Abstract
Objectives
Accumulating evidence show that histone demethylases play important roles in various types of cancers, including non-small cell lung cancer (NSCLC). In the current study, we evaluated the diagnostic value of lysine(K)-specific demethylase 6B (KDM6B) in NSCLC.
Methods
Serum KDM6B expression levels of 115 NSCLC patients and 88 healthy volunteers were detected by reverse transcription quantitative real-time polymerase chain reaction (qRT-PCR). The relationship between KDM6B and clinical characteristics was assessed by chi-square test. Receiver operating characteristic (ROC) analysis was applied to evaluate diagnostic efficacy.
Results
Serum KDM6B was significantly lower in NSCLC patients than that in healthy controls (p < .001). Moreover, low KDM6B expression was significantly associated with the high clinical stage (p = .028) and positive lymph node metastasis (p = .031). Besides, we found that the expression of KDM6B mRNA was also significantly different among healthy controls, NSCLC early stage and later stage patients (p < .05). ROC curve indicated that KDM6B could serve as a diagnostic marker for NSCLC with the cut-off value of 0.955. The AUC was 0.897 with a sensitivity of 79.5% and specificity of 84.3%.
Conclusion
Down-regulation of KDM6B is significantly associated with aggressive progression of NSCLC and KDM6B may be a tool of early detection of NSCLC.
Introduction
Non-small cell lung cancer (NSCLC) is the most common type of lung cancer with high morbidity and mortality in the world [Citation1–3]. What is worse, the occurrence rate of NSCLC is increasing in recent years [Citation4]. Although great progress has been made in NSCLC treatments, the clinical outcomes of the patients, especially those with advanced NSCLC, remain poor [Citation5]. Unfortunately, more than 65% of the patients with NSCLC are diagnosed in advanced stages with locally advanced or metastasis [Citation6]. Therefore, the markers for early detection of NSCLC are vitally important for the prognosis of the patients. In previous studies, several molecular markers were identified for NSCLC detection, including epidermal growth factor receptor, TP53, carcinoembryonic antigen (CEA), and cancer antigen-125 (CA125) [Citation7–9]. However, due to the low sensitivity and specificity, the clinical application of these markers is limited [Citation10,Citation11]. Therefore, novel biomarkers with high sensitivity and specificity are urgently needed for the early diagnosis of NSCLC, which may also improve the treatments of cancer.
Histone methylation, as a mechanism of epigenetic regulation, plays important roles in the development of several diseases, as well as cancers [Citation12]. Accumulating evidence have proved that histone methyltransferases and demethylases are significantly associated with various cells and tumor progression, including cell proliferation, invasion and metastasis [Citation13–15]. It was reported that the genes associated with histone methylation could serve as diagnostic and prognostic markers for cancer [Citation16]. Lysine(K)-specific demethylase 6B (KDM6B, also known as JMJD3), an H3K27 demethylase, is a histone lysine demethylase. KDM6B plays importantly functional roles in tumor progression. A study carried out by Tokunaga et al. proved that KDM6B could regulate colorectal cancer cell line progression, moreover, its expression level could indicate the outcomes of the patients, which may act as a prognostic marker in the patients [Citation17]. In the study of Ene et al., KDM6B was proved to involve in the gliomagenesis via regulating the p53 pathway [Citation18]. The effects of KDM6B on NSCLC cell lines were also investigated in the previous studies, suggesting KDM6B can promote cell apoptosis [Citation19]. However, the diagnostic value of KDM6B in NSCLC had been rarely reported in the previous studies.
In the present study, we aimed to evaluate the diagnostic significance of KDM6B in NSCLC. Patients pathologically diagnosed with NSCLC and healthy individuals were enrolled in the study. We compared the expression level of KDM6B between the participants and evaluated the diagnostic value of the gene. The present study may exploit a novel diagnostic marker for NSCLC, which may be helpful for cancer detection and therapy.
Methods and materials
Patients and samples
This study was approved by the Ethics Committee of the Hospital. Informed consent was obtained from each participant.
The study was scheduled in Shanghai East Hospital, Tongji University School of Medicine. One hundred fifteen patients pathologically diagnosed with NSCLC were finally enrolled, including 73 with squamous carcinoma and 42 with adenocarcinoma. The control group included 88 healthy volunteers. None of the control patients had formerly been diagnosed with any malignancy. Blood specimens were collected from all the participants in the morning after fasting for 8–10 h.
About 5 ml peripheral blood was collected in PAXgene Blood RNA Tubes (Qiagen, PreAnalytiX, Hombrechtikon, Switzerland ) specifically designed for the collection and stabilization of cellular RNA from whole blood. Serum samples were obtained by being centrifuged at 2500 × rpm for 20 min. The serum samples were stored at -80 °C until RNA extraction, which was performed within 6 months of collection.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from serum using TRIzol LS reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA quantity and quality were assessed with NanoDrop 2000 (Thermo Scientific, Wilmington, DE, USA). cDNA was generated by reverse transcription of 100 ng RNA from each sample in triplicate, using PrimeScript RT kit (Tahara, Dalian, China), according to the manufacturer’s instructions. The resulting cDNA was pooled and amplified by qRT-PCR, which was performed with SYBR green I Master Mix kit (Invitrogen). GAPDH served as internal control and the relative expression of KDM6B was calculated by 2-ΔΔCt method. The primer sequences were as followed: KDM6B forward: 5′-TGGTCTGTTGTACCCCACTG-3′, reverse: 5′-CGACAATGACTCCGCCTCTG-3′; GAPDH forward: 5′-TGCACCACCAACTGCTTAGC-3′, reverse: 5′-GGCATGCACTGTGGTCATGAG-3′.
The protein level of KDM6B was evaluated by Western blot according to the standard operation. The primary antibody was anti-JMJD3 (ab38113, Abcam, Cambridge, UK) and the protein levels were detected by ECL Prime Western Blotting Detection System (Amersham/GE, USA).
Statistical analysis
All statistical analyses were carried out in SPSS 18.0 software (SPSS, Chicago, IL, USA). Graphs were plotted by GraphPad Prism 5 (GraphPad, San Diego, CA, USA). The continuous data were presented as mean ± SD and compared between two groups by Student's t test. Chi-square test was used to analyze the association between KDM6B expression and clinicopathologic features. ROC curve was applied to evaluate the diagnostic value of KDM6B in NSCLC. p values less than .05 was considered statistically significant.
Results
Serum KDM6B expression in NSCLC patients
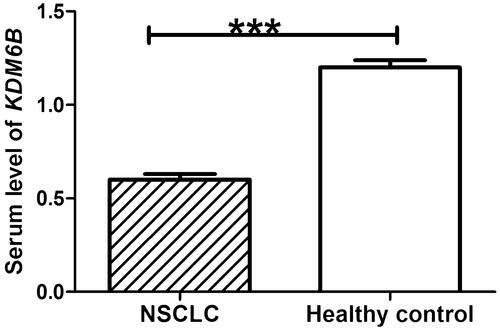
QRT-PCR was used to detect the relative expression of KDM6B in collected serum specimens. The results showed that serum KDM6B expression was significantly decreased in NSCLC, compared with healthy control (p < .001) (). The results of Western blot suggested that the protein level of KDM6B was markedly down-regulated in NSCLC patients, compared with healthy control, which was in accordance with the results of mRNA level.
Figure 1. Relative expression levels of KDM6B mRNA levels in NSCLC patients and healthy control. Student’s t test indicated that serum level of KDM6B was significantly decreased in NSCLC patients, compared with healthy control. ***: indicated p < .001.
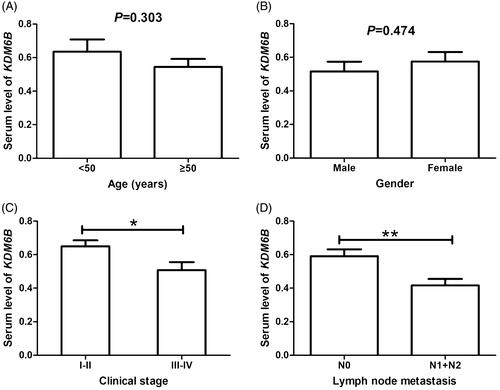
In addition, we also analyzed the expression differences of KDM6B mRNA among NSCLC patients according to different age, gender, clinical stage, and lymph node metastasis. As displayed in , KDM6B expression was not significantly influenced by patients’ age (p = .303) or gender (p = .474), but clinical stage (p = .009) and lymph node metastasis (p = .004) showed close association with KDM6B mRNA levels in NSCLC patients.
The association of KDM6B expression with clinicopathologic features of NSCLC
In order to evaluate the relationship between KDM6B expression and clinicopathologic variables, the patients were divided into two groups according to their average expression of KDM6B. Sixty-two patients were classified as low-KDM6B group and 53 patients were classified in high-KDM6B group. Chi-square test was carried out to assess the association between KDM6B expression levels and different clinicopathologic factors. The result showed that low KDM6B expression was significantly correlated with high clinical stage (p = .028) and positive lymph node metastasis (p = .031). In addition, there was no marked relationship between KDM6B expression and other clinical features, such as age, gender, smoking index, histological type, differentiation, or tumor size (p>.05 for all) ().
Table 1. Association between KDM6B expression and clinicopathological characteristics of NSCLC patients.
The diagnostic value of KDM6B expression for NSCLC patients
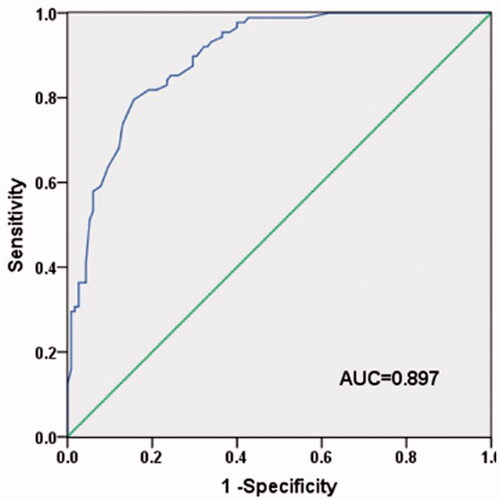
To evaluate the diagnostic value of serum KDM6B mRNA level in NSCLC patients, we performed ROC curve analysis. Analysis results indicated that KDM6B could distinguish the NSCLC patients from healthy individuals with the optimal cut-off value of 0.955. The area under the ROC curve (AUC) was 0.897, with the sensitivity of 79.5% and the specificity of 84.3% ().
Figure 3. ROC curve analyses for the diagnostic value of KDM6B in NSCLC. Analysis results suggested that KDM6B could serve as a diagnostic marker for NSCLC with the sensitivity of 79.5% and the specificity of 84.3%. The cut-off value was 0.955 and the AUC was 0.897.
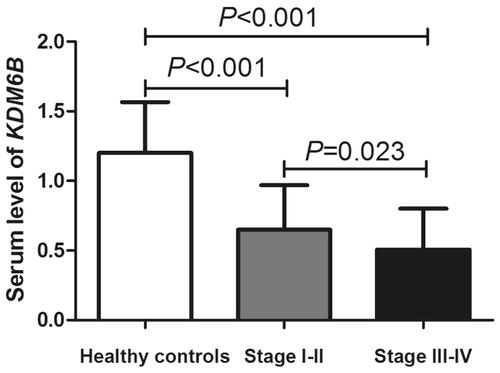
Moreover, we explored the expression differences of serum KDM6B mRNA among healthy controls, NSCLC early stage (stage I–II) and later stage patients (stage III–IV), the results indicated the difference was significant between any compared two groups (p < .05, ). So, KDM6B might be a tool of early detection of NSCLC.
Discussion
NSCLC is one of the most lethal and aggressive neoplasms. The low-survival rate of cancer may be contributed to the poor early detection and the lack of effective treatments for advanced stage [Citation20]. The novel biomarkers with high sensitivity and specificity for early diagnosis of NSCLC may significantly improve the outcomes of the patients. In the present study, we investigated the diagnostic value of KDM6B in NSCLC. Analysis results indicated that KDM6B may be a potential biomarker for early detection of NSCLC.
Histone methylation plays a key role in the regulation of gene expression during the cell cycle and its abnormality may be related to tumor occurrence [Citation21]. It was reported that site-specific histone methyltransferases and demethylases could regulate histone methylation, which is a reversible process [Citation22]. Abnormal expression or mutation of histone methyltransferases and demethylases related genes were frequently associated with cancer progression [Citation23]. KDM6B encoding an enzyme which can catalyze histone H3K27me3 demethylation. Abnormal H3K27me3 methylation was reported to be correlated with several cancers, including melanoma, colon, gastric, stomach, ovarian, breast and kidney cancers [Citation24,Citation25]. Thus, KDM6B may also involve in tumor progression.
In this study, we investigated the clinical significance of KDM6B in NSCLC. We firstly detected KDM6B expression levels in the NSCLC serum samples. Our results showed that serum KDM6B expression levels were significantly lower in NSCLC patients compared with healthy controls. Moreover, the down-regulated level of KDM6B was significantly correlated with high clinical stage and positive lymph nodes metastasis. Ma et al. had reported that KDM6B was a tumor suppressor gene in NSCLC, which can promote cell apoptosis [Citation19]. The conclusion supported our results. However, some studies hold different opinions. Tian et al. had proved that the decreased expression level of KDM6B was significantly associated with high TNM stage and positive lymph node metastasis, however, the cell experiments indicated that KDM6B can promote NSCLC cell growth, inhibit cell apoptosis and had no effects on cell migration [Citation26]. The differences may reveal that KDM6B can influence the progression of the NSCLC by different pathways. In addition, the effects of KDM6B on NSCLC cell lines were needed to be identified in the next study.
Various biomarkers are confirmed to be used in the diagnosis of NSCLC, of which the most widely used are CEA and CK19 [Citation27]. However, the low sensitivity and specificity limit their clinical application, especially for the detection of early NSCLC. Therefore, the novel diagnostic markers with high sensitivity and specificity for early detection of NSCLC were urgently needed. In the present study, we evaluated the diagnostic value of KDM6B in patients with NSCLC. ROC curve indicated that KDM6B could distinguish patients with NSCLC from healthy individuals, with high sensitivity and specificity. In the previous studies, KDM6B was reported to be markedly correlated with various tumor progression, such as pancreatic cancer, colon cancer, Hodgkin’s Lymphoma, clear cell renal cell carcinoma [Citation28–31]. The tumor progression gene may be effective for early detection of NSCLC, which can significantly improve the outcomes of the patients.
Although we proved the diagnostic value of KDM6B in NSCLC, there were still several limitations in the present study. Firstly, the number of patients collected in the study was small and further studies with larger numbers of patients are needed to validate the results. Secondly, the patients, in this study, all came from one hospital and the study results may differ according to the techniques used. Therefore, multi-center studies are needed to confirm the diagnostic value of KDM6B in NSCLC. In addition, the mechanisms for KDM6B regulating tumor progression of NSCLC were needed to be identified further. Given the mentioned limitations, the application of serum KDM6B for early diagnosis of NSCLC might cause diagnostic errors. The final diagnosis of NSCLC should be made according to the histopathological examinations. Serum KDM6B might be employed as an auxiliary tool for NSCLC screening. The combined application of KDM6B with other indicators may have the ability to improve the early diagnosis rate of NSCLC.
In conclusion, serum KDM6B mRNA level is markedly lower in patients with NSCLC than that in the healthy controls. Moreover, the decreased level is significantly associated with aggressive clinical characteristics in NSCLC. KDM6B may be applied for early screening of NSCLC, which may markedly improve the management of cancer.
Disclosure statement
We declare that we do not have any conflict of interest.
Additional information
Funding
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29.
- Wang J, Yang B, Han L, et al. Demethylation of miR-9-3 and miR-193a genes suppresses proliferation and promotes apoptosis in non-small cell lung cancer cell lines. Cell Physiol Biochem. 2013;32:1707–1719.
- Li H, Zhang Y, Bai X, et al. TRIM31 is downregulated in non-small cell lung cancer and serves as a potential tumor suppressor. Tumor Biol. 2014;35:5747–5752.
- Sun M, Song J, Zhou Z, et al. Comparison of serum microRNA21 and tumor markers in diagnosis of early non-small cell lung cancer. Dis Markers. 2016;2016:3823121
- Wink KC, Roelofs E, Solberg T, et al. Particle therapy for non-small cell lung tumors: where do we stand? A systematic review of the literature. Front Oncol. 2014;4:292.
- Reck M, Heigener DF, Mok T, et al. Management of non-small-cell lung cancer: recent developments. Lancet. 2013;382:709–719.
- Arya SK, Bhansali S. Lung cancer and its early detection using biomarker-based biosensors. Chem Rev. 2011;111:6783–6809.
- Cabrera-Alarcon JL, Carrillo-Vico A, Santotoribio JD, et al. CYFRA 21-1 as a tool for distant metastasis detection in lung cancer. Clin Lab. 2011;57:1011–1014.
- Gridelli C, Rossi A, Maione P. 2010 Consensus on lung cancer, new clinical recommendations and current status of biomarker assessment–first-line therapy. Eur J Cancer. 2011;47: S248–S57.
- Cho JY, Sung HJ. Proteomic approaches in lung cancer biomarker development. Expert Rev Proteomics. 2009;6:27–42.
- Wang J, Wang K, Xu J, et al. Prognostic significance of circulating tumor cells in non-small-cell lung cancer patients: a meta-analysis. PloS One. 2013;8:e78070.
- Hoffmann I, Roatsch M, Schmitt ML, et al. The role of histone demethylases in cancer therapy. Mol Oncol. 2012;6:683–703.
- Yamane K, Tateishi K, Klose RJ, et al. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–812.
- Kim JH, Sharma A, Dhar SS, et al. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2014;74:1705–1717.
- Teng YC, Lee CF, Li YS, et al. Histone demethylase RBP2 promotes lung tumorigenesis and cancer metastasis. Cancer Res. 2013;73:4711–4721.
- Pires-Luis AS, Vieira-Coimbra M, Vieira FQ, et al. Expression of histone methyltransferases as novel biomarkers for renal cell tumor diagnosis and prognostication. Epigenetics. 2015;10:1033–1043.
- Tokunaga R, Sakamoto Y, Nakagawa S, et al. The prognostic significance of histone lysine demethylase JMJD3/KDM6B in colorectal cancer. Ann Surg Oncol. 2016;23:678–685.
- Ene CI, Edwards L, Riddick G, et al. Histone demethylase Jumonji D3 (JMJD3) as a tumor suppressor by regulating p53 protein nuclear stabilization. PloS One. 2012;7:e51407.
- Ma J, Wang N, Zhang Y, et al. KDM6B elicits cell apoptosis by promoting nuclear translocation of FOXO1 in non-small cell lung cancer. Cell Physiol Biochem. 2015;37:201–213.
- Taverna S, Giallombardo M, Gil-Bazo I, et al. Exosomes isolation and characterization in serum is feasible in non-small cell lung cancer patients: critical analysis of evidence and potential role in clinical practice. Oncotarget. 2016;7(19):28748–28760.
- Campbell MJ, Turner BM. Altered histone modifications in cancer. Adv Exp Med Biol. 2013;754:81–107.
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727.
- Varier RA, Timmers HT. Histone lysine methylation and demethylation pathways in cancer. Biochim Biophys Acta. 2011;1815:75–89.
- Ke XS, Qu Y, Rostad K, et al. Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PloS One. 2009;4:e4687.
- Rada-Iglesias A, Enroth S, Andersson R, et al. Histone H3 lysine 27 trimethylation in adult differentiated colon associated to cancer DNA hypermethylation. Epigenetics. 2009;4:107–113.
- Tian C, Deng H, Tang X, et al. Effect of Jumonji domain-containing protein-3 on the proliferation and migration of lung cancer cell line. Sheng wu yi Xue Gong Cheng Xue za Zhi = J Biomed Eng = Shengwu Yixue Gongchengxue Zazhi. 2012;29:514–518.
- Holdenrieder S, Nagel D, Stieber P. Estimation of prognosis by circulating biomarkers in patients with non-small cell lung cancer. CBM. 2010;6:179–190.
- Yamamoto K, Tateishi K, Kudo Y, et al. Loss of histone demethylase KDM6B enhances aggressiveness of pancreatic cancer through down-regulation of C/EBPalpha. Carcinogenesis. 2014;35:2404–2414.
- Pereira F, Barbachano A, Silva J, et al. KDM6B/JMJD3 histone demethylase is induced by vitamin D and modulates its effects in colon cancer cells. Human Mol Genet. 2011;20:4655–4665.
- Anderton JA, Bose S, Vockerodt M, et al. The H3K27me3 demethylase, KDM6B, is induced by Epstein–Barr virus and over-expressed in Hodgkin's lymphoma. Oncogene. 2011;30:2037–2043.
- Li Q, Hou L, Ding G, et al. KDM6B induces epithelial-mesenchymal transition and enhances clear cell renal cell carcinoma metastasis through the activation of SLUG. Int J Clin Exp Pathol. 2015;8:6334–6344.