Abstract
This research planned to dig the impacts and potential principles of long noncoding RNA RP4 onH9c2 cell injury induced by hypoxia. The H9c2 cardiac muscle cells were cultured under 3% O2 concentration to induce hypoxia injury, followed by detection of RP4 expression. RP4 was then overexpressed and silenced to investigate its effects on cell injury induced by hypoxia. The potential correlation between RP4 and miR-939, between miR-939 and Bnip3, and between RP4/miR-939/Bnip3 axis and Wnt/β-catenin pathway activation were explored. Biological processes (suppressed cell viability, migration and invasion, but enhanced cell apoptosis) were changed by hypoxia. Upregulation of RP4 enhanced hypoxia-produced damage in H9c2 cells. Additionally, miR-939 expression was opposite regulated by RP4, and miR-939 mimic abrogated the influences of pc-RP4 on enhanced hypoxia damage in H9c2 cells. Moreover, Bnip3 was targeted by miR-939 and their correlation is negative. Furthermore, upregulation of RP4 exacerbated hypoxia-produced injury in H9c2 cells by sensitizing Wnt/β-catenin signals in H9c2 cells, which was regulated by miR-939/Bnip3 axis. Our findings reveal that RP4 is highly expressed in the hypoxia-resulted H9c2 cells. Enhanced expression of RP4 may exacerbate hypoxia injury in cardiomyocytes through regulating miR-939/Bnip3 axis-mediated briskness of Wnt/β-catenin signals. Our study will offer a fresh theoretical basis for the treatment of ischemic myocardial injury.
Introduction
Myocardial ischemia occurs when arterial blood flow to the myocardium decreases or stops to a part of the heart, functioning as a major cause of several heart diseases, including myocardial infarction [Citation1]. Although substantial improvements have been made in prognosis over the past decade, acute myocardial infarction (AMI) is considered as a major factor of morbidity and mortality worldwide [Citation2,Citation3]. Current treatment strategies that prevent myocardial ischemia-reperfusion (I/R) injury is beneficial to the management of AMI [Citation4,Citation5]. Therefore, it is still important to deepen the mechanisms underlying myocardial ischemia.
Long noncoding RNAs (lncRNAs), is pointed out to be a kind of transcripts with no longer than 200 bp and lack of protein-coding ability, are emerged as pivotal players in plenty of biological and pathological processes [Citation6–8]. LncRNAs are also involved in the process of ischemic heart diseases [Citation9–11]. Moreover, angiogenic lncRNAs have been considered as possible therapeutic targets for ischemic heart diseases [Citation12]. Therefore, expounding of crucial lncRNAs involved in ischemic heart diseases will promote the process of effective therapies for halting these diseases.
Previous studies showed that the dysregulation of lncRNA RP4 is pivotal in the process of human colorectal cancer [Citation13]. However, the role of lncRNA RP4 has not been reported in heart-related diseases. The complete function mechanisms of lncRNA are usually linked by miRNAs/genes/proteins in the biology of diseases [Citation10,Citation11,Citation14]. miR-939 is pointed out to be widely involved in the process and progression of various diseases including myocardial ischemia [Citation11,Citation15,Citation16]. Otherwise, the correlation between RP4 and miR-939 in myocardial ischemia remains highly unknown.
In this study, the H9c2 cardiac muscle cells were cultured under 3% O2 concentration to induce hypoxia injury, followed by detection of RP4 expression. The function and mechanism of abnormal level of RP4 in ischemic myocardial injury (IMI) were explored from the perspective of molecular levels, in order to offer a fresh theoretical basis for the targeted therapy of ischemic heart diseases.
Materials and methods
Cell culture and disposes
The H9c2 cardiac muscle cell line (Sigma–Aldrich, St. Louis, MO, USA) were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Sangon Biotech, Shanghai, China), added with 1% penicillin/streptomycin (100 U/mL:100 mg/mL), 10% fetal bovine serum (FBS) and 1% GlutaMAX (Life Technologies) and maintained at 37 °C under 5% CO2. The hypoxia and normoxia culture conditions were created by filling with the 3 and 21% O2 concentration.
Cell transfection
The coding sequences of RP4 were inserted into the pcDNA3.1 vector to produce the vector pc-RP4. Short-hairpin RNA binding RP4 was combined into the U6/GFP/Neo plasmid (Sangon Biotech) to form the depression vector sh-RP4. The full-length sequence of Bnip3 was inserted into pEX-2 vector to establish the up-expressed vector pEX-Bnip3. The short-interfering RNA directed against Bnip3 was combined with U6/GFP/Neo plasmids (Sangon Biotech) to construct the knockdown vector si-Bnip3. To investigate the function of miR-939, miR-939 mimics, miR-939 inhibitors and their respective NC were synthesized in Sangon Biotech. Cells transfection was then carried out using lipofectamine 3000 (Sangon Biotech). Afterwards, cells were harvested for the next experiments in post 72 h.
Cell proliferation assay
H9c2 cells at a density of 1 × 105 were incubated in duplicate in 60-mm dishes. Cells were then stained with trypan blue (Sigma–Aldrich) according to the indicated time periods. The number of live cells was calculated by trypan blue exclusion.
Transwell assay
Transwell assay was used for determining the abilities of cell migration and invasion. For migration, cells were cultured in serum-free DMEM medium containing .01% BSA (Sigma–Aldrich) in post 48 h of transfection. The serum-free medium was mixed with the superstratum of the Transwell chamber (8 μm) and then dried at air condition at 4 °C. After sweeping the medium from the cell cultures, 50 μL fresh serum-free medium containing BSA (10 g/L) was mixed for 30 min at 37 °C. Then, the cells were seeded in the 24-well plates and mixed with 10% FBS. After 48 h, cells were washed with PBS buffer to remove the upper cells on the microporous membrane and fixed in ice-cold 75% methanol at 4 °C for 10 min. Finally, cells were stained with crystal violet (0.1%) for 30 min at room temperature, then decoloured with acetic acid (33%), and counted microscopically. For cell invasion, 90 μL Matrigel was added in the upper chamber during experiments.
Apoptosis test
We carried out the apoptotic cell analysis by flow cytometry. Briefly, H9c2 cells were forested and fixed in 70% ethanol. Afterwards, the fixed cells were treated with PBS buffer for 2 times washing. Then, the washed cells were subjected to PI and FITC-conjugated Annexin V staining mixed with RNase A (50 μg/mL; Sigma-Aldrich). Then, cells were cultured for another 1 h in dark at room temperature, FACS can (Beckman Coulter, Fullerton, CA, USA) was chosen for the analysis of cell apoptosis, while FlowJo software (Tree Star, San Carlos, CA, USA) was used for data calculation.
Quantitative PCR
We isolated the total RNA from cells in post-cell disposes using Trizol reagent (TaKaRa Biotech, Dalian, China). Real-time quantitative PCR analysis was then detected to test the levels of RP4 and Bnip3 using the One Step SYBR® PrimeScript®PLUS RT-RNA PCR Kit (TaKaRa) and RNA PCR Kit (AMV) Ver.3.0 (TaKaRa), respectively. Meanwhile, miR-939 was also evaluated by the Taqman MicroRNA Reverse Transcription Kit and Taqman Universal Master Mix II with the TaqMan MicroRNA Assay (TaKaRa) of miR-939. We thereby chose β-actin and U6 for the internal control of genes and miRNA, respectively. The relative gene expression was evaluated by relative quantification (2−ΔΔCt) method.
Luciferase reporter test
The Bnip3 fragment along with the predicted miR-939 binding site was produced by PCR and then cloned into a pmirGlO Dual-luciferase miRNA Target Expression Vector (Promega, Madison, WI, USA) to establish the Bnip3 reporter vector Bnip3-wild-type (Bnip3-wt). The fragment of ATG3 contained in the mutant binding site of miR-939 was synthesized by Sangon (Shanghai, China), and also linked with pmirGlO vector to form reporter vector ATG3-mutant-type (Bnip3-mt). The reporter vector Bnip3-wt or Bnip3-mt and miR-939 mimics or mimic NC were co-transfected into HEK 293 T cells. Then we chose Dual-Luciferase Reporter Assay System (Promega) to test the luciferase activity in each group.
Western blotting test
Total protein in H9c2 cells was extracted using RIPA lysis buffer (TaKaRa, Japan), and protease inhibitors (Roche, Guangzhou, China) were mixed with the isolated protein for further quantitation through the BCA™ Protein Assay Kit (Pierce, Appleton, WI, USA). Afterwards, protein samples (50 μL per lane) were added into the Tris Gel system for further experiment. Primary antibodies Bcl-2, Bax, pro- and cleaved-Caspase-3, pro- and cleaved-Caspase-9, Bnip3, Wnt3a, Wnt5a, β-Catenin and β-actin were disposed in 5% blocking buffer with a dilution of 1:1,000, and then used to incubate the Polyvinylidene Difluoride (PVDF) membrane (Millipore) overnight at 4 °C. After rinsing three times, the membranes were mixed with secondary antibody symbol by horseradish peroxidase for 1 h at room temperature. After rinsing, the membranes taking along blots and antibodies were transferred into the Bio-Rad ChemiDoc™ XRS system, followed by incubation with 200 μL Immobilon Western Chemiluminescent HRP Substrate (Millipore). The protein signals were captured, followed by quantification of the intensity of the bands using Image Lab™ Software (Bio-Rad, Shanghai, China). All antibodies were gained from Abcam (Cambridge, MA, USA).
Statistical evaluation
We carried out all multiple experiments with repeated 3 times independently. Data were displayed as the mean ± standard deviation (SD). The statistical difference among groups was evaluated using ANOVA. p < .05 was chosen to be statistically significant. Statistical analyses were completed using SPSS 16.0 (San Diego, CA, USA).
Results
Hypoxia induces H9c2 cell damage and RP4 is increased in H9c2 cells under hypoxia
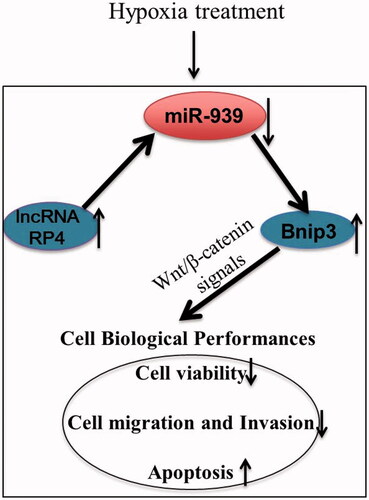
Hypoxia-induced damage in H9c2 cells was firstly investigated. We found that hypoxia treatment remarkably inhibited H9c2 cell viability (), migration () and invasion () but induced apoptosis (; all p < .05) relative to control. Moreover, western blot also exhibited consistent results that hypoxia dramatically increased the levels of Bax/Bcl-2 ratio, cleaved-caspase-3 and cleaved-caspase-9 (). Besides, we further analyzed the level of RP4, and found that hypoxia significantly accelerated the level of RP4 in H9c2 cells (p < .01, ), indicating that RP4 may be involved in myocardial injury.
Figure 1. Hypoxia induced injury in H9c2 cells. (A) Cell viability, (B) cell migration; (C) cell invasion; (D) cell apoptosis and the expression of apoptosis-related proteins. The experiments were repeated three times. (E) Hypoxia promoted the expression of RP4 in H9c2 cells. The experiment was repeated three times. Data are expressed as mean ± SD. **p < .01 compared to control.
Influences of RP4 abnormal expression on hypoxia-disposed damage in H9c2 cells
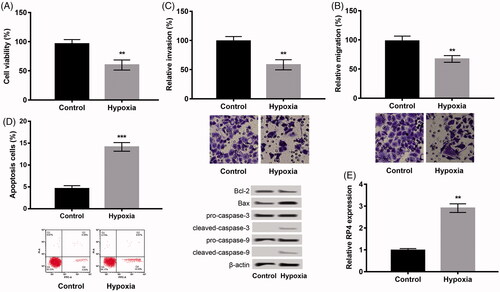
We subsequently overexpressed and silenced RP4 to detect the effects of RP4 on hypoxia-disposed damage in H9c2 cells. RP4 was successfully up-expressed or under-expressed in H9c2 cells because the RP4 was significantly up-expressed in pc-RP4 group and remarkably under-expressed in sh-RP4 relative to that in their respective NC group (p < .01, ). Moreover, further experiments uncovered that pc-RP4 distinctly aggravated the damage induced by hypoxia in H9c2 cells by decreasing cell viability (), suppressing migration (), inhibiting invasion (), but promoting apoptosis (; all p < .05), whereas silencing of RP4 had opposite effects.
Figure 2. Overexpression of RP4 aggravated hypoxia-induced injury in H9c2 cells, while suppression of RP4 relieved the injury. (A) The expression of RP4 in H9c2 cells after transfection with pc-RP4, sh-RP4 and their NC. (B–E) H9c2 cells were transfected with pc-RP4, sh-RP4 and their NC under hypoxia condition. (B) Cell viability of different treatment groups; (C) cell migration of different treatment groups; (D) cell invasion of different treatment groups; (E) cell apoptosis and the expression of apoptosis-related proteins in different treatment groups. The experiments were repeated three times. Data are expressed as mean ± SD. *p < .05; **p < .01 and ***p < .001 compared to control.
miR-393 is involved in the process of RP4 on hypoxia-disposed damage in H9c2 cells
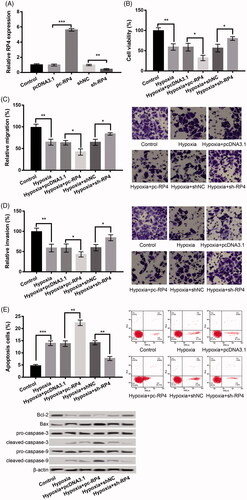
We further analyzed the correlation between RP4 and miR-939. In comparison to their respective NC group, miR-939 was markedly decreased in the pc-RP4 group and obviously increased in the sh-RP4 group (p < .01, ), implying a negative correlation between miR-939 and RP4. Subsequently, we found that miR-939 was dramatically increased in a miR-939 mimic group or dramatically inhibited in miR-939 inhibitor group (p < .01, ). Subsequently, further experiments revealed that co-transfection of pc-RP4 and miR-939 mimic in hypoxia-resulted H9c2 cells significantly reversed the influences of pc-RP4 transfection alone on hypoxia-induced injury by increasing cell viability (), promoting migration (), enhancing invasion (), but inhibiting apoptosis (; all p < .05), indicating that upregulation of miR-939 abrogated the influences of pc-RP4 on hypoxia-induced injury.
Figure 3. RP4 negatively regulated the expression of miR-939 in H9c2 cells, and overexpression of miR-939 abrogated the effects of overexpression of RP4 on hypoxia-induced injury. (A) The expression of miR-939 in H9c2 cells after transfection with pc-RP4, sh-RP4 and their NC. (B) The expression of miR-939 in H9c2 cells after transfection with miR-939 mimic, miR-939 inhibitor and their NC. (C–F) H9c2 cells were co-transfected with pc-RP4, miR-939 mimic and/or their respective NC under hypoxia condition. (C) Cell viability of different treatment groups; (D) cell migration of different treatment groups; (E) cell invasion of different treatment groups; (F) cell apoptosis and the expression of apoptosis-related proteins in different treatment groups. The experiments were repeated three times. Data are expressed as mean ± SD. *p < .05; **p < .01 and ***p < .001 compared to control.
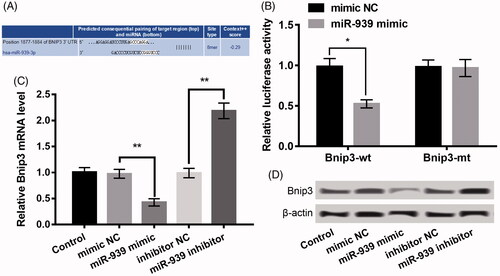
Bnip3 is targeted by miR-939, and miR-939 negatively regulates Bnip3
Previous evidence showed the pivotal roles of Bnip3 in regulating oxidative stress during myocardial I/R [Citation17] and hypoxia may induce cardiac myocyte death via regulating Bnip3 [Citation18]. Notably, Bnip3 was proved as a latent target of miR-939 on the basis of the information of Tagetscan online tool and the binding sequence of them was shown in . Moreover, luciferase analysis revealed that only the luciferase activity of Bnip3-wt was markedly suppressed by miR-939 mimic (p < .05, ). In addition, transfection with miR-939 mimic significantly decreased the Bnip3 expression, whereas transfection with miR-939 inhibitor enhanced Bnip3 (p < .05, ). These data indicated that there exited a negative correlation between Bnip3 and miR-939 in H9c2 cells.
Figure 4. Bnip3 was a target of miR-939, and miR-939 negatively regulated Bnip3 expression. (A) The binding sequence between miR-939 and Bnip3. (B) Luciferase reporter assay revealed the target relationship between miR-939 and Bnip3. (C–D) The mRNA and protein expression of Bnip3 in H9c2 cells after transfection with miR-939 mimic, miR-939 inhibitor and their NC. The experiments were repeated three times. Data are expressed as mean ± SD. *p < .05 and **p < .01 compared to control.
miR-939 prevents hypoxia-disposed damage in H9c2 cells by targeting Bnip3
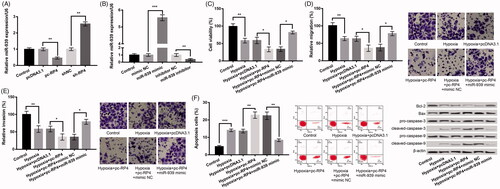
We further verified whether miR-939 protected against cell injury induced by hypoxia by targeting Bnip3. As revealed in , Bnip3 was dramatically enhanced in the pEX-Bnip3 group relative to that in pEX group, and remarkably depressed in si-Bnip3 group relative to the si-NC group (p < .001), pointing out that the transfection was successful. In relative to hypoxia + pc-RP4 + miR-939 mimic + pEX group, pc-Bnip3 in hypoxia + pc-RP4 + miR-939 mimic + pEX-Bnip3 group markedly changeover the impacts of RP4 overexpression and miR-939 overexpression on H9c2 cell viability (), migration (), invasion () and apoptosis (; all p < .05), indicating that miR-939 mimic functioned as a protector in cell injury induced by hypoxia by targeting Bnip3.
Figure 5. miR-939 prevented hypoxia-induced injury in H9c2 cells by targeting Bnip3. (A) Bnip3 expression in H9c2 cells after transfection with pEX-Bnip3, si-Bnip3 and their NC. (B–E) H9c2 cells were co-transfected with pc-RP4, miR-939 mimic, pEX-Bnip3 and/or their respective NC under hypoxia condition. (B) Cell viability of different treatment groups; (C) cell migration of different treatment groups; (D) cell invasion of different treatment groups; (E) cell apoptosis and the expression of apoptosis-related proteins in different treatment groups. The experiments were repeated three times. (F) The expression of Wnt/β-catenin pathway-related proteins in H9c2 cells that were co-transfected with pc-RP4, miR-939 mimic, pEX-Bnip3 and/or their respective NC under hypoxia condition. The experiments were repeated three times. Data are expressed as mean ± SD. *p < .05; **p < .01 and ***p < .001 compared to control.
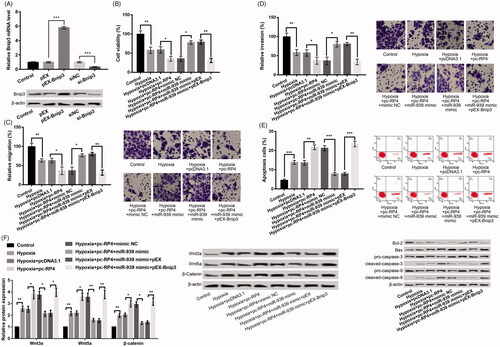
RP4 up-expression aggravates hypoxia-disposed damage by brisking Wnt/β-catenin signals in H9c2 cells
The association of RP4/miR-939/Bnip3 axis and activation of Wnt/β-catenin signals was further investigated to grab the downstream regulatory reason of RP4 on hypoxia-disposed damage in H9c2 cells. The results uncovered that the levels of Wnt3a, Wnt5a and β-Catenin were all significantly increased in H9c2 cells after hypoxia, which were further increased after overexpression of RP4 (). In addition, co-transfection of RP4 and miR-939 mimic markedly reversed the influences of RP 4 overexpression alone on these protein expression levels, which was further reversed by overexpression of Bnip3 concurrently ( and ).
Discussion
Abnormal expression of lncRNAs has been pointed out to be crucial in the pathogenesis of cardiovascular diseases [Citation14,Citation19,Citation20]. Scholars also revealed that lncRNAs have been discovered to be implicated in myocardial I/R injury. Apoptosis has been reported to be induced under hypoxic stress in ischemic injury [Citation21], and lncRNA UCA1 can regulate cardiomyocyte apoptosis in myocardial I/R injury [Citation15]. LncRNA CARL is found to inhibit anoxia-induced cardiomyocyte apoptosis [Citation22] and suppression of lncRNA TTTY15 can protect against hypoxia-induced cardiomyocyte apoptosis [Citation23], suggesting the latent role of these lncRNAs in IMI. Currently, we investigated the possible impacts of lncRNA RP4 on the hypoxia-disposed damage in H9c2 cells. The results uncovered that upregulation of RP4 was associated with hypoxia. Overexpression of RP4 significantly enhanced hypoxia damage in H9c2 cells, whereas downregulation of RP4 remits the injury. All of these results indicated that RP4 was pivotal in regulating myocardial ischemic injury (MII).
In addition, miR-939 expression was testified as a target of RP4. And miR-939 mimic changeover the impacts of pc-RP4 on the aggravated hypoxia-disposed damage in H9c2 cells. A recent study has revealed that the miR-939-mediated Nitric Oxide signals are a possible mechanism to modulate the effect of coronary serum exosomes from patients with myocardial ischemia in angiogenesis [Citation16]. Mao et al. demonstrated that circ-SATB2 regulates the cell proliferation and differentiation of vascular smooth muscle cells through the miR-939/STIM1 axis, thus, playing important roles in coronary heart disease [Citation24]. Given the potential impact of miR-939 in heart diseases, we speculated that RP4 may enhance myocardial ischemia injury via negative regulation of miR-939.
Moreover, we found that Bnip3 was targeted by miR-939. Bnip3 has been reported to regulate oxidative stress during myocardial I/R [Citation17], and hypoxia may induce cardiac myocyte death via regulating Bnip3 [Citation18]. Bnip3-related mitophagy is also confirmed as one of key mechanism to mediate the role of dual-specificity protein phosphatase1in alleviating cardiac I/R injury [Citation25]. In addition, miR-210 is shown to alleviate oxidative stress-associated cardiomyocyte apoptosis through targeting Bnip3 [Citation26]. In this study, Bnip3 overexpression significantly reversed the impacts of pc-RP4 and miR-939 mimic on hypoxia-disposed damage in H9c2 cells. We, therefore, speculate that overexpression of miR-939 may protect against myocardial ischemia injury by regulating Bnip3.
Furthermore, overexpression of RP4 exacerbated hypoxia injury in H9c2 cells by brisking Wnt/β-catenin pathway in H9c2 cells, which was regulated by miR-939/Bnip3 axis. Wnt/β-catenin signals have been reported to be involved in cardiac injury response or cardiac development [Citation27–29]. Also, lncRNA TALNEC2 is shown to be crucial in regulating MII in H9c2 cells via activation of Wnt/β-catenin signals [Citation30]. Our study preliminarily revealed that pc-RP4 further brisked Wnt/β-catenin signals in hypoxia-disposed H9c2 cells. Combination of pc-RP4 and miR-939 mimic markedly changeover the impacts of pc-RP4 alone on the pathway, which was further reversed by overexpression of Bnip3 concurrently. Taken together, we speculated that briskness of Wnt/β-catenin signals may be a downstream mechanism in regulating the impact of RP4 in MII.
To sum up, our data discover that RP4 is increased expressed in hypoxia-induced H9c2 cells. Enhanced expression of RP4 may exacerbate hypoxia injury in cardiomyocytes through regulating miR-939/Bnip3 axis-mediated briskness of Wnt/β-catenin signals (). Our study will offer a fresh theoretical basis for the treatment of IMI. However, we did not perform experiments at clinical and in vivo levels to further confirm the impact of RP4 in MII, more studies are still required to confirm our observation.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Hashmi S, Al-Salam S. Acute myocardial infarction and myocardial ischemia-reperfusion injury: a comparison. Int J Clin Exp Pathol. 2015;8:8786.
- Mythili S, Malathi N. Diagnostic markers of acute myocardial infarction. Biomed Rep. 2015;3:743–748.
- Reed GW, Rossi JE, Cannon CP. Acute myocardial infarction. Lancet. 2017;389:197–210.
- Ruiz-Meana M, García-Dorado D. Pathophysiology of ischemia-reperfusion injury: new therapeutic options for acute myocardial infarction. Rev Esp Cardiol. 2009;62:199–209.
- Hausenloy DJ, Yellon DM. Myocardial ischemia-reperfusion injury: a neglected therapeutic target. J Clin Invest. 2013;123:92.
- Piccoli M-T, Gupta SK, Thum T. Noncoding RNAs as regulators of cardiomyocyte proliferation and death. J Mol Cell Cardiol. 2015;89:59–67.
- Simion V, Haemmig S, Feinberg MW. LncRNAs in vascular biology and disease. Vascul Pharmacol. 2019;114:145–156.
- Yan B, Wang Z-H, Liu J-Y. Long noncoding RNAs: versatile players in biologcial processes and human disorders. Epigenomics. 2014;6:375–379.
- Greco S, Zaccagnini G, Perfetti A, et al. Long noncoding RNA dysregulation in ischemic heart failure. J Transl Med. 2016;14:183.
- Li N, Ponnusamy M, Li M-P, et al. The role of microRNA and LncRNA–MicroRNA interactions in regulating ischemic heart disease. J Cardiovasc Pharmacol Ther. 2017;22:105–111.
- Yu S-y, Dong B, Tang L, et al. LncRNA MALAT1 sponges miR-133 to promote NLRP3 inflammasome expression in ischemia-reperfusion injured heart. Int J Cardiol. 2018;254:50.
- Xu Z-M, Huang F, Huang W-Q. Angiogenic lncRNAs: a potential therapeutic target for ischaemic heart disease. Life Sci. 2018;211:157.
- Liu M-L, Zhang Q, Yuan X, et al. Long noncoding RNA RP4 functions as a competing endogenous RNA through miR-7-5p sponge activity in colorectal cancer. World J Gastroenterol. 2018;24:1004–1012.
- Thum T, Condorelli G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ Res. 2015;116:751–762.
- Yu S-Y, Dong B, Zhou S-H, et al. LncRNA UCA1 modulates cardiomyocyte apoptosis by targeting miR-143 in myocardial ischemia-reperfusion injury. Int J Cardiol. 2017;247:31.
- Li H, Liao Y, Gao L, et al. Coronary serum exosomes derived from patients with myocardial ischemia regulate angiogenesis through the miR-939-mediated nitric oxide signaling pathway. Theranostics. 2018;8:2079–2093.
- Kubli DA, Quinsay MC. Bnip3 functions as a mitochondrial sensor of oxidative stress during myocardial ischemia and reperfusion. Am J Physiol. 2008;295:H2025–H2031.
- Kubasiak LA, Hernandez OM, Bishopric NH, et al. Hypoxia and acidosis activate cardiac myocyte death through the Bcl-2 family protein BNIP3. Proc Natl Acad Sci USA. 2002;99:12825–12830.
- Uchida S, Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res. 2015;116:737–750.
- Lorenzen JM, Thum T. Long noncoding RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol. 2016;12:360.
- Liu H, Jing X, Dong A, et al. Overexpression of TIMP3 protects against cardiac ischemia/reperfusion injury by inhibiting myocardial apoptosis through ROS/Mapks pathway. Cell Physiol Biochem. 2017;44:1011–1023.
- Wang K, Long B, Zhou L-Y, et al. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun. 2014;5:3596.
- Huang S, Tao W, Guo Z, et al. Suppression of long noncoding RNA TTTY15 attenuates hypoxia-induced cardiomyocytes injury by targeting miR-455-5p. Gene. 2019;701:1.
- Mao Y-Y, Wang J-Q, Guo X-X, et al. Circ-SATB2 upregulates STIM1 expression and regulates vascular smooth muscle cell proliferation and differentiation through miR-939. Biochem Biophys Res Commun. 2018;505:119–125.
- Jin Q, Li R, Hu N, et al. DUSP1 alleviates cardiac ischemia/reperfusion injury by suppressing the Mff-required mitochondrial fission and Bnip3-related mitophagy via the JNK pathways. Redox Biol. 2018;14:576–587.
- Diao H, Liu B, Shi Y, et al. MicroRNA-210 alleviates oxidative stress-associated cardiomyocyte apoptosis by regulating BNIP3. Biosci Biotechnol Biochem. 2017;81:1712–1720.
- Bergmann MW. WNT signaling in adult cardiac hypertrophy and remodeling: lessons learned from cardiac development. Circ Res. 2010;107:1198–1208.
- Gessert S, Kühl M. The multiple phases and faces of wnt signaling during cardiac differentiation and development. Circ Res. 2010;107:186–199.
- Buikema J, Zwetsloot P-P, Doevendans P, et al. Wnt/β-catenin signaling during cardiac development and repair. J Cardiovasc Dev Dis. 2014;1:98–110.
- Hao L, Wang J, Liu N. Long noncoding RNA TALNEC2 regulates myocardial ischemic injury in H9c2 cells by regulating miR‐21/PDCD4‐medited activation of Wnt/β‐catenin pathway. J Cell Biochem. 2019;120:12912.