Abstract
IL-1α is closely related to the development and metastasis of cancer, and its polymorphisms have been reported affecting the susceptibility of malignancy tumors, yet the conclusions are controversial. Present an overall meta-analysis was performed to reach more general findings. Relevant literature was searched from Google Scholar, Web of Science, PubMed, EMBASE, CNKI database and Wanfang database, and the association among polymorphism and cancer risk was appraised by counting ORs and 95% CIs of overall and stratification analysis. The date from 15,586 cases and 18,430 controls in 40 publications were enrolled. There is significantly upregulated risk leads by rs3783553 in all genetic models, while the same tendency was found in all cancer types. The results also suggest a high risk of cancer susceptibility in patients carried rs1800587 polymorphism, of which draw out form allelic and homozygote models in overall studies, especially for cervical cancer. However, there are no significant results in analysis of rs17561. In a word, IL1A SNPs play an essential role in upregulating cancer susceptibility, and current analysis provides detail date for further study in the future.
Introduction
Worldwide, the incidence and mortality of cancer continue to rise year by year. Due to many intricate reasons, of which are related to the growth of population, aging, and environmental implication, cancer became the primary reason for death and resulted in more than 9.6 million people lost their lives in 2018. Across all cancer ranges, the incidence of malignancy tumor is about 20% higher in male than in female. Lung cancer is associated with the first incidence and mortality in male, and prostate cancer, the most widespread malignancy tumor among countries, is not far behind. It is estimated that in 2019, 174,650 people will newly suffer from prostate cancer in the United States, and there are about 31,620 confirmed patients meet the finally death. For female, the incidence of breast cancer is head and shoulder above any other cancers, followed by colorectal and cervical cancer, nonetheless, lung cancer is still the primary strength which contributes to the most of cancer-death[Citation1,Citation2].
Interleukin-1(IL-1), cognized firstly in all Interleukins, has been studied for a long history. It is well known to us all, as indispensable members of cytokines in our body, Interleukin-1 contains IL-1 alpha(IL-1α), IL-1 beta(IL-1β) and IL-1 receptor antagonist(IL-1Ra), which are edited by different genes and interact with each other to influence innate immunity and acquired inflammation[Citation3]. In the past few years, a lot of researches pay attention to the function of IL-1α. IL-1α is mainly produced by monocytes and macrophages, which play the role of proproteins in the inflammation, and may contribute to tumor growth, invasion and distant metastasis in different cancers [Citation4]. Previous studies have demonstrated that IL-1α is significantly related to the risk of prostate cancer, lung cancer, colon cancer, cervical cancer, and breast cancer, etc. [Citation4–13]. These come to conclusions that IL-1α may be responsible for cancer development and treatment.
IL1A, which encodes IL-1α protein, locates at chromosome 2q and consists of 7 exons and 6 introns. rs17561, rs1800587, and rs3783553 are three SNPs on IL1A gene, which are believed as functional roles. rs17561 is a mutation at +4845 of 5th exon of IL1A, which leads to the G-to-T variation, as well as the change of amino acid[Citation14]. Located at -889 of IL1A promoter region, rs1800587(C-to-T) was reported might affect the expression of IL-1α[Citation15,Citation16]. In addition, SNP occurred within the IL1A 3’-untranslated regions (UTR) may affects the expression of IL-1α by influencing the binding of miR-122. That’s an insertion/deletion variation, what we called rs3783553.
Recently, relationship between the polymorphisms of IL1A and different types of cancer has been reported by many researchers, but the conclusions are still not consistent. Hence, an update mete-analysis among all cancers has been carried out, to further discover the effects of IL1A polymorphism on tumors.
Methods
Retrieval of literature
As of May 20, 2019, comprehensive searches of relevant literatures were carried out from several online databases: Web of Science, Google Scholar, PubMed, EMBASE, CNKI database, and Wanfang database. For purpose of hitting more accurate studies mentioning the IL1A polymorphisms and cancer risks, we used “IL1A OR (IL-1 Alpha) OR (Interleukin 1 Alpha) OR (Interleukin-1 Alpha) OR Pro-Interleukin-1-Alpha OR (Preinterleukin 1 Alpha) OR Hematopoietin-1” AND “tumor OR cancer OR carcinoma OR neoplasm” AND “SNP OR variant OR mutation OR polymorphism” as the search items. Moreover, references from selected articles were also searched and viewed for adding any other relevant studies. All the processes were completed along with the PRISMA 2009 statement[Citation17].
Criteria for identification
Inclusions should meet the following criteria after checking the full-text: (1) English or Chinese articles, (2) case-control studies concerning the link between cancer susceptibility and IL1A polymorphism, (3) the studies performed in patients diagnosed with cancer, (4) the data of genotype frequency from cases and controls are sufficient reported or (5) can be calculated from other information in the article.
Otherwise, if the above conditions were not met, or had the following characteristics: (1) repeated studies using overlapping data, (2) the studies aim at animals or cell lines, (3) genotype frequency cannot be obtained, the article would be excluded.
Quality assessment
Researchers assessed the quality of study by putting into force the Newcastle–Ottawa Scale (NOS)[Citation18]. It can be explained by the following standard rules: (1) “high” quality choices were identified with a “star”, (2) each numbered item within the Selection and Exposure categories will acquire up to 1 star maximumly, (3) each study of case-control comparability could be awarded a maximum of 2 stars, (4) “*” means “Yes”, while “NA” means “not applicable”. Two authors give a to unified dissents after comprehensive discussion.
Date extraction
When the articles passed the selection criteria, two authors extracted the date we need from these articles, to ensure the accuracy of the recorded data. The extracted items include: the year of publication, last name of the first author, numbers and genotype frequency of controls and cases, cancer type, genotyping method, ethnicity and source of control. If the date was inconsistent or controversial, the two authors would check the articles, and determine the final data after discussion.
Statistical methods
Firstly, we generated 5 analysis models based on the allele frequency in case group and control group, including allele contrast model, homozygote comparison model, heterozygote comparison model, dominant comparison model, and recessive comparison model. They were (G vs. T), (G/G vs. T/T), (G/T vs. T/T), (G/G + G/T vs. T/T), (G/G vs. G/T + T/T) for rs17561; (C vs. T), (C/C vs. T/T), (C/T vs. T/T), (C/C + C/T vs. T/T), (C/C vs. C/T + T/T) for rs1800587 and (Ins vs. Del), (Ins/Ins vs. Del/Del), (Ins/Del vs. Del/Del), (Ins/Ins + Ins/Del vs. Del/Del), (Ins/Ins vs. Ins/Del + Del/Del) for rs3783553. Then, STATA software (Stata 12.0, College Station, Texas) was run to execute statistical calculations, and the entire inspection process were all two-tailed. We also performed χ2 test to checkout Hardy–Weinberg equilibrium (HWE) of control group, which can compare actual frequencies of genotype with expected value. We used odds ratios (ORs) with 95% confidence intervals (CIs) to demonstrate the computation result of each genic model in overall and stratified groups to appraise the correlation between IL1A polymorphism and cancer susceptibility. The subgroups were stratified by cancer type, source of control, ethnicity and whether comfort to HWE. Meanwhile, the chi-squared (χ2)-based Q test was employed to assess heterogeneity, which was regarded as significant when P < 0.1[Citation19]. For each group stratified date, if existed heterogeneity (P < 0.1)[Citation20], we used random-effects model to pool the date, otherwise fixed-effects model would be applied[Citation21]. Furthermore, p values from Egger’s regression test and symmetrical feature from Begg’s funnel plot were obtained to evaluate whether there is a significant bias. Above all analysis, if p values ≤ 0.05, the results were considered as statistically significant.
Results
Literature searches and characteristics
exhibits the course of screening process from searching to including. 317 relevant articles were downloaded from all database, as well as 23 from their references were also enrolled. We initially obtained 323 studies from getting rid of the duplicated, and then, through screening and checking the titles and abstracts, we continued to remove 260 articles which were significantly different from our objective. Finally, we included 40 eligibilities by applying the selection criteria after browsing and reading the full text of the rest 63 articles.
The characteristics of the enrolled researches on IL1A polymorphism and cancers were showed in . There were 5 studies about rs17561, 22 about rs1800587, and 18 about rs1783553(one article can contain more than one study about IL1A polymorphisms). Among all studies, the results of heterogeneity test were showed in . For studies on rs17561, they respectively investigated the ovarian cancer, glioma, cervical cancer, nasopharyngeal cancer, and lung cancer [Citation22–26]. As for rs1800587, there are 2 studies for each type of cancer on ovarian cancer, cervical cancer, breast cancer, odontogenic tumor, nasopharyngeal cancer, lymphoma, and lung cancer. The rest studies put research centers on kaposi sarcoma, vulvar carcinoma, multiple myeloma, gastric cancer, prostate cancer, esophageal cancer, and glioma[Citation22,Citation25–44]. In term of rs3783553, it is a contention of a hundred schools of thought. It includes researches on multiple systems of cancer diseases, such as hepatocellular carcinoma, nasopharyngeal cancer, glioma, prostate cancer, gastric cancer, thyroid carcinoma, oral squamous cell carcinoma, cervical cancer, ovarian cancer, colorectal cancer, colorectal cancer, and endometrial cancer[Citation37,Citation45–61]. Analyzed by ethnicity, most of studies focus on Asians, yet researches direct to Caucasians are also quite a lot. Additionally, Bushley et al, Foster et al, and Rothman et al did the study from mixed races[Citation25,Citation33,Citation35]. On the whole, all the cases add up to 15,586 for patients (case group), as well as 18,430 for healthy subjects (control group). The whole sample size is large, and we could make our study get results more accurately and convincingly.
Table 1. Characteristics of the enrolled studies on IL-1A polymorphism and cancer.
Results of NOS and HWE checking
We performed Newcastle–Ottawa Scale in evaluating the overall quality of enrolled studies. Table S1 showed that all included studies scored more than 5 stars, and more than 70% of them were higher than 7 stars. The maximum score was 8 stars, with 5 cases. Therefore, the overall quality of the study is at a high level. For three IL1A polymorphisms, showed in , apart from 4 studies on rs1800587 from Grimm et al, Hefler et al, Abazis-Stamboulieh et al, and Eshghyar et al[Citation32,Citation34,Citation40,Citation42], all studies gone through the test of HWE. Mentionable, the source of control in Grimm et al ’s study was based on hospital, the other three were patient based. These four studies all received high stars in NOS, which got 7 stars, 7 stars, 8 stars and 6 stars respectively. We consider that although they did not meet the HWE, their quality may still be high to ensure their reliability and significance.
Association between IL1A +4845- G > T (rs17561) and cancer susceptibility
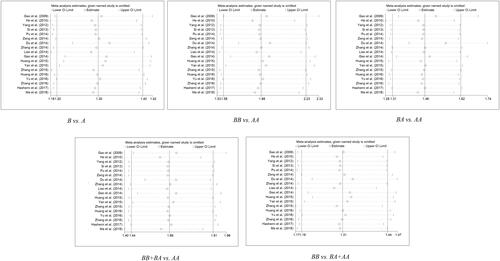
As showed in Figure S1 and , the overall p values and ORs demonstrate no significant rising or declining link between IL1A rs17561 - G > T SNP and cancer risk, whatever the genotype (G vs. T: P = 0.995 OR= 1, 95% CI= 0.77– 1.3; G/G vs. T/T: P = 0.855, OR= 0.97, 95% CI= 0.71– 1.33; G/T vs. T/T: P = 0.955, OR= 1.01, 95% CI= 0.66– 1.54; G/G + G/T vs. T/T: P = 0.923, OR= 1.02, 95% CI= 0.69– 1.5; G/G vs. G/T + T/T: P = 0.647, OR= 0.94, 95% CI= 0.7– 1.24). And further stratification by cancer type, ethnicity, and control of source showed meaningless results, too.
Table 2. Results of pooled analysis for IL-1A polymorphism and cancer susceptibility.
Association between IL1A -889- C > T (rs1800587) and cancer susceptibility
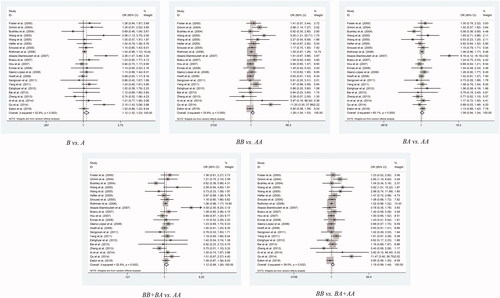
When it comes to rs1800587 polymorphism, we found that there is a statistically significant uplifted overall risk between it and cancer susceptibility only in allelic contrast model (C vs. T: P = 0.018, OR= 1.12, 95% CI= 1.02– 1.23) and homozygote comparison model (CC vs. TT: P = 0.021, OR= 1.26, 95% CI= 1.04– 1.53) (the others: CT vs. TT: P = 0.294, OR= 1.08, 95% CI= 0.94– 1.24; C/C + C/T vs. T/T: P = 0.075, OR= 1.12, 95% CI= 0.99– 1.26; C/C vs. C/T + T/T: P = 0.06, OR = 1.19, 95% CI= 0.99– 1.44; , ).
What is more, the studies stratified by cancer type showed statistical significances in homozygote and recessive two type models (CC vs. TT: P = 0.017, OR= 3.4, 95% CI= 1.24– 9.29; CC vs. CT + TT: P = 0.013, OR= 3.44, 95% CI = 1.3– 9.07) in Cervical Cancer, but it suggested few link with the SNP to breast cancer, lung cancer, lymphoma, odontogenic and ovarian cancer. Another finding showed a high cancer risk of rs1800587 SNP in allelic comparison model of population-based studies (C vs. T: P = 0.049, OR= 1.06, 95% CI = 1– 1.12), while the same trend risk of same mutation was found in recessive comparison model in hospital-based studies (CC vs. CT + TT: P = 0.046, OR= 1.69, 95% CI= 1.01– 2.83).
We have also expounded there are four studies did not pass the Hardy–Weinberg equilibrium in rs1800587 polymorphism. According to whether they were accord with the HWE, we divided them into two parts. Notable, climbing risks were discovered in homozygote comparison model of those four who did not conform to Hardy–Weinberg equilibrium (C/C vs. T/T: P = 0.037, OR = 1.63, 95% CI= 1.03–2.57). (.)
Association between IL1A 3’UTR- ins > del (rs3783553) and cancer susceptibility
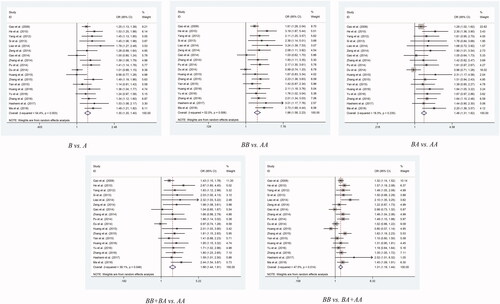
As for rs3783553, the IL1A 3’UTR-ins/del SNP, studies showed strongly significantly links with positive risk to overall cancers in all genetic models: allelic contrast model (Ins vs. Del: P < 0.001, OR = 1.3, 95% CI = 1.21– 1.4), homozygote comparison model (Ins/Ins vs. Del/Del: P < 0.001, OR= 1.88, 95% CI= 1.58– 2.23), heterozygote comparison model (Ins/Del vs. Del/Del: P = P < 0.001, OR = 1.46, 95% CI = 1.31– 1.62), dominant comparison model (Ins/Ins + Ins/Del vs. Del/Del: P < 0.001, OR = 1.66, 95% CI = 1.44– 1.91), and recessive comparison model (Ins/Ins vs. Ins/Del + Del/Del: P < 0.001, OR = 1.31, 95% CI = 1.19– 1.44) (, ).
After that, tumor type classification analysis revealed an interesting finding. We found risk of cancer is upregulated in homozygote, heterozygote and dominant models (Ins/Ins vs. Del/Del: P = 0.001, OR = 1.98, 95% CI = 1.31– 2.98; Ins/Del vs. Del/Del: P = 0.003, OR = 1.84, 95% CI = 1.22– 2.76; Ins/Ins + Ins/Del vs. Del/Del: P = 0.001, OR = 1.91, 95% CI = 1.29– 2.82) in cervical cancer, and there were the similar trend in cerival cancer in homozygote and recessive models of rs1800587 as well. Then, when we analyzed rs3783553 separately, except cervical cancer, there were other statistically significant rising risk between it and different type of cancer, including colorectal cancer for all comparison models(Ins vs. Del: P = 0.042, OR = 1.31, 95% CI = 1.01– 1.69; Ins/Ins vs. Del/Del: P = 0.037, OR = 1.98, 95% CI = 1.04– 3.74; Ins/Del vs. Del/Del: P = 0.001, OR = 1.77, 95% CI = 1.26– 2.48; Ins/Ins + Ins/Del vs. Del/Del: P = 0.025, OR = 1.85, 95% CI = 1.08– 3.18; Ins/Ins vs. Ins/Del + Del/Del: P = 0.02, OR = 1.28, 95% CI = 1.04– 1.58), gastric cancer for all comparison models(Ins vs. Del: P < 0.001, OR = 1.32, 95% CI = 1.14– 1.52; Ins/Ins vs. Del/Del: P < 0.001, OR = 2.03, 95% CI = 1.44– 2.87; Ins/Del vs. Del/Del: P = 0.002, OR = 1.72, 95% CI = 1.23– 2.41; Ins/Ins + Ins/Del vs. Del/Del: P < 0.001, OR = 1.86, 95% CI = 1.34– 2.57; Ins/Ins vs. Ins/Del + Del/Del: P = 0.009, OR = 1.31, 95% CI = 1.07– 1.59), hepatocellular carcinoma for 2 comparison models(Ins vs. Del: P = 0.049, OR = 1.23, 95% CI = 1– 1.51; Ins/Ins vs. Ins/Del + Del/Del: P = 0.038, OR = 1.27, 95% CI = 1.01– 1.58) and prostate cancer for 4 models(Ins vs. Del: P < 0.001, OR = 1.66, 95% CI = 1.3– 2.11; Ins/Ins vs. Del/Del: P < 0.001, OR = 3.14, 95% CI = 1.67– 5.9; Ins/Ins + Ins/Del vs. Del/Del: P = 0.005, OR = 1.76, 95% CI = 1.19– 2.6; Ins/Ins vs. Ins/Del + Del/Del: P < 0.001, OR = 2.18, 95% CI = 1.47– 3.23).
Another result that caught our attention is, for source of control stratification analysis, population-based subgroup analysis found a greatly increased risk in all five comparison models (C vs. T: P < 0.001, OR = 1.29, 95% CI = 1.2– 1.4; CC vs. TT: P < 0.001, OR = 1.86, 95% CI = 1.56– 2.21; CT vs. TT: P < 0.001, OR = 1.46, 95% CI = 1.31– 1.63; C/C + C/T vs. T/T: P < 0.001, OR = 1.67, 95% CI = 1.44–1.94; C/C vs. C/T + T/T: P < 0.001, OR = 1.3, 95% CI = 1.18– 1.43). But noticeable, the hospital-based subgroup is missing in this part. (.)
Analysis using ethnicity as subgroup hinted a closely association, in both Asian and Caucasian, between ins/del mutation and high cancer susceptibility risk in all five models. For Asian, there were 16 case-controls consisting of 6493 cases and 7528 controls, whose significant genotype models were: Ins vs. Del: P < 0.001, OR = 1.28, 95% CI = 1.18– 1.39; Ins/Ins vs. Del/Del: P < 0.001, OR = 1.84, 95% CI = 1.53– 2.2; Ins/Del vs. Del/Del: P < 0.001, OR = 1.46, 95% CI = 1.3– 1.63; Ins/Ins + Ins/Del vs. Del/Del: P < 0.001, OR = 1.67, 95% CI = 1.42– 1.95; Ins/Ins vs. Ins/Del + Del/Del: P < 0.001, OR = 1.28, 95% CI = 1.16– 1.41. In contrast, 2 studies about Caucasian included 475 cases and 490 controls, with significant models as following: Ins vs. Del: P < 0.001, OR = 1.5, 95% CI = 1.24– 1.81; Ins/Ins vs. Del/Del: P < 0.001, OR = 2.38, 95% CI = 1.55– 3.66; Ins/Del vs. Del/Del: P = 0.022, OR = 1.48, 95% CI = 1.06– 2.06; Ins/Ins + Ins/Del vs. Del/Del: P = 0.001, OR = 1.7, 95% CI = 1.24– 2.34; Ins/Ins vs. Ins/Del + Del/Del: P < 0.001, OR = 1.71, 95% CI = 1.27– 2.3.
Sensitivity analysis and bias evaluate
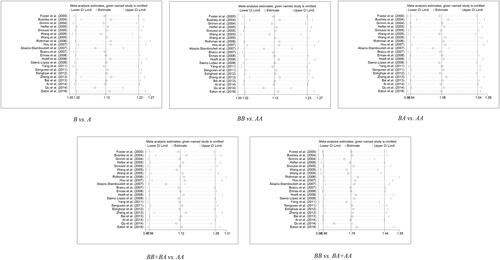
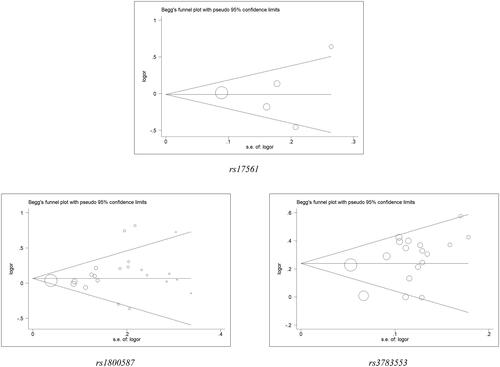
Revealed from sensitivity analysis, see Table S2, , there had no significant impacts of any individual study on the pooled ORs. Besides, Egger’s test and Begg’s funnel plot showed that in each allelic contrast and genotype model, there had no publication bias all over the comparison of models (Table S3, ).
Figure 6. Begg’s funnel plot for IL1A polymorphisms and cancer risk. The x-axis is log (OR), and the y-axis is natural logarithm of OR. The horizontal line in the figure represents the overall estimated log (OR). The two diagonal lines indicate the pseudo 95% confidence limits of the effect estimate.
Summary and discussion
IL1A, together with other members of IL-1 family, locates on chromosome 2 and form a cluster of cytokines. There are 7 exons and 6 introns in IL1A, the 1st exon encodes the 5’-untranslated regions, the 2nd, 3rd and 4th exons encode the enzymatic hydrolysis fragment of the pro-proteins, the 5th, 6th, 7th exons encode IL-1α, and the 7th exon encode 3’-untranslated regions[Citation62]. The promoter of IL1A lacks of typical regulatory areas, but can be regulated by variable transcription factors through the binding sites, such as AP1, Sp1, NF- κB, etc.[Citation63,Citation64].
IL-1α, the production of IL1A, can be rapidly up-regulated by multiple molecular factors, and enhance the pro-inflammatory effects, accompany with the other interleukin family molecules[Citation65]. Furthermore, IL-1α can reverse the transport of transcriptase form cytoplasm to nucleus and independently regulate gene expression, which may have anti-apoptotic effects on non-inflammatory cells[Citation4]. Researches also illustrated that the existing of IL-1α in the tumor microenvironment can enhance growth, invasion and metastasis of cancer cell, and its necrosis production could promote the vessel formation and invasion risk of tumor through boosting the expression of vascular endothelial growth factor (VEGF)[Citation4,Citation66]. On the other way, there are also some findings suggested that the immune response mediated by IL1α would build up an immune memory for tumor cells, so as to continuously play an anti-tumor role by guiding the immune system to attack tumor cells[Citation67]. Therefore, IL1α may have both cancer-causing and anti-cancer effects, this kind of mysterious and powerful molecule is worth for a further investigating.
SNPs have a great value in cancer susceptibility, especially for the mutations which do alter cancer risk but are difficult to cause apparent symptoms. When it happens to IL1A, it may increase or decrease the risk of cancers through affecting gene transcription rates, mRNA stability, expression or activity of IL1α, etc.[Citation14–16,Citation68,Citation69]. Previous studies had revealed 51 unique variations in IL1A, many of them are exact functional. In the 5th exon region +4845 site, IL1A has a G > T transform SNP (rs17561), of which could lead to the conversion of encoding amino acid at position 144 from alanine (Ala) to serine (Ser), and such change could increase the release of IL-1α through enhancing the efficiency of specific enzymes which process IL-1α precursors, thereby enhancing the inflammatory response[Citation14]. Also, IL1A owns a C > T SNP at -889 site of 5’-regulatory region (rs1800587). Accumulating evidence suggested that this mutation may affect gene expression by affecting transcriptional activity[Citation15,Citation16]. Besides, previous studies had shown that a 4-bp insertion/deletion (rs3783553) polymorphism happening in the gene 3’-UTR could alter the strength of miRNA binding, thereby affecting cancer risks[Citation68,Citation69]. In the past decades, lots of researches stated the connection between cancer risk and IL1A polymorphisms, but the conclusions were contradictory. Some case-control studies reported a high risk of cancer susceptibility with IL1A polymorphism[Citation47,Citation61]. On the contrary, others suggested a low risk in the same polymorphism and the same kind cancer[Citation45,Citation60]. According to the law of statistics, limitation of sample size, difference of methods, ethnicity, control’s source, systematic and sampling errors, any of them may cause the conflicting results. In order to eliminate potential errors to obtain a more accurate conclusion, we performed an overall meta-analysis in current study.
In the current work, we enrolled 45 eligible case-control studies, containing 15,586 cancer cases and 18, 430 healthy controls. The five genetic model of three SNPs were under analyzing, and comparison distributions of alleles were pooled to get a more precise result by the larger sample size. Evidence from NOS and sensitivity analysis ensured a high quality and credibility of each enrolled study. Final conclusion drew a vivid, specific and accurate picture. It revealed a prominent relationship between IL1A rs3783553 ins/del SNP and rising risk of overall cancer susceptibility. The similar findings were found in all cancer type we enrolled, including cervical cancer, colorectal cancer, gastric cancer, HCC, and prostate cancer. This conclusion shows the importance of IL1A ins/del SNP for tumor susceptibility, which means that this mutation may be used as an early screening marker for cancer, or even help explore a touchable and useful way to treat cancer in the future. The final result also suggested IL1A rs1800587 SNP may influence the overall risk with an increased trend, particularly for cervical cancer. This finding can only be found in allele and homozygote model for overall study, while it can be found in homozygote and dominant model in cervical cancer. And there is an interesting founding that we cannot ignore, rs1800587 and rs3783553 both have pointed to a link with high risk to cervical cancer. It reminds us that different genetic variations may indeed affect gene regulation in a certain link, separately and mutually. From this, the gynecologists and scholars can set out to explore the mechanism at deeper levels. Nevertheless, IL1A rs17561 SNP was found to have no effect on risk of overall cancer susceptibility. Likewise, results of pooled analysis did not demonstrate any low risk of cancer susceptibility to IL1A SNP in neither of three type mutation.
Stratification analysis based on ethnicity was done in Asian and Caucasian, it showed that IL1A rs3783553 ins/del SNP promotes the risk of cancer in both subgroups. We can make the explanation as the allele frequency of the SNP is similar between 2 kind ethnicities. In terms of OR-values, the link of this kind SNP to high cancer risk may be a little stronger among Caucasians, but the difference is not huge. This result can reinforce the idea that the ins/del mutation could enhance cancer susceptibility. Yet, we cannot extend this result to overall ethnicity because we only included Asian and Caucasian, and the allele frequency may change in other ethnicity, such as Africans.
Admittedly, there are still some limitations such as the ethnicity problem mentioned above in our study. On the one hand, the result from the source of control stratification analysis of IL1A rs1800587 SNP suggested possible bias. On the other hand, the result from the HWE based stratification analysis of same SNP implied the same condition. Although we have conducted a quality assessment on the use of NOS for those who do not comply with HWE to ensure the credibility of the results, it need to be supported by more case-control studies. Another important thing to note is, according to the result of meta-analysis, the conclusion about how IL1A rs1800587 SNP influence cancer susceptibility remain cautious. What need to ponder is the meaningless result in allele contract model in cervical cancer. It requires to be interpreted with caution that IL1A -889 T may be a potential risk allele for cancer and it is worthy of further study. Then, all SNP considered, we only enrolled English and Chinese articles, this may exist underlying selection biases. Besides, we cannot exclude influence of some confounding factors, such as gender, age, life habits like drinking and smoking, other diseases, etc. Because of that, we have no way to take potential interactions of these factors to gene into consideration. Furthermore, the development of cancer is an extremely complex process, various members of interleukin family may be involved in. Therefore, in addition to the study of SNP, we need to establish a more comprehensive study of the interaction between different genes to verify the influence of gene SNP on cancer susceptibility.
Limitations cannot belittle virtues, there does have many merits in recent research. First, by retrieving, we found that no similar study about IL1A-SNP in overall cancer susceptibility have been published before. In present study we included a large number of case-controls, so that it can supplement powerful and accurate evidence for overall study to help understand the role of how IL1A-SNPs could impact the development of cancer. Second, we evaluated the quality of enrolled studies on using NOS scale, and results from it ensure every enrolled research is with a high-quality to fulfill the pre-set criteria. Third, sensitivity analysis was preformed to test for bias, meanwhile, stratification analysis was done by cancer type, ethnicity, source of controls. All these measures are designed to avoid potential biases and lower heterogeneities so as to get the most precise conclusion in the end.
Conclusion
Taken as a whole, our study had confirmed in detail that IL1A rs3783553 SNP play an important role in cancer development process, it can significant up-regulation risk of cancer susceptibility. Also, the mutation link with high risk of cancer both in Caucasians and Asians. By comparison, the link seemed stronger among Caucasians, but the difference was not huge. Another implication is that IL1A rs1800587 SNP may be responsible for high cancer risks, especially for cervical cancer. However, this conclusion needs to be carefully explained due to its limitations. Lastly, there is no evidence that IL1A rs17561 SNP associates with cancer risk, no matter high or low. Above, present study suggests the closely relationship of IL1A SNP and risk of cancers. We are pleased to be able to contribute to the search for mechanisms and treatments for cancers, at the same time, we look forward to further researches and more comprehensive conclusions in the future.
Author contributions
Haoran Xia and Yiding Chen performed the literature search, data extraction, and statistical analysis and wrote the manuscript. Haoran Xia and Jialin Meng supervised the literature search, data extraction, analysis, Jialin Meng and ChaozhaoLiang reviewed the manuscript.
Supplementary_data.pdf
Download ()Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA A Cancer J Clin. 2019;69:7–34.
- Dinarello CA. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol Rev. 2018;281:8–27.
- Malik A, Kanneganti TD. Function and regulation of IL-1α in inflammatory diseases and cancer. Immunol Rev. 2018;281:124–137.
- Kuan EL, Ziegler SF. A tumor-myeloid cell axis, mediated via the cytokines IL-1alpha and TSLP, promotes the progression of breast cancer. Nat Immunol. 2018;19:366–374.
- Matsuo Y, Sawai H, Ma J, et al. IL-1alpha secreted by colon cancer cells enhances angiogenesis: the relationship between IL-1alpha release and tumor cells' potential for liver metastasis. J Surg Oncol. 2009;99:361–367.
- Seddighzadeh M, Larsson P, Ulfgren AC, et al. Low IL-1alpha expression in bladder cancer tissue and survival. Eur Urol. 2003;43:362–368.
- Song Z, Lin Y, Ye X, et al. Expression of IL-1alpha and IL-6 is Associated with Progression and Prognosis of Human Cervical Cancer. Med Sci Monit. 2016;22:4475–4481.
- Sorrentino R, Terlizzi M, Di Crescenzo VG, et al. Human lung cancer-derived immunosuppressive plasmacytoid dendritic cells release IL-1alpha in an AIM2 inflammasome-dependent manner. The American Journal of Pathology. 2015;185:3115–3124.
- Xu D, Matsuo Y, Ma J, et al. Cancer cell-derived IL-1alpha promotes HGF secretion by stromal cells and enhances metastatic potential in pancreatic cancer cells. J Surg Oncol. 2010;102:469–477.
- Schmidt SV, Seibert S, Walch-Ruckheim B, et al. RIPK3 expression in cervical cancer cells is required for PolyIC-induced necroptosis, IL-1alpha release, and efficient paracrine dendritic cell activation. Oncotarget 2015;6:8635–8647.
- Ma J, Sun X, Guo T, et al. Interleukin-1 receptor antagonist inhibits angiogenesis via blockage IL-1alpha/PI3K/NF-kappabeta pathway in human colon cancer cell. Cmar. 2017;Volume 9:481–493.
- Cheng J, Li L, Liu Y, et al. Interleukin-1alpha induces immunosuppression by mesenchymal stem cells promoting the growth of prostate cancer cells. Molecular Medicine Reports. 2012;6:955–960.
- Kawaguchi Y, Tochimoto A, Hara M, et al. Contribution of single nucleotide polymorphisms of the IL1A gene to the cleavage of precursor IL-1alpha and its transcription activity. Immunogenetics 2007;59:441–448.
- McDowell TL, Symons JA, Ploski R, et al. A genetic association between juvenile rheumatoid arthritis and a novel interleukin-1 alpha polymorphism. Arthritis and rheumatism 1995;38:221–228.
- Dominici R, Cattaneo M, Malferrari G, et al. Cloning and functional analysis of the allelic polymorphism in the transcription regulatory region of interleukin-1 alpha. Immunogenetics 2002;54:82–86.
- Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol. 2009;62:1006–1012.
- Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605.
- Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;1;127:820–826.
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188.
- Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. Journal of the National Cancer Institute. 1959;22:719–748.
- Eaton KD, Romine PE, Goodman GE, et al. Inflammatory Gene Polymorphisms in Lung Cancer Susceptibility. Journal of Thoracic Oncology: official Publication of the International Association for the Study of Lung Cancer. 2018;13:649–659.
- Sousa H, Mesquita L, Ribeiro J, et al. Polymorphisms in host immune response associated genes and risk of nasopharyngeal carcinoma development in Portugal. Immunobiology 2016;221:145–152.
- Zidi S, Sghaier I, Zouidi F, et al. Interleukin-1 Gene Cluster Polymorphisms and its Haplotypes may Predict the Risk to Develop Cervical Cancer in Tunisia. Pathol Oncol Res. 2015;21:1101–1107.
- Bushley AW, Ferrell R, McDuffie K, et al. Polymorphisms of interleukin (IL)-1alpha, IL-1beta, IL-6, IL-10, and IL-18 and the risk of ovarian cancer. Gynecol Oncol. 2004;95:672–679.
- Ai L, Shu Y. Association between interleukin- 1A polymorphisms and risk of glimoa. Chinese Journal of Laboratory Diagnosis 2014;18:1446–1448.
- Zheng L, Yin J, Wang L, et al. Interleukin 1B rs16944 G > A polymorphism was associated with a decreased risk of esophageal cancer in a Chinese population. Clinical Biochemistry. 2013;46:1469–1473.
- Hoeft B, Becker N, Deeg E, et al. Joint effect between regular use of non-steroidal anti-inflammatory drugs, variants in inflammatory genes and risk of lymphoma. Cancer Causes Control. 2008;19:163–173.
- Saenz-Lopez P, Carretero R, Cozar JM, et al. Genetic polymorphisms of RANTES, IL1-A, MCP-1 and TNF-A genes in patients with prostate cancer. BMC Cancer. 2008;8:382.
- Ennas MG, Moore PS, Zucca M, et al. Interleukin-1B (IL1B) and interleukin-6 (IL6) gene polymorphisms are associated with risk of chronic lymphocytic leukaemia. Hematol Oncol. 2008;26:98–103.
- Hou L, El-Omar EM, Chen J, et al. Polymorphisms in Th1-type cell-mediated response genes and risk of gastric cancer. Carcinogenesis 2007;28:118–123.
- Abazis-Stamboulieh D, Oikonomou P, Papadoulis N, et al. Association of interleukin-1A, interleukin-1B and interleukin-1 receptor antagonist gene polymorphisms with multiple myeloma. Leuk Lymphoma. 2007;48:2196–2203.
- Rothman N, Skibola CF, Wang SS, et al. Genetic variation in TNF and IL10 and risk of non-Hodgkin lymphoma: a report from the InterLymph Consortium. The Lancet Oncology 2006; Jan7:27–38.
- Grimm C, Berger I, Tomovski C, et al. A polymorphism of the interleukin-1 receptor antagonist plays a prominent role within the interleukin-1 gene cluster in vulvar carcinogenesis. Gynecologic Oncology. 2004;92:936–940.
- Foster CB, Lehrnbecher T, Samuels S, et al. An IL6 promoter polymorphism is associated with a lifetime risk of development of Kaposi sarcoma in men infected with human immunodeficiency virus. Blood 2000; Oct 196:2562–2567.
- Bai L, Yu H, Wang H, et al. Genetic single-nucleotide polymorphisms of inflammation-related factors associated with risk of lung cancer. Med Oncol. 2013;30:414.
- Yang ZH, Dai Q, Zhong L, et al. Association of IL-1 polymorphisms and IL-1 serum levels with susceptibility to nasopharyngeal carcinoma. Mol Carcinog. 2011;50:208–214.
- Qu YL, Yu H, Chen YZ, et al. Relationships between genetic polymorphisms in inflammation-related factor gene and the pathogenesis of nasopharyngeal cancer. Tumor Biol. 2014;35:9411–9418.
- Senguven B, Oygur T. Investigation of interleukin-1 alpha and interleukin-6 expression and interleukin-1 alpha gene polymorphism in keratocystic odontogenic tumors and ameloblastomas. Med Oral. 2011;16:e467–72.
- Eshghyar N, Nikbin B, Amirzargar A, et al. Gene polymorphism of interleukin-1 alpha and beta in keratocystic odontogenic tumors. Journal of Oral Pathology & Medicine: official Publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology. 2012;41:697–701.
- Snoussi K, Strosberg AD, Bouaouina N, et al. Genetic variation in pro-inflammatory cytokines (interleukin-1beta, interleukin-1alpha and interleukin-6) associated with the aggressive forms, survival, and relapse prediction of breast carcinoma. European Cytokine Network 2005; Dec16:253–260.
- Hefler LA, Grimm C, Lantzsch T, et al. Interleukin-1 and interleukin-6 gene polymorphisms and the risk of breast cancer in caucasian women. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2005;11:5718–5721.
- Wang HM. Relationship between interleukin- 1 gene cluster gene polymorphisms and susceptibility to cervical cancer : Dalian Medical University; 2005.
- Ioana Braicu E, Mustea A, Toliat MR, et al. Polymorphism of IL-1alpha, IL-1beta and IL-10 in patients with advanced ovarian cancer: results of a prospective study with 147 patients. Gynecol Oncol. 2007;104:680–685.
- Liao H, Zhang L, Chen P, et al. Insertion/Deletion Polymorphism of IL1A 3'- UTR Associated with the Susceptibility of Prostate Cancer. Journal of Sichuan University (Medical Science Education) 2014; 45: 956–959.
- Ma Q, Mao Z, Du J, et al. Association between an insertion/deletion polymorphism in the interleukin-1alpha gene and the risk of colorectal cancer in a Chinese population. The International Journal of Biological Markers. 2018;33:401–406.
- Hashemi M, Bahari G, Sarhadi S, et al. 4-bp insertion/deletion (rs3783553) polymorphism within the 3'UTR of IL1A contributes to the risk of prostate cancer in a sample of Iranian population. J Cell Biochem. 2018;119:2627–2635.
- Zhang J, Shi H, Xue M, et al. An insertion/deletion polymorphism in the interleukin-1A 3'untranslated region confers risk for gastric cancer. Cbm. 2016;16:359–365.
- Yu X, Zhou B, Zhang Z, et al. Insertion/deletion polymorphism in IL1A 3'-UTR is associated with susceptibility to endometrial cancer in Chinese Han women. J Obstet Gynaecol Res. 2016;42:983–989.
- Huang X, Yang Y, Cui ZW, et al. A functional insertion/deletion polymorphism in the IL1A gene is associated with decreased risk of breast cancer. Genet Mol Res. 2016;15: gmr7486.
- Yan H, Sun R, Pan X, et al. Lack of association between an insertion/deletion polymorphism in IL1A and risk of colorectal cancer. Genet Mol Res. 2015;14:8490–8495.
- Zhang Y, Sturgis EM, Sun Y, et al. A functional variant at miRNA-122 binding site in IL-1α 3' UTR predicts risk and HPV-positive tumours of oropharyngeal cancer. Eur J Cancer. 2015;51:1415–1423.
- Huang J, Ni S, Li D, et al. An insertion/deletion polymorphism at miRNA-122 binding site in the IL1A is associated with a reduced risk of cervical squamous cell carcinoma. Genet Test Mol Biomarkers. 2015;19:331–334.
- Du Y, Han X, Pu R, et al. Association of miRNA-122-binding site polymorphism at the interleukin-1 α gene and its interaction with hepatitis B virus mutations with hepatocellular carcinoma risk. Front Med. 2014;8:217–226.
- Pu Y, Zhang Z, Zhou B, et al. Association of an insertion/deletion polymorphism in IL1A 3'-UTR with risk for cervical carcinoma in Chinese Han Women. Hum Immunol. 2014;75:740–744.
- Zhang Z, Zhou B, Gao Q, et al. A polymorphism at miRNA-122-binding site in the IL-1alpha 3'UTR is associated with risk of epithelial ovarian cancer. Familial Cancer. 2014;13:595–601.
- Gao L, Zhu X, Li Z, et al. Association between a functional insertion/deletion polymorphism in IL1A gene and risk of papillary thyroid carcinoma. Tumor Biol. 2014;35:3861–3865.
- Zeng XF, Li J, Li SB. A functional polymorphism in IL-1A gene is associated with a reduced risk of gastric cancer. Tumor Biol. 2014;35:265–268.
- Sima XT, Liu H, Yang YW, et al. Association between an insertion/deletion polymorphism at miRNA- 122 binding site in the 3' untranslated region of interleukin- 1A and risk of glioma Chinese. Journal of Cancer Prevention and Treatment 2013;20:580–582.
- He Y. Screening and Functional Analysis of Polymorphisms Residing in the MicroRNA Binding Sites of Hepatocellular Carcinoma- related Genes. Soochow University. 2010.
- Gao Y, He Y, Ding J, et al. An insertion/deletion polymorphism at miRNA-122-binding site in the interleukin-1alpha 3' untranslated region confers risk for hepatocellular carcinoma. Carcinogenesis 2009;30:2064–2069.
- Nicklin MJ, Weith A, Duff GW. A physical map of the region encompassing the human interleukin-1 alpha, interleukin-1 beta, and interleukin-1 receptor antagonist genes. Genomics 1994;19:382–384.
- Bailly S, Fay M, Israel N, et al. The transcription factor AP-1 binds to the human interleukin 1 alpha promoter. Eur Cytokine Netw. 1996; Apr-Jun7:125–128.
- McDowell TL, Symons JA, Duff GW. Human interleukin-1 alpha gene expression is regulated by Sp1 and a transcriptional repressor. Cytokine 2005;30:141–153.
- Weber A, Wasiliew P, Kracht M. Interleukin-1 (IL-1) pathway. Science Signaling. 2010;3:cm1.
- Feldmeyer L, Werner S, French LE, et al. Interleukin-1, inflammasomes and the skin. Eur J Cell Biol. 2010;89:638–644.
- Voronov E, Weinstein Y, Benharroch D, et al. Antitumor and immunotherapeutic effects of activated invasive T lymphoma cells that display short-term interleukin 1alpha expression. Cancer Research 1999; Mar 159:1029–1035.
- Wu M, Jolicoeur N, Li Z, et al. Genetic variations of microRNAs in human cancer and their effects on the expression of miRNAs. Carcinogenesis 2008;29:1710–1716.
- Yu Z, Li Z, Jolicoeur N, et al. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Research. 2007;35:4535–4541.