (30 mg/mL) for 20 min at room temperature in the dark. Finally, cell cycle was analyzed by flow cytometry with FACS calibers (BD Biosciences, San Jose, CA, USA) using CellQuest software (version 3.3, BD Biosciences, San Jose, CA, USA).
Luciferase reporter gene assays
The 3’-UTR of human Naa10p was amplified and subcloned into the pGL3-luciferase reporter plasmid (Promega). The miR-342 target of Naa10p 1 3’-UTR was mutated to the built pGL3- Naa10p-mut vector. The HEK293T cells were transfected with the plasmids and miR-342 mimics and then assayed the luciferase activity using the Dual-Luciferase Reporter Assay System (Promega) 48 h later.
Western blotting analysis
We used RIPA (Thermo Fisher Scientific) to get the whole cell extracts, and then, the protein samples were separated by SDS-PAGE (10%) and incubated with polyclonal (rabbit) anti-Naa10p antibody (Santa Cruz Bio-technology, Santa Cruz, CA, USA) overnight at 4 °C. Goat anti-rabbit IgG (Pierce, Rockford, IL, USA) secondary antibody for 1 h at room temperature and ECL detection systems (SuperSignal West Femto, Pierce) were used for detection.
Statistical analysis
Data are expressed as means ± standard errors of at least 3 independent experiments. Student’s t-test and one-way ANOVA were used to determine significance between groups. Statistical analysis was carried out using SPSS software (version 17.0). p-values <.05 were considered to be statistically significant.
Results
miR‑342 expression downregulates in patients with AML
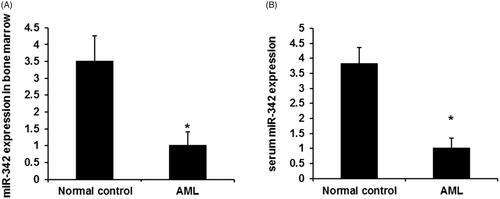
To reveal the potential role of miR-342 in AML, we firstly performed the qPCR assay to detect the expression levels of miR-342 in AML patient samples and normal control samples. Consistent with the reported upregulation of miR-342 in non-small cell lung cancer and cervical cancer [Citation12,Citation24], our data demonstrated a significant and global downregulation of miR-342 in both bone marrow and serum in AML patients compared with that in normal controls (). In conclusion, this result indicated that miR-342 may function as tumour suppressor in AML.
Figure 1. The expression levels of miR‑342 expression in AML. (A) The expression levels of miR‑342 expression in bone marrow AML patients and control groups. (B) The expression levels of miR-342 in patients’ sera and healthy controls. p-values were calculated using ANOVA. *p < .01 compared with the normal control group.
MiR‑342 represses cell proliferation and G1/S transition of AML
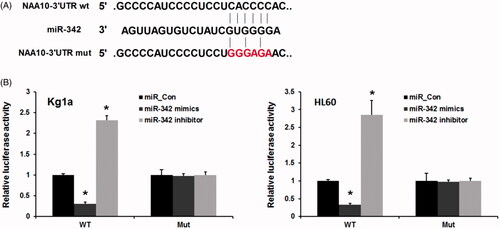
To elucidate the potential anti-tumour function of miR-183 in the progression of pediatric AML, we use miR-342 mimics and miR-342 inhibitor to overexpress and knockdown miR-342 expression. As shown in , miR-342 mimics evidently induces miR-342 expression, while miR-342 inhibitors evidently dramatically downregulated miR-342 expression in both two leukemia cells Kg1a and HL60. Next, we detected the cell proliferation ability of leukemia cells using the MTT assay. As shown in , forced expression of miR-342 also almost completely inhibited cell proliferation (p < .01), conversely, miR-342 knockdown significantly promoted AML cell proliferation (p < .01).
Figure 2. MiR-342 upregulation repress cell proliferation and G1/S transition of human leukemia cells. (A) MiR-342 mimics induced miR-342 expression in Kg1a and HL60 cells. (B) MTT assay revealed the effects of miR-342 on cell proliferation in Kg1a and HL60 cells. (C) The effects of MiR-342 on G1/S transition in Kg1a and HL60 cells. p < .01 vs. normal controls.
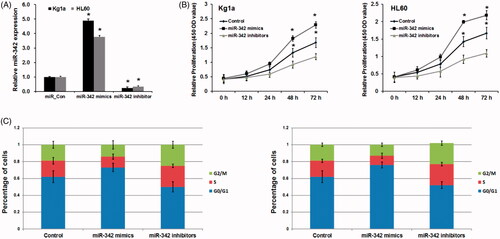
To further support these findings we examined the effects of miR-342 on the cell cycle of leukemia cells. As shown in , miR-342 mimic remarkably promoted ratio of G0-/G1-phase cells and significantly decreased the ratio of S-phase and G2-/M-phase cells, compared with the control group (p < .01). Inversely, miR-342 inhibitor promoted cell population of the S-phase and G2-/M- phase but decreased cell population of G0-/G1-phase of leukemia cell lines compared with the control group (p < .01). Collectively, our findings demonstrate that miR-342 is a pivotal anti-tumour gatekeeper in the progression of AML.
Naa10p is a direct target of MiR‑342 in AML
Recently, miR-342 was identified as an upstream regulator of several genes in various cancers, including AEG-1 [Citation25], E2F1 [Citation14], TIAM1 [Citation26] and BRCA1 [Citation27]. We then sought to investigate the downstream gene of miR-342 in AML. In this study, a target prediction analysis (TargetScan and the Miranda database) was used and revealed that Naa10p may contain putative binding sites of miR-342 at its 3’UTR ((A)). We next performed a luciferase 3′-UTR reporter assay to further confirm our speculation. As expected, we found that overexpressed miR-342 markedly inhibited the mRNA levels of Naa10p. Furthermore, inhibitors of miR-342 significantly increased the expression level of Naa10p. Consistently, miR-342 did not affect the luciferase activities of the Naa10p 3′-UTR with mutant-binding sites. Our results indicate that the binding sites may be involved in miR-342 regulation, and miR-342 directly binds to the 3′-UTR of Naa10p in human leukemia cells.
Naa10 is upregulated in AML and regulated by miR-342
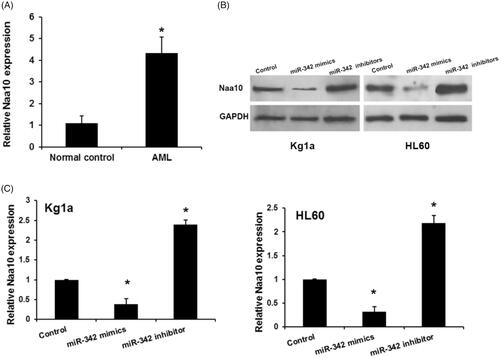
Given that Naa10p is known as the catalytic subunit of N-acetyltransferase A (NatA) and plays important role in several biological progression including tumourigenesis, we reasoned that Naa10 may function as an oncogene and be dysregulated in AML. We then sought to investigate the expression levels of Naa10 in AML. As shown in , Naa10 mRNA expression levels in serum in AML patients were significantly upregulated compared with that in normal controls. Then we analyzed the effects of miR-342 on Naa10 expression in human leukemia cells. As shown in , forced expression of miR-342 decreased the expression of endogenous Naa10 at both the mRNA and protein levels in Kg1a and HL60 cells, conversely, miR-342 knockdown significantly promoted Naa10p mRNA and protein expression in HL60 and K562 cells. Taken together, these data demonstrated that miR-342 could attenuate the expression of Naa10p by directly targeting its 3′-UTR.
Figure 4. Naa10 is upregulatd in AML and regulated by miR-342. (A) Expression levels of Naa10 in ALM. *p < .05, comparison with normal control groups B and C, miR-342 remarkably regulated Naa10 protein and mRNA expression in Kg1a and HL60 cells, respectively. Each sample was examined in triplicate. *p < .05, comparison with control groups.
Discussion
Although microRNAs (miRNAs) are increasingly linked to various physiologic processes, including tumourigenesis, the potential role of miRNAs in the myeloid development is poorly understood. In this study, we defined a novel tumour suppressor miRNA in AML, and confirm that miR-342 are key regulators of normal myeloid proliferation and their reduced expression is involved in acute myeloid leukemia development by target Naa10p directly.miR-342 was reported as a tumour suppressor and downregulated in various human cancers [Citation11,Citation28]. Li, X. R. reported that miR-342 is down-regulated in human cervical cancer tissues compared to the adjacent normal tissues [Citation12]. Smid, D. found high expression of miR-342 was relate to shorter time to progression and overall survival of gastric cancer [Citation29]. Decreased miR-342 also observed in osteosarcoma tissues and cell lines [Citation25]. In the present study, our clinical data showed that the expression levels of miR-342 decreased in both bone marrow and serum in AML patients compared with that in normal controls. The functional assay revealed the suppression of miR-343 on cell proliferation and G1/S transition. More recently, E2F1 and RAP2B were identified as a novel direct target of miR-342 in the progression of human lung cancer [Citation14,Citation24]. TIAM1 also regard as a direct target of miR-342 in extranodal NK/T-cell lymphoma [Citation26]. These studies showed that miR-342 may elicit its biological function by regulating numerous target genes expression during various pathogenesis. However, the target genes of miR-342 in AML remains unknown. In this study, we investigated the downstream gene of miR-342 in AML using TargetScan and the Miranda database. Combined with bioinformatics algorithms of miRNA target prediction and luciferase reporter assay, Naa10p was detected as the target gene of miR-342.
Naa10p first identified as the catalytic subunit of N-acetyltransferase A (NatA), was one of the major N-terminal acetyltransferase complexes in eukaryotes. Studies showed that Naa10p plays an important role in several biological progression [Citation15,Citation16] including tumourigenesis [Citation17,Citation23]. In this study, we found that Naa10p mRNA expression levels in serum in AML patients were significantly upregulated compared with that in normal controls. miR-342 decreased the expression of endogenous Naa10p at both the mRNA and protein levels, indicating that miR-342 could attenuate the expression of Naa10p by directly targeting its 3’-UTR in AML.
In conclusion, the dysregulation of miR-342- Naa10p axis may be involved in the aggressive progression of pediatric AML, implying a novel prognostic biomarker and potential therapeutic target for AML.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Zeisig BB, Kulasekararaj AG, Mufti GJ, et al. SnapShot: acute myeloid leukemia. Cancer Cell. 2012;22:698–698 e691.
- Wang XS, Gong JN, Yu J, et al. MicroRNA-29a and microRNA-142-3p are regulators of myeloid differentiation and acute myeloid leukemia. Blood. 2012;119:4992–5004.
- Deng L, Jiang L, Lin XH, et al. The PI3K/mTOR dual inhibitor BEZ235 suppresses proliferation and migration and reverses multidrug resistance in acute myeloid leukemia. Acta Pharmacol Sin. 2017;38:382–391.
- Schoof EM, Lechman ER, Dick JE. Global proteomics dataset of miR-126 overexpression in acute myeloid leukemia. Data Brief. 2016;9:57–61.
- Lu F, Zhang J, Ji M, et al. miR-181b increases drug sensitivity in acute myeloid leukemia via targeting HMGB1 and Mcl-1. Int J Oncol. 2014;45:383–392.
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96.
- Hu Y, Dong X, Chu G, et al. miR-137 downregulates c-kit expression in acute myeloid leukemia. Leuk Res. 2017;57:72–77.
- Krowiorz K, Ruschmann J, Lai C, et al. MiR-139-5p is a potent tumour suppressor in adult acute myeloid leukemia. Blood Cancer J. 2016;6:e508.
- Zhang TJ, Wang YX, Yang DQ, et al. Down-regulation of miR-186 correlates with poor survival in de novo acute myeloid leukemia. Clin Lab. 2016;62:113–120.
- Si X, Zhang X, Hao X, et al. Upregulation of miR-99a is associated with poor prognosis of acute myeloid leukemia and promotes myeloid leukemia cell expansion. Oncotarget. 2016;7:78095–78109.
- Gao Y, Zhang SG, Wang ZH, et al. Down-regulation of miR-342-3p in hepatocellular carcinoma tissues and its prognostic significance. Euro Rev Med Pharmacol Sci. 2017;21:2098–2102.
- Li XR, Chu HJ, Lv T, et al. miR-342-3p suppresses proliferation, migration and invasion by targeting FOXM1 in human cervical cancer. FEBS Lett. 2014;588:3298–3307.
- Zhao L, Zhang Y. miR-342-3p affects hepatocellular carcinoma cell proliferation via regulating NF-kappaB pathway. Biochem Biophys Res Commun. 2015;457:370–377.
- Tai MC, Kajino T, Nakatochi M. et al. miR-342-3p regulates MYC transcriptional activity via direct repression of E2F1 in human lung cancer. Carcinogenesis. 2015;36:1464–1473.
- Lim JH, Park JW, Chun YS. Human arrest defective 1 acetylates and activates beta-catenin, promoting lung cancer cell proliferation. Cancer Res. 2006;66:10677–10682.
- Seo JH, Cha JH, Park JH, et al. Arrest defective 1 autoacetylation is a critical step in its ability to stimulate cancer cell proliferation. Cancer Res. 2010;70:4422–4432.
- Arnesen T, Gromyko D, Pendino F, et al. Induction of apoptosis in human cells by RNAi-mediated knockdown of hARD1 and NATH, components of the protein N-alpha-acetyltransferase complex. Oncogene. 2006;25:4350–4360.
- Lee MN, Lee SN, Kim SH, et al. Roles of arrest-defective protein 1(225) and hypoxia-inducible factor 1alpha in tumour growth and metastasis. J Nat Cancer Inst. 2010;102:426–442.
- Ohkawa N, Sugisaki S, Tokunaga E, et al. N-acetyltransferase ARD1-NAT1 regulates neuronal dendritic development. Genes to Cells: Devoted to Mol Cell Mech. 2008;13:1171–1183.
- Kuo HP, Lee DF, Chen CT, et al. ARD1 stabilization of TSC2 suppresses tumourigenesis through the mTOR signaling pathway. Sci Signal. 2010;3:ra9.
- Zeng Y, Zheng J, Zhao J, et al. High expression of Naa10p associates with lymph node metastasis and predicts favorable prognosis of oral squamous cell carcinoma. Tumour Biol. 2016;37:6719–6728.
- Arnesen T, Thompson PR, Varhaug JE, et al. The protein acetyltransferase ARD1: a novel cancer drug target? Curr Cancer Drug Targ. 2008;8:545–553.
- Zeng Y, Min L, Han Y, et al. Inhibition of STAT5a by Naa10p contributes to decreased breast cancer metastasis. Carcinogenesis. 2014;35:2244–2253.
- Xie X, Liu H, Wang M, et al. miR-342-3p targets RAP2B to suppress proliferation and invasion of non-small cell lung cancer cells. Tumour Biol. 2015;36:5031–5038.
- Zhang S, Liu L, Lv Z, et al. MicroRNA-342-3p inhibits the proliferation, migration, and invasion of osteosarcoma cells by targeting astrocyte-elevated gene-1 (AEG-1). Oncol Res. 2017;09:1505–1515.
- Huang H, Fan L, Zhan R, et al. Expression of microRNA-10a, microRNA-342-3p and their predicted target gene TIAM1 in extranodal NK/T-cell lymphoma, nasal type. Oncol Lett. 2016;11:345–351.
- Crippa E, Lusa L, De Cecco L, et al. miR-342 regulates BRCA1 expression through modulation of ID4 in breast cancer. PloS One. 2014;9:e87039.
- Weng C, Nguyen T, Shively JE. miRNA-342 Regulates CEACAM1-induced lumen formation in a three-dimensional model of mammary gland morphogenesis. J Biol Chem. 2016;291:16777–16786.
- Smid D, Kulda V, Srbecka K, et al. Tissue microRNAs as predictive markers for gastric cancer patients undergoing palliative chemotherapy. Int J Oncol. 2016;48:2693–2703.