Abstract
Enterovirus 71 (EV71) which commonly caused the hand-foot-mouth disease (HFMD) has become one of public health challenges worldwide. However, no effective vaccines or drugs for this disease has been developed. Thus, there is an urgent need to find a new strategy for treating the EV71 infection. Oseltamivir (OT) is an effective antiviral agent, but continuous use of oseltamivir leads to a diminished therapeutic effect in the clinic. In order to improve the antiviral activity of oseltamivir, oseltamivir was loaded onto surfaces of selenium nanoparticles (SeNPs) to fabricate a functionalized antiviral nanoparticles SeNPs@OT. The size of SeNPs@OT was tested by TEM and dynamic light scattering. The chemical structure and elemental composition of SeNPs@OT were analyzed by FT-IR and EDX, respectively. SeNPs@OT exhibited good stability and effective drug release in serum and PBS. SeNPs@OT efficiently entered into human astrocyte U251 cells (host cells) via clathrin-associated endocytosis and inhibited EV71 proliferation, which could protect EV71-infected U251 cells from apoptosis through mitochondrial pathway. Furthermore, SeNPs@OT inhibited EV71 activity probably by reducing the generation of reactive oxygen species in EV71-infected U251 cells. Interestingly, SeNPs obviously enhanced antiviral activity of oseltamivir in the anti-EV71 cell model. Taken together, SeNPs@OT is a promising antiviral drug candidate for EV71 infection.
Introduction
Enterovirus 71 (EV71) is a non-enveloped, positive-sense, single-stranded RNA virus, which is one of the Picornaviridae family [Citation1,Citation2]. This virus was originally isolated and identified from the feces of an infant suffering from encephalitis in 1969 in California, USA [Citation3]. After that, outbreaks of EV71 infection have periodically occurred worldwide, particularly in the Asia-Pacific region [Citation4–6]. EV71 is the most common pathogens of hand, foot and mouth disease (HFMD) in infants and children under 6 years old, which usually leads to bad clinical consequence and even death in severe cases [Citation7]. To date, the pathogenesis of EV71 has not been clarified and no vaccine or specific antiviral agent is available for treating EV71 infection [Citation8]. Thus, the development of non-toxic and effective antiviral agents against EV71 is imminently needed [Citation9].
Several antiviral drugs such as oseltamivir and amantadine have been approved by the FDA in the U.S [Citation10]. In the previous report, oseltamivir was widely investigated as one of the inhibitors against influenza which is a segmented RNA virus [Citation11]. Thus, in this study, oseltamivir was chosen to explore the antiviral action of EV71-infected cells. However, the emergence of drug resistance weakens the antiviral activity of antivirus drugs [Citation12]. Therefore, new technology is needed to improve this situation. Recently, nanomaterials have gradually developed to be an ideal choice for the treatment of virus [Citation13,Citation14]. The design of effective nanoscale antiviral drugs should consider a strategy to control the infection and replication of virus as well as the cytotoxicity against host cells. Vonnemann et al. [Citation15] reported that the polyvalent nanoparticles with various sizes could inhibit the virus. Khanal et al. [Citation16] reported that phenylboronic acid-modified nanoparticles exhibited potent antiviral activity in the treatment of the virus. Wang et al. [Citation17] reported that the broad spectrum antiviral polyoxometalate may become a novel antiviral drug. In our previous research, selenium nanoparticles (SeNPs) have been proven to exhibit significant antiviral activity [Citation18,Citation19]. Selenium as an essential trace element control some key biological processes containing specific enzyme modulation and reactive oxygen species (ROS) elimination [Citation19]. A deficiency in selenium could cause susceptibility to the virus infection [Citation20]. Cheng et al. [Citation21] reported that SeNPs could inhibit the replication and transcription of the hepatitis B virus.
In the current study, the antiviral drug oseltamivir was loaded onto the surface of SeNPs to fabricate the nano-scale functionalized antiviral drug system SeNPs@OT. We look forward to proving that SeNPs@OT has the ability to exhibit great anti-EV71 activity in human astrocytoma cell (U251) model. Interestingly, compared to oseltamivir, SeNPs@OT exhibited a stronger ability to inhibit EV71 activity and prevent the host U251 cells from apoptosis. The antiviral mechanisms of SeNPs@OT are further investigated in this paper.
Materials and methods
Materials
The human astrocytoma (U251) cells were purchased from ATCC. Enterovirus 71 (EV71) was provided by the Virus Laboratory, Guangzhou Women and Children’s Medical Center. DMEM and FBS were obtained from Thermo Fisher. Na2SeO3, vitamin C (Vc), oseltamivir (OT) and MTT were obtained from Sigma-Aldrich Co.
Preparation and characterization of SeNPs@OT
SeNPs was prepared as following: Fresh 0.1M Na2SeO3 solution, 40 mM Vc solution and 1 μM oseltamivir solution was prepared first. 0.1 ml of Na2SeO3 solution was gradually dropped to 1 ml of Vc. Then, 0.1 ml of oseltamivir solution was dropped to the mixed solution. The excess oseltamivir, Na2SeO3 and Vc were removed by dialysis for 12 h. SeNPs@OT was filtered through a 0.2 μm filter. The concentrations of oseltamivir and SeNPs was tested via inductively coupled plasma mass spectrometry (ICP-MS). The morphology of SeNPs@OT was characterized by TEM. Elemental compositions of SeNPs@OT were examined via EDX. Size distributions of SeNPs@OT were observed by Malvern Zetasizer.
The uptake pathway of SeNPs@OT
In order to study the cellular uptake pathway, U251 cells were co-incubated with different endocytosis inhibitors. Subsequently, the cell was exposed to SeNPs@OT at 4 °C for 6 h without inhibitors, or at 37 °C containing inhibitors. Intracellular SeNPs@OT was tested by ICP-MS [Citation22].
In vitro oseltamivir release
The in vitro release profile of oseltamivir from SeNPs@OT was investigated by dialysis method [Citation23]. In brief, 2 mg SeNPs@OT was dissolved in water and introduced into the dialysis tube. Afterwards, the dialysis tube was put in PBS and stirred at 100 rpm. The released oseltamivir was examined by ICP-MS during 30 h. The amounts of the payload were confirmed by the standard curve.
Determination of cell viability
Cell viabilities were tested by MTT method. Briefly, U251 cells were incubated in 96-well plate overnight and infected with EV71. Then the cells were treated with SeNPs@OT containing 9.8 μM SeNPs and 20 nM oseltamivir, 9.8 μM SeNPs and 20 nM oseltamivir for 24 or 48 h. Then the cells were examined as reported previously [Citation24].
qRT-PCR quantification
U251 cells were incubated in 6-well plate overnight and infected with EV71. Then the cells were treated with SeNPs@OT containing 9.8 μM SeNPs and 20 nM oseltamivir, 9.8 μM SeNPs and 20 nM oseltamivir for 24 or 48 h, respectively. The total RNA was extracted by using an RNA isolation kit (Qiagen) according to the manufacturer’s instruction. The target mRNA (VP1) were amplified using primer sets shown in . The relative mRNA level of VP1 was evaluated against β-actin mRNA by comparative Ct (2−△△Ct) method [Citation25].
Table 1. The sequence primer used for quantitative real-time PCR.
Cell apoptosis analysis
The apoptosis of U251 cells was examined by flow cytometry [Citation26]. In brief, U251 cells were incubated overnight to reach about 60% confluence and then infected with EV71. Subsequently, the EV71-infected U251 cells were treated with SeNPs@OT containing 9.8 μM SeNPs and 20 nM oseltamivir, 9.8 μM SeNPs and 20 nM oseltamivir for 24 h, respectively. The cells were collected for staining with Annexin V-FITC/PI for 15 min and then examined by FACS flow cytometer (BD Biosciences, USA).
Detection of mitochondrial membrane potential (△Ψm)
Mitochondria depolarization was tested by JC-1 monomers to assess the status of △Ψm in U251 cells as previously described [Citation27]. Briefly, the cells were infected with EV71 and exposed to SeNPs@OT containing 9.8 μM SeNPs and 20 nM oseltamivir, 9.8 μM SeNPs and 20 nM oseltamivir for 24 h, respectively. Then the cells were collected for quantitative analysis by flow cytometry.
The test of caspase-3 activity
The EV71-infected U251 cells were exposed to SeNPs@OT containing 9.8 μM SeNPs and 20 nM oseltamivir, 9.8 μM SeNPs and 20 nM oseltamivir for 24 h, respectively. The activity of caspase-3 was examined as previously reported [Citation27].
The assessment of reactive oxygen species (ROS)
The production of ROS in U251 cells was detected as described previously [Citation27]. In brief, after EV71-infected U251 cells treatment with SeNPs@OT containing 9.8 μM SeNPs and 20 nM oseltamivir, 9.8 μM SeNPs and 20 nM oseltamivir for 24 h, respectively, the treated cells were stained with 10 μM of DCFH-DA for 20 min and the fluorescence intensity of cells was detected by a fluorescence plate reader.
Statistical analysis
The data were expressed as mean ± standard deviations (SD). Statistical differences were evaluated by one-way analysis of variance (ANOVA). Differences with *p < .05 and **p < .01 were considered statistically significant.
Results and discussion
Preparation and characterization of SeNPs@OT
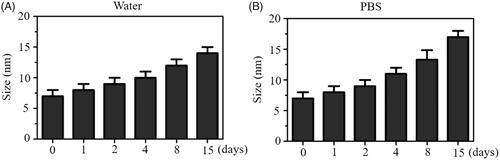
In this paper, we constructed a chemical method to prepare selenium nanoparticles (SeNPs) coated with oseltamivir to synthesize functionalized SeNPs (SeNPs@OT). The morphology of SeNPs@OT was presented via transmission electron microscope (TEM). Seen from , SeNPs@OT presented small and sphere particles. Size distribution of SeNPs@OT confirmed that the average sizes of SeNPs@OT were about 10 nm (). Fourier transform infrared (FTIR) spectra of SeNPs, SeNPs@OT and oseltamivir was used to further characterize the chemical binding between oseltamivir and SeNPs (). FTIR spectrum of SeNPs@OT demonstrated the similar FTIR spectrum of oseltamivir, and the characteristic peak of oseltamivir at 3248, 1248, and 733 cm−1 corresponding to –CH2, –NH2, and C–N were also observed in spectrum of SeNPs@OT, indicating the effectual formation of SeNPs@OT. As shown in , several key signals, including Se atom signal from SeNPs as well as O and C that from oseltamivir were observed in the EDX analysis of SeNPs@OT. These results verified the successful preparation of SeNPs@OT. Furthermore, the size distribution of SeNPs@OT in water solution and phosphate buffer saline (PBS) were observed, and the results showed that SeNPs@OT could keep small size less 20 nm at least 15 days (), indicating that SeNPs@OT was stable in water solution and PBS.
Figure 1. Characterization of SeNPs@OT. (A) Particle size distribution of SeNPs@OT. (B) TEM image of SeNPs@OT. (C) FTIR spectra of SeNPs, SeNPs@OT and oseltamivir. (D) EDX analysis of SeNPs@OT.
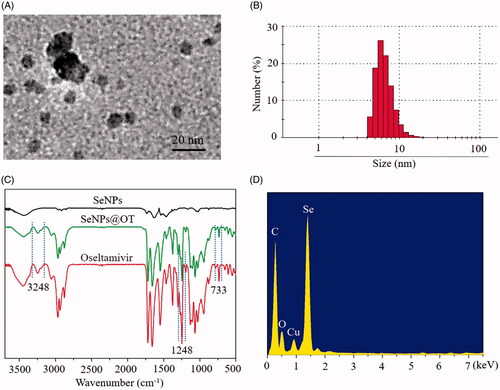
Figure 2. (A) Stability analysis of SeNPs@OT in aqueous solution. (B) Stability analysis of SeNPs@OT in PBS.
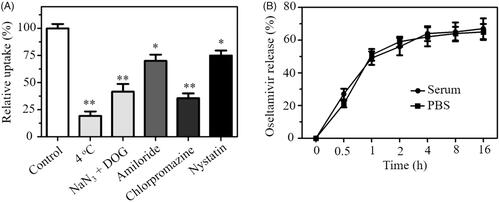
Endocytosis is an important transport mechanism for nanoparticles across the membrane [Citation28]. An uptake inhibition assay was carried out to explore the uptake pathways of SeNPs@OT. Pretreatment with NaN3/DOG or incubation at 4 °C obviously reduced the uptake of SeNPs@OT in U251 cells (), suggesting that the endocytosis of SeNPs@OT is an energy-dependent process. Various endocytosis inhibitor was used to study the endocytosis mechanism of SeNPs@OT. Chlorpromazine, amiloride and nystatin were the inhibitors of clathrin-associated endocytosis, macropinocytosis and caveolae-mediated endocytosis, respectively. As shown in , after pretreatment with nystatin or amiloride, uptake of SeNPs@OT was reduced by 25.2 and 30.6%, respectively. However, chlorpromazine-pretreatment resulted in 64.3% reduction in the uptake of SeNPs@OT, indicating that SeNPs@OT entered U251 cells mainly by clathrin-associated endocytosis way.
In vitro release of oseltamivir
In vitro release of oseltamivir from SeNPs@OT in serum and PBS at 37 °C was investigated by dialysis method. In this assay, two conditions in serum and PBS were applied to simulate the intravascular environment and the normal physiological environment, respectively. The obtained results () showed that SeNPs@OT exhibited similar time-dependent release pattern in serum and PBS, and the release of oseltamivir from SeNPs@OT showed an initial burst release during initial 1 h and then reached a steady state at 4 h. The release rates of oseltamivir in serum and PBS at 16 h were about 67.6 and 65.2%, respectively. The ideal drug release in serum and PBS exhibited great potential for in vivo treatment.
SeNPs@OT enhances viability of EV71-infected host cells and inhibits viral proliferation
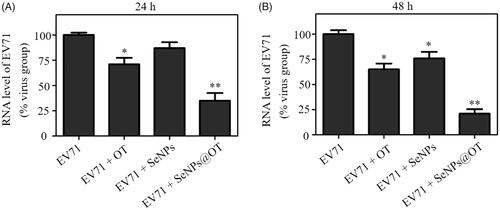
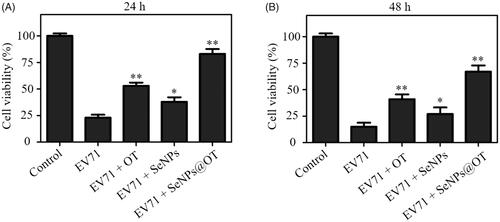
The viabilities of EV71-infected U251 cells at 24 and 48 h postinfection (p.t.) were examined and the antiviral activity of oseltamivir, SeNPs and SeNPs@OT was studied via MTT method. As shown in , viability of EV71-infected U251 cells without any treatment was 23.5% at 24 h p.t., while the viability of EV71-infected cells exposed to oseltamivir and SeNPs were about 53.3% and 38.6%, respectively. However, the treatment with SeNPs@OT increased the viability of EV71-infected cells to 83.2%, which was obviously higher than that of the untreated cells. At 48 h p.t., the similar antiviral activity of oseltamivir, SeNPs and SeNPs@OT was observed (). These results indicated that antiviral ability of oseltamivir was efficaciously increased by loading oseltamivir onto the surfaces of SeNPs. Total RNA of cells was extracted and mRNA level of EV71 in different groups was examined. As presented, the infected cells exposed to SeNPs@OT for 24 and 48 h both obtained lower mRNA level of EV71 than oseltamivir, suggesting that SeNPs@OT showed higher efficacy than oseltamivir to inhibit the proliferation of EV71.
Figure 4. (A) The viability of uninfected and EV71-infected U251 cells exposed to SeNPs, oseltamivir and SeNPs@OT for 24 h. (B) The viability of uninfected and EV71-infected U251 cells exposed to SeNPs, oseltamivir and SeNPs@OT for 48 h. *p < .05, **p < .01 vs. infected cells without treatment group.
SeNPs@OT protects host cells from apoptosis
The regulation of cells apoptosis is a common characteristic of animal virus infection, and it also contributes to the pathogenesis process. Many animal viruses including EV71 were reported to induce host cells apoptosis [Citation29]. The apoptotic cells percentages of U251 cells after EV71 infection were quantitatively analyzed by flow cytometry. As shown in , the percentage of the apoptotic cell was dramatically elevated from 4.37% (uninfected control group) to 67.7% (EV71-infected group). After oseltamivir-, SeNPs- and SeNPs@OT-treatment, the apoptotic cell population of U251 cells were obviously decreased to 48.4, 57.4 and 26.8%, respectively, indicating that SeNPs@OT exhibited the strongest antivirus ability in EV71-infected U251 cells. These results revealed that SeNPs@OT could heighten the antivirus ability of oseltamivir to prevent U251 cells from apoptosis by loading oseltamivir onto the surface of SeNPs.
Figure 6. (A) Effects of SeNPs, oseltamivir and SeNPs@OT on the apoptosis of EV71-infected U251 cells. (B) Mitochondrial membrane potential of uninfected and EV71-infected U251 cells exposed to SeNPs, oseltamivir and SeNPs@OT. (C) Inhibition of caspase-3 activity by SeNPs, oseltamivir and SeNPs@OT in EV71-infected U251 cells. *p < .05, **p < .01 vs. infected cells without treatment group.
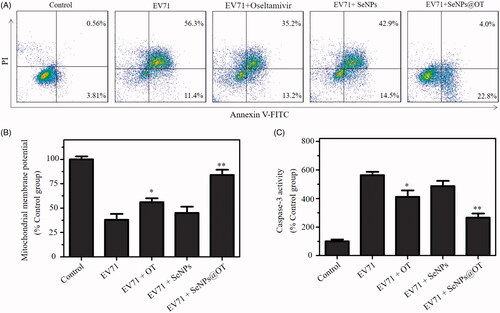
The mitochondrial pathway of apoptosis is a key process in EV71-induced apoptosis [Citation30]. Thus, mitochondrial membrane potentials (△Ψm) were detected by flow cytometry to investigate the initiation of cell apoptosis. As shown in , EV71-infection in U251 cells led to the decrease of mitochondrial membrane potential, indicating that EV71-infection could trigger U251 cells apoptosis by inducing mitochondrial dysfunction. The percentage of △Ψm was markedly increased in SeNPs@OT-treatment group in comparison with SeNPs- and oseltamivir-treatment groups. These results demonstrated that SeNPs@OT prevented U251 cells from infection with EV71, which triggered cells apoptosis through inducing mitochondrial dysfunction.
The cell apoptosis was further examined by caspase-3 activity [Citation31]. As shown in , the infection with EV71 significantly enhanced the caspase-3 activity of U251 cells to 564.3%. The caspase-3 activity of U251 cells treated with SeNPs and oseltamivir was 487.3 and 412.7%, respectively. Nevertheless, SeNPs@OT obviously reduced the caspase-3 activity to 267.4%. This result showed that SeNPs@OT obviously inhibited EV71 activity.
SeNPs@OT inhibits the ROS generation
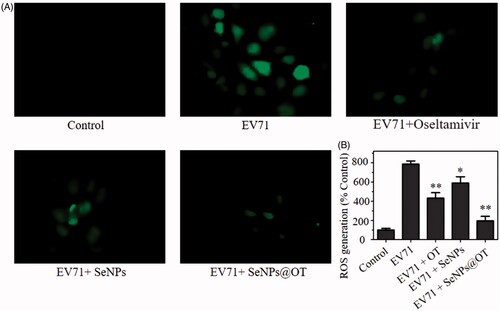
DCF assay was used to detect the ROS generation of U251 cells to further investigate the action mechanism of SeNPs@OT. Seen from , the fluorescent intensity of DCF in U251 cells infected with EV71 was significantly stronger in comparison with uninfected cells (control group). However, compared to oseltamivir- and SeNPs-treatment groups, SeNPs@OT showed stronger ability to inhibit the ROS generation of EV71-infected U251 cells. Seen from , the quantitative analysis showed that ROS generation of U251 cells infected by EV71 was increased to 786.4%. SeNPs- and oseltamivir-treatment slightly suppressed ROS generation to 589.3 and 432.6%, respectively. Nevertheless, SeNPs@OT dramatically reduced the ROS generation rate (196.5%). These results showed that SeNPs@OT could inhibit the ROS level in the antiviral action.
Conclusion
In conclusion, the antiviral drug oseltamivir (OT) was loaded onto the surface of SeNPs to synthesize a functionalized selenium nanoparticles SeNPs@OT. SeNPs@OT could effectively release oseltamivir in serum and PBS, and the antiviral activity of oseltamivir in human astrocyte U251 cells model showed that SeNPs obviously enhanced the antiviral ability of oseltamivir to restrain the proliferation of EV71 and protect the host U251 cells from apoptosis. Altogether, this study elaborated that SeNPs loaded with oseltamivir prevented the host cells apoptosis induced by EV71 infection through the mitochondrial pathway and reducing the generation of reactive oxygen species.
Disclosure statement
The authors declare that they have no conflict of interest regarding this publication.
Additional information
Funding
References
- Zhang F, Liu Y, He X, et al. RASSF4 promotes EV71 replication to accelerate the inhibition of the phosphorylation of AKT. Biochem Biophys Res Commun. 2015;458:810–815.
- Hou YH, Wang JJ, Jiang YZ, et al. A colorimetric and electrochemical immunosensor for point-of-care detection of enterovirus 71. Biosens Bioelectron. 2018;99:186–192.
- Yen M-H, Huang C-I, Lee M-S, et al. Artemisia capillaris inhibited enterovirus 71-induced cell injury by preventing viral internalization. Kaohsiung J Med Sci. 2018;34:150–159.
- Fu Y, Xu W, Chen D, et al. Enterovirus 71 induces autophagy by regulating has-miR-30a expression to promote viral replication. Antiviral Res. 2015;124:43–53.
- Pan H, Yao X, Chen W, et al. Dissecting complicated viral spreading of enterovirus 71 using in situ bioorthogonal fluorescent labeling. Biomaterials 2018;181:199–209.
- Luo W, Zhong J, Zhao W, et al. Proteomic analysis of human brain microvascular endothelial cells reveals differential protein expression in response to enterovirus 71 infection. Biomed Res Int. 2015;2015:1–8.
- Chen D, Su A, Fu Y, et al. Harmine blocks herpes simplex virus infection through downregulating cellular NF-kappaB and MAPK pathways induced by oxidative stress. Antivir Res. 2015;123:27–38.
- Prabowo BA, Wang RYL, Secario MK, et al. Rapid detection and quantification of Enterovirus 71 by a portable surface plasmon resonance biosensor. Biosens Bioelectron. 2017;92:186–191.
- Dewangan HK, Pandey T, Singh S. Nanovaccine for immunotherapy and reduced hepatitis-B virus in humanized model. Artif Cells Nanomed Biotechnol. 2018;46:2033–2042.
- Li Y, Lin Z, Guo M, et al. Inhibition of H1N1 influenza virus-induced apoptosis by functionalized selenium nanoparticles with amantadine through ROS-mediated AKT signaling pathways. Int J Nanomed. 2018;13:2005–2016.
- Yan Z-L, Liu A-Y, Wei X-X, et al. Divalent oseltamivir analogues as potent influenza neuraminidase inhibitors. Carbohydr Res. 2019;477:32–38.
- Antipov EA, Pokryshevskaya EB. The effects of adverse drug reactions on patients’ satisfaction: evidence from publicly available data on Tamiflu (oseltamivir). Int J Med Inform. 2019;125:30–36.
- Guo M, Li Y, Lin Z, et al. Surface decoration of selenium nanoparticles with curcumin induced HepG2 cell apoptosis through ROS mediated p53 and AKT signaling pathways. RSC Adv. 2017;7:52456–52464.
- Negahdari B, Darvishi M, Saeedi AA. Gold nanoparticles and hepatitis B virus. Artif Cells Nanomed Biotechnol. 2019;47:469–474.
- Vonnemann J, Sieben C, Wolff C, et al. Virus inhibition induced by polyvalent nanoparticles of different sizes. Nanoscale 2014;6:2353–2360.
- Khanal M, Vausselin T, Barras A, et al. Phenylboronic-acid-modified nanoparticles: potential antiviral therapeutics. ACS Appl Mater Interfaces. 2013;5:12488–12498.
- Wang J, Liu Y, Xu K, et al. Broad-spectrum antiviral property of polyoxometalate localized on a cell surface. ACS Appl Mater Interfaces. 2014;6:9785–9789.
- Lin Z, Li Y, Gong G, et al. Restriction of H1N1 influenza virus infection by selenium nanoparticles loaded with ribavirin via resisting caspase-3 apoptotic pathway. Int J Nanomed. 2018;13:5787–5797.
- Li Y, Lin Z, Guo M, et al. Inhibitory activity of selenium nanoparticles functionalized with oseltamivir on H1N1 influenza virus. Int J Nanomed. 2017;12:5733–5743.
- Lin Z, Li Y, Guo M, et al. Inhibition of H1N1 influenza virus by selenium nanoparticles loaded with zanamivir through p38 and JNK signaling pathways. RSC Adv. 2017;7:35290–35296.
- Cheng Z, Zhi X, Sun G, et al. Sodium selenite suppresses hepatitis B virus transcription and replication in human hepatoma cell lines. J Med Virol. 2016;88:653–663.
- Xia Y, Lin Z, Li Y, et al. Targeted delivery of siRNA using RGDfC-conjugated functionalized selenium nanoparticles for anticancer therapy. J Mater Chem B. 2017;5:6941–6952.
- Xia Y, Xu T, Wang C, et al. Novel functionalized nanoparticles for tumor-targeting co-delivery of doxorubicin and siRNA to enhance cancer therapy. Int J Nanomed. 2018;13:143–159.
- Li Y, Guo M, Lin Z, et al. Multifunctional selenium nanoparticles with Galangin-induced HepG2 cell apoptosis through p38 and AKT signalling pathway. R Soc Open Sci. 2018;5:180509.
- Wang H, Li K, Ma L, et al. Berberine inhibits enterovirus 71 replication by downregulating the MEK/ERK signaling pathway and autophagy. J Virol. 2017;14:2.
- Chen Q, Fang X, Jiang C, et al. Thrombospondin promoted anti-tumor of adenovirus-mediated calreticulin in breast cancer: relationship with anti-CD47. Biomed Pharmacother. 2015;73:109–115.
- Wang J, Zhu R, Sun X, et al. Intracellular uptake of etoposide-loaded solid lipid nanoparticles induces an enhancing inhibitory effect on gastric cancer through mitochondria-mediated apoptosis pathway. Int J Nanomed. 2014;9:3987–3998.
- Xia Y, Guo M, Xu T, et al. siRNA-loaded selenium nanoparticle modified with hyaluronic acid for enhanced hepatocellular carcinoma therapy. Int J Nanomed. 2018;13:1539–1552.
- Xu T, Lin Z, Wang C, et al. Heat shock protein 70 as a supplementary receptor facilitates enterovirus 71 infections in vitro. Microb Pathogenesis. 2019;128:106–111.
- Li Y, Guo M, Lin Z, et al. Polyethylenimine-functionalized silver nanoparticle-based co-delivery of paclitaxel to induce HepG2 cell apoptosis. Int J Nanomed. 2016;11:6693–6702.
- Xia Y, Zhong J, Zhao M, et al. Galactose-modified selenium nanoparticles for targeted delivery of doxorubicin to hepatocellular carcinoma. Drug Deliv. 2019;26:1–11.