Abstract
Background
Prostate cancer (PC) fearfully impacts men's health. We explored the efficacy and mechanism of circular RNA circZNF609 (circZNF609) on colony formation, viability, apoptosis, migration and invasion and in PC cells.
Methods
Colony formation, CCK-8, flow cytometry, migration and invasion assay were respectively used to detect the functions of circZNF609 and microRNA (miR)-186-5p on cell colony ability, viability, apoptosis, migration and invasion. circZNF609 and miR-186-5p expression were changed by cell transfection and tested by RT-qPCR. Moreover, Cleaved-Caspase-3, Cleaved-Caspase-9, matrix metalloproteinase-9 (MMP-9), Vimentin and relate-proteins of cell pathways were examined through Western blot.
Results
circZNF609 was highly expressed at PC tissues. circZNF609 declined cell colony ability, viability, migration and invasion and caused apoptosis. Furthermore, circZNF609 negatively regulated miR-186-5p, miR-186-5p inhibitor could reverse impacts of circZNF609. Finally, circZNF609 restrained the YAP1 and AMPK pathways by up-regulating miR-186-5p.
Conclusion
Silencing circZNF609 restrained growth, migration and invasion of PC cells by up-regulating miR-186-5p via YAP1 and AMPK pathways.
circZNF609 is highly expressed in PC tissues;
circZNF609 restrains cell growth, migration and invasion;
circZNF609 exerts its function by up-regulating miR-186-5p;
circZNF609 exerts its function by YAP1 and AMPK signaling pathways.
Highlights
Introduction
Prostate cancer (PC) is a most familiar male tumour in Western countries [Citation1]. Although China is a lower incidence area for PC, studies present that the incidence of PC in China is rising linearly in recent years, which fearfully menaces men's health [Citation2]. Compared with other malignant tumours, PC cells proliferate rate is slower and it is discovered and diagnosed when urine retention and other symptoms occur [Citation3]. In recent years, the arise of blood prostate specific antigen (PSA) has enhanced the detection rate of inchoate PC [Citation4], but studies have demonstrated that certain organs of human can produce PSA under certain circumstances, the blood PSA lacks the specificity for diagnosing PC [Citation5]. Therefore, there is an urgency to find new indicators for detecting inchoate PC.
circRNA is a kind of endogenous long non-coding RNA molecules with looping structure [Citation6]. circRNAs are structurally more stable than linear RNAs, so it has more preponderances in developing new indicators for clinical diagnosis [Citation7]. Delightfully, there were studies that had pointed out that circRNAs may act as tumour markers [Citation8]. circZNF609 first appeared in muscle cell disparateiation [Citation9], Later studies demonstrated that circZNF609 highly expressed in nasopharynx tumour and other tumours [Citation10]. In addition, the efficacy of circZNF609 in PC has not been reported until now.
miRNA is another kind of non-coding RNA, whose length is approximately 22 nt [Citation11]. miRNAs are initiated with the deletion of miRNA in leukemia patients [Citation12]. Since then, increasing evidence demonstrated that miRNAs play a vital part in tumour progression and mechanisms [Citation13,Citation14]. A research pointed out miR-186-5p restrained colorectal tumour [Citation15]. Besides, further studies discovered that miR-186-5p restrained the development of kidney tomor, also of the urinary system [Citation16]. In addition, there was a study that had demonstrated that miR-186-5p is highly expressed and plays a part on PC [Citation17], so miR-186-5p could be an underlying treatment for PC.
We focused on the efficacy of circZNF609 at PC in this experiment. We first investigated the level of circZNF609 in PC tissues. Furthermore, circZNF609 and miR-186-5p expressions were examined in PC3 and LNCaP cell colony, viability, apoptosis, migration and invasion in cellular and molecular levels. At last, we explored the possible molecular mechanisms adjusted by circZNF609 and miR-186-5p. This study may help to comprehend the part of circZNF609 and furnish fresh thought to the diagnosis of PC.
Materials and methods
Clinical specimens
Human PC tissues and adjacent tissues (n = 25) were acquired from The First Affiliated Hospital of Harbin Medical University (Harbin, China). The patients did not acquire any therapies before surgery. Every patient agreed with joining the research and writing informed consent and this research was ratified via the Medical Ethics Committee of The First Affiliated Hospital of Harbin Medical University.
Cell culture
Human PC cell (PC 3) and lymph node carcinoma of the prostate (LNCaP) were furnished from Riken Cell Bank (Tsukuba, Japan). Cells were cultured at Roswell Park Memorial Institute-1640 medium (RPMI-1640, Gibco, Grand Island, NY, US). The medium contained 10% fetal bovine serum (FBS, Gibco) 100 U/mL penicillin (Beyotime, Haimen, China) and 100 μg/mL streptomycin (Beyotime, Haimen, China). The culture medium was replaced every 2 days.
CCK-8 assay
Cells were inoculated in 96-well plates (5 × 103 cells/well). After the dosing treatment, CCK-8 solution (Bioswamp, Wuhan, China) was appended on cells in light of instructions, the plates were incubated 1 h (dark, 5% CO2, 37 °C). The absorbance was quantitated at 450 nm by a microplate reader (Bio-Rad, Sunnyvale, CA, US).
Flow cytometry
PI and FITC-conjugated annexin V staining (Bioswamp, Wuhan, China) were used for flow cytometry, 1 × phosphate-buffered saline (PBS) washed and 70% ethanol fixed cells. Then, washed clean and PI/FITC-Annexin V stained in 50 μg/ml RNase A (Sigma, St. Louis, MO, US). After that, we incubated the cells 1 h (dark, 25 °C). Results were performed through FACS can (Beckman Coulter, Fullerton, CA, USA). Data were analyzed by FlowJo software (Tree Star Software, San Carlos, CA, USA).
Colony formation assay
Cells were inoculated in 6-well plates 24 h (5 × 102 cells/well). PBS washed and cells were cultured at complete medium for 14 days, fixed and stained the colonies. Each treatment run in triplicate and microscope used (Olympus, Tokyo, Japan) to count colonies.
Migration and invasion assay
A 24-well millicell hanging cell culture chamber with an 8 µm PET membrane (Millipore, Bedford, MA, USA) were used to investigate cell migration and invasion. In this step, the only difference was a layer of matrigel that was placed on the upper side to mimic the extracellular matrix in vivo in invasion experiment. The cells secreted matrix metalloproteinase to degrade the matrigel, which can enter the nether chamber. PC cells (5.0 × 104 cells/well) transfected with different vectors were seeded in 200 μl transwell serum-free medium while 600 μl complete medium was appended in the nether chamber for migration experiments. The intruding chamber was treated for 48 h (37 °C, 5% CO2) to specification. Non-invasive cells were taken out, methanol fixed, crystal violet stained invading and microscope (Olympus, Japan) counted the cells. Each group was randomly selected 5 cells of fixed cells for counting the average.
Reverse transcription quantitative PCR (RT-qPCR)
Trizol was used to extract RNA (Molecular Research Center, Cincinnati, OH, USA). The Taqman MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific, Runcorn, UK) and Taqman Universal Master Mix II within TaqMan MicroRNA Assay of miR-186-5p and U6 (Thermo Fisher Scientific, Runcorn, UK) were used to detect miR-186-5p expression. SYBR® PrimeScript®PLUS RT-RNA PCR Kit (TaKaRa,Beijing, China) was used to test the circZNF609 level, β-actin was an internal parameter. Samples were run in triplicate. The 2−ΔΔCt equation was used to quantify the data.
Transfection
circRNA sequence was attached to PLCDH-circle vector (Ribobio, Guangzhou, China) to manufacture circZNF609. The negative control (NC) inhibitor and miR-186-5p inhibitor were compounded via GenePharma Co. (GenePharma, Shanghai, China). The transfection of circZNF609 was investigated by Lipofectamine TM2000 (Life Technologies, Carlsbad, CA, US) and the transfection of miR-186-5p was investigated by Lipofectamine 3000 (Life Technologies). G418 medium (Solarbio, Shanghai, China) was used to select stably transfected cells. After about 4 weeks, successfully transfected cells were found.
Western bolt
Proteins were extracted from cells by RIPA lysis buffer (Beyotime) plus protease inhibitor (Beyotime). We used protein assay kit (Bioswamp) quantified the proteins. This experiment was established by Bio-Rad. Primary antibodies were prepared in 5% blocking buffer (1:1000). The primary antibodies were cultured at 4 °C overnight and secondary antibodies of goat, anti-rabbit IgG (ab6721, Abcam, Cambridge, UK, 1:5000) cultured 1 h at 25 °C. After rinsing, the polyvinylidene fluoride (PVDF) membrane of the antibody was shifted to the system. Finally, 200 μL horseradish peroxidase (HRP, Millipore Corp., Billerica, MA, US) was appended on the surface. Captured signal was quantified by Image Lab™ Software (Bio-Rad). Above experiments used β-actin as internal parameters.
Primary antibodies (Abcam) contained anti-Cleaved-Caspase-3 antibody (ab2302), anti-Cleaved-Caspase-9 antibody (ab2324), anti-matrix metalloproteinase-9 (MMP-9) antibody (ab38898), anti-Vimentin antibody (ab137321), anti-β-actin antibody (ab8227), anti-YAP1 antibody (ab217693), anti-AMPK antibody (ab218136) and anti-p-AMPK antibody (ab92701).
Statistical analysis
All experiments were repeated three times. Data were expressed as mean ± standard deviation (SD). Statistical analyses were performed by using Graphpad 6.0 (Graph Pad Software, CA, US). The p values were calculated by one-way analysis of variance (ANOVA). p < .05 was considered statistically significant.
Results
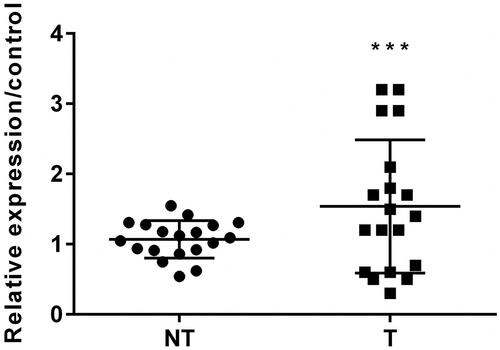
circZNF609 was highly expressed in PC tissues
circZNF609 expression was examined by RT-qPCR in PC patient tissues and corresponding normal lung tissues. The results demonstrated that the level of circZNF609 was conspicuously highly expressed in PC tissues (, p < .001).
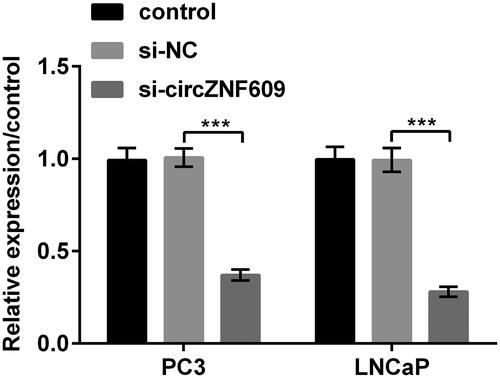
circZNF609 was silenced in PC3 and LNCaP cells
result pointed out that the levels of circZNF609 were conspicuously declined in PC3 and LNCaP cells when circZNF609 was silenced (p < .001). This manifested that the transfection efficiency was high.
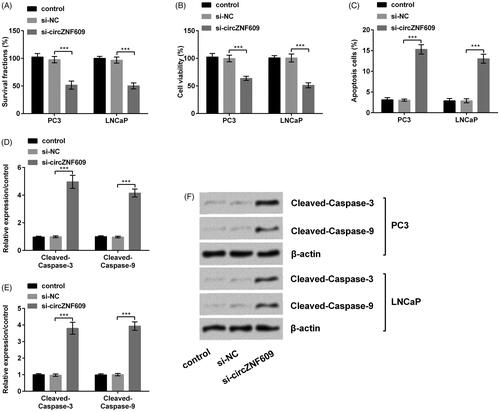
si-circZNF609 restrained cell viability and colony formation and caused apoptosis
had demonstrated that we successfully transfect circZNF609 into cells. On this basis, we investigated the functions of circZNF609 in PC3 and LNCaP cells at the cellular and molecular levels. si-circZNF609 meaningfully restrained cell colony formation and viability and enhanced apoptosis (, p < .001). At the molecular level, when circZNF609 was silenced, Western blot results demonstrated the expressions of Cleaved-Caspase-3 and Cleaved-Caspase-9 were conspicuously heightened (), p < .001). In conclusion, silencing circZNF609 restrained cell viability and colony formation and caused apoptosis in PC.
Figure 3. Silencing circular RNA circZNF609 (circZNF609) restrained cell viability and colony formation and caused apoptosis. (A) Silencing circZNF609 meaningfully restrained PC3 and LNCaPcell colony ability. (B) Silencing circZNF609 meaningfully restrained PC3 and LNCaPcell viability. (C) Silencing circZNF609 meaningfully enhanced PC3 and LNCaPcell apoptosis. (D–F) Silencing circZNF609 meaningfully heightened the expressions of Cleaved-Caspase-3 and Cleaved-Caspase-9. (***p < .001).
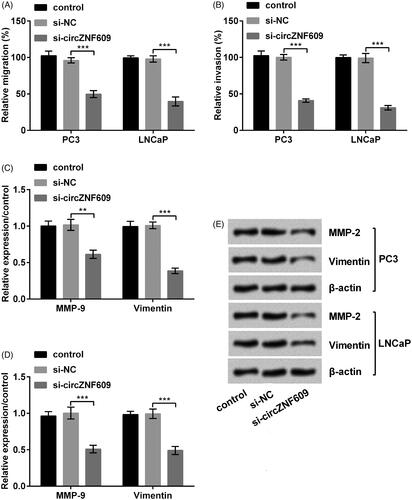
Silencing circZNF609 restrained cell migration and invasion
si-circZNF609 restrained PC3 cell migration and invasion (, p < .001). Moreover, the expressions of MMP-9 and Vimentin was meaningfully declined after silencing circZNF609 (), p < .01 or p < .001). A similar result was demonstrated at LNCaP cells. The result manifested that silencing circZNF609 restrained cell migration and invasion in PC.
Figure 4. Silencing circular RNA circZNF609 (circZNF609) restrained cell migration and invasion. (A) Silencing circZNF609 restrained cell migration in PC3 and LNCaP cells. (B) Silencing circZNF609 restrained cell invasion in PC3 and LNCaP cells. (C–E) Silencing circZNF609 restrained the levels of matrix metalloproteinase-9 (MMP-9) and Vimentin in PC3 and LNCaP cells. (**p < .01; ***p < .001).
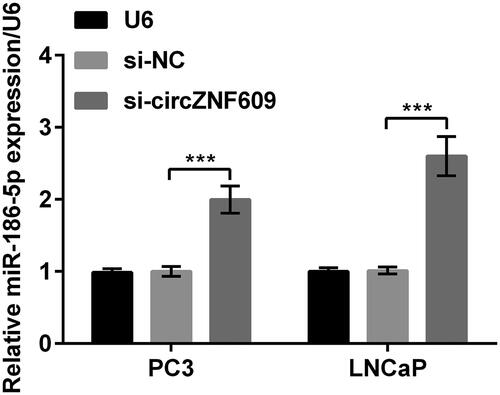
Silencing circZNF609 up-regulated miR-186-5p
After silencing circZNF609 in PC3 and LNCaP cells, the level of miR-186-5p was meaningfully enhanced (, p < .001). The results manifested that circZNF609 negatively regulated miR-186-5p.
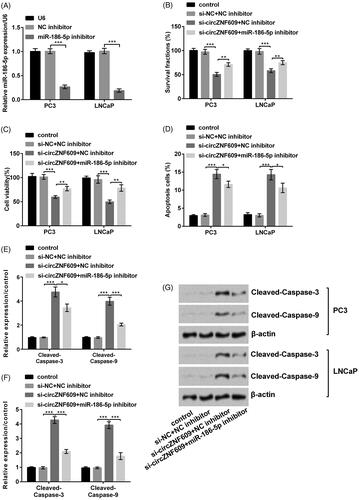
Silencing circZNF609 restrained cell growth by up-regulating miR-186-5p
We firstly transferred NC inhibitor and miR-186-5p inhibitor at PC3 and LNCaP cells. The result demonstrated that miR-186-5p inhibitor statistically declined miR-186-5p expression, this manifested that we successfully transfected miR-186-5p inhibitor into the cell (, p < .001). Further results put forward that miR-186-5p inhibitor reversed the circZNF609-induced decline in cell colony and viability (, p < .01) and rise in cell apoptosis (, p < .05) in PC3 and LNCaP cells. On the other side, Western blot results () demonstrated that the expressions of Cleaved-Caspase-3 (p < .05) and Cleaved-Caspase-9 (p < .001) were conspicuously declined when transfected with miR-186-5p inhibitor, compared to silence circZNF609 alone in PC3 cells. The similar result of Western blot was demonstrated in LNCaP cells. In short, silencing circZNF609 restrained cell growth by up-regulating miR-186-5p in GC.
Figure 6. Silencing circular RNA circZNF609 (circZNF609) restrained cell viability by up-regulating microRNA (miR)-186-5p. (A) miR-186-5p inhibitor conspicuously declined the level of miR-186-5p. (B) miR-186-5p inhibitor conspicuously enhanced the circZNF609-induced decline in PC3 and LNCaP cell colony. (C) miR-186-5p inhibitor conspicuously enhanced the circZNF609-induced decline in PC3 and LNCaP cell viability. (D) miR-186-5p inhibitor conspicuously declined the circZNF609-induced rise in PC3 and LNCaP cell apoptosis. (E–G) Silencing circZNF609 meaningfully declined expressions of Cleaved-Caspase-3 and Cleaved-Caspase-9, miR-186-5p inhibitor conspicuously reversed this expression. (*p < .05; **p < .01; ***p < .001).
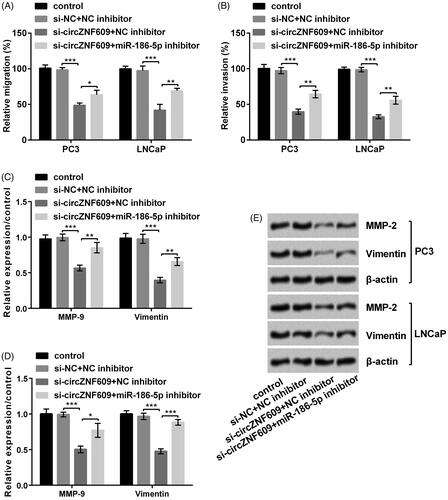
Silencing circZNF609 restrained cell migration and invasion by up-regulating miR-186-5p
results pointed out that miR-186-5p inhibitor reversed the circZNF609-induced decline in cell migration (, p < .05 or p < .01) and invasion (, p < 0.01) at PC3 and LNCaP cells. Besides, Western blot results () demonstrated the circZNF609-induced levels of MMP-9 (p < .05 or p < .01) and Vimentin (p < .01 or p < .001) were meaningfully reversed after silencing circZNF609. In short, Silencing circZNF609 restrained cell migration and invasion by up-regulating miR-186-5p in GC.
Figure 7. Silencing circular RNA circZNF609 (circZNF609) restrained cell migration and invasion through up-regulating microRNA (miR)-186-5p. (A) miR-186-5p inhibitor conspicuously enhanced circZNF609-induced decline in PC3 and LNCaP cell migration. (B) miR-186-5p inhibitor conspicuously enhanced the circZNF609-induced decline in PC3 and LNCaP invasion. (C–E) Silencing circZNF609 meaningfully declined levels of matrix metalloproteinase-9 (MMP-9) and Vimentin, and miR-186-5p inhibitor conspicuously reversed this expression. (*p < .05; **p < .01; ***p < .001).
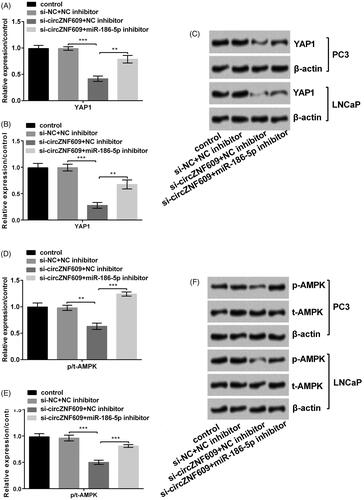
Silencing circZNF609 restrained YAP1 and AMPK signaling pathways by up-regulating miR-186-5p
We investigated the circZNF609 expression in the signaling pathways. From the results of , si-circZNF609 declined the level of YAP1 (p < .001) and miR-186-5p inhibitor (p < 0.01) enhanced the circZNF609-induced change in PC3 and LNCaP cells. On the AMPK pathway, si-circZNF609 and miR-186-5p inhibitor played the same part in PC3 and LNCaP cells (), p < .01 or p < .001). The above results manifested that si-circZNF609 restrained YAP1 and AMPK signaling pathways by up-regulating miR-186-5p.
Figure 8. Silencing circular RNA circZNF609 (circZNF609) restrained YAP1 and AMPK signaling pathways through up-regulating miR-186-5p. (A) Silencing circZNF609 restrained YAP1 pathway through up-regulating miR-186-5p in PC3 cells. (B) Silencing circZNF609 restrained YAP1 pathway through up-regulating miR-186-5p in LNCaP cells. (C) Silencing circZNF609 restrained AMPK pathway through up-regulating miR-186-5p in PC3 cells. (D) Silencing circZNF609 restrained AMPK pathway through up-regulating miR-186-5p in LNCaP cells. (**p < .01; ***p < .001).
Discussion
PC is a tumour that fearfully menaces men's health [Citation18]. In our research, firstly, we presented that circZNF609 was highly expressed in PC tissues. In addition, firstly, we discovered that silencing circZNF609 restrained cell colony, viability, migration and invasion and caused apoptosis in PC3 and LNCaP cells. Besides, the results demonstrated that circZNF609 negatively regulated miR-186-5p and miR-186-5p inhibitor reversed the impacts of circZNF609 on cell colony, viability, migration, invasion and apoptosis. Finally, we discovered that circZNF609 exerted its efficacy by restraining the YAP1and AMPK signaling pathways
circZNF609 is first spotted in 2017. In particular, the study noted that circZNF609 can translate proteins [Citation9]. Moreover, more and more studies pointed out that circZNF609 played a promoting tumour efficacy in cancer. For instance, there was a study that pointed out that circZNF609 enhanced the progress of nasopharyngeal tumour [Citation10]. Besides, other study pointed out that circZNF609 enhanced breast tumour [Citation19]. Surprisingly, we also discovered the tumour-promoting function of circZNF609 in the paper, silencing circZNF609 conspicuous restrained cell colony, viability, migration and invasion and caused apoptosis in PC3 and LNCaP cells.
Increasing studies demonstrated that circRNAs can sponge miRNA exerting efficacy in disparate species and play a very vital part in disease regulation [Citation20]. On the other side, Dai et al. had verified that circRNA Myosin Light Chain Kinase (MYLK) enhanced PC progress by miR-29a [Citation21]. Xiong et al. had verified that CircZNF609 exerted its functions by miR-138-5p in renal tumour [Citation22]. Therefore, we hypothesized that circZNF609 may also be achieved by regulating miRNAs in PC. In this paper, we were pleasantly surprised to find that circZNF609 negatively regulated miR-186-5p, miR-186-5p inhibitor reversed the impacts of circZNF609 in cell colony, viability, migration, invasion and apoptosis.
miR-186-5p was cut from miR-186. Many researchers had put forward that miR-186 conspicuously restrained the occurrence of tumour. For instance, Tian et al. had verified that miR-186 restrained squamous cell tumour [Citation23] and Hua et al. had verified that miR-186 restrained PC cell viability [Citation24]. In addition, Jones et al. had verified that miR-186-5p restrained PC [Citation17]. In this paper, miR-186-5p also presented the protective efficacy in PC. This was surprisingly consistent with the results of the above studies.
The YAP1 and AMPK signaling pathways play momentous parts in tumour [Citation25,Citation26]. Previous studies had reported that YAP1 and AMPK pathways are activated in PC and the two pathways are involved in the pathogenesis of PC [Citation27,Citation28]. Research demonstrated circ0023404 exerted its efficacy via YAP1 signaling pathways [Citation29] and other research demonstrated that circ0008274 exerted its efficacy by AMPK signaling pathway [Citation30]. On the other hand, miR-590 involved in the tumour through YAP1 and mi-451 was involved in tumour through AMPK signaling pathways [Citation31]. Fortunately, we also discovered that the ratio of p/t-AMPK and the level of YAP1 were declined when silenced sh-circZNF609 and these level were reversed when transfected with miR-186-5p inhibitor in this system. In short, silencing of circZNF609 restrained YAP1 and AMPK-catenin signaling pathways through up-regulating miR-186-5p.
In summary, silencing of circZNF609 restrained PC cells growth, metastasis and YAP1 and AMPK pathways by up-regulating miR-186-5p. This article might afford a new thought for the clinical treatment of PC.
Ethical approval
All procedures performed in the studies involving human participants were in accordance with the ethical standards of the institutional committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Every patient agreed with joining the research and writing informed consent and this research was ratified via the Medical Ethics Committee of The First Affiliated Hospital of Harbin Medical University.
Informed consent was obtained from all individual participants included in the study.
Author contributions
Conceived and designed the experiments: Chengjun Jin, Wanpeng Liu
Performed the experiments and analyzed the data: Chengjun Jin, Weiming Zhao, Zijian Zhang
Wrote the manuscript: Chengjun Jin, Wanpeng Liu
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Disclosure statement
All authors approved this manuscript for publication.
Additional information
Funding
References
- Noguchi, M, Koga, N, Igawa, T, et al. Clinical development of immunotherapy for prostate cancer. Int J Urol. 2017;24:675–680.
- Ren SC, Chen R, Sun YH. Prostate cancer research in China. Asian J Androl. 2013;15:350–353.
- Grozescu T, Popa F. Prostate cancer between prognosis and adequate/proper therapy. J Med. Life. 2017;10:5–12.
- Castillejos-Molina RA, Gabilondo-Navarro FB. Prostate cancer. Salud Publica Mex. 2016;58:279–284.
- Kwiatkowski MK, Recker F, Piironen T, et al. In prostatism patients the ratio of human glandular kallikrein to free PSA improves the discrimination between prostate cancer and benign hyperplasia within the diagnostic “gray zone” of total PSA 4 to 10 ng/mL. Urology. 1998;52:360–365.
- Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388.
- Hamam R, Hamam D, Alsaleh KA, et al. Circulating microRNAs in breast cancer: novel diagnostic and prognostic biomarkers. Cell death Dis. 2017;8:e3045.
- Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94.
- Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol cell. 2017;6;66:22–37.e9.
- Zhu L, Liu Y, Yang Y, et al. CircRNA ZNF609 promotes growth and metastasis of nasopharyngeal carcinoma by competing with microRNA-150-5p. Eur Rev Med Pharmacol Sci. 2019;23:2817–2826.
- Cai Y, Yu X, Hu S, et al. A brief review on the mechanisms of miRNA regulation. GPB. 2009;7:147–154.
- Calin GA, Liu CG, Sevignani C, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101:11755–11760.
- Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol Mech Dis. 2014;9:287–314.
- Acunzo M, Romano G, Wernicke D, et al. MicroRNA and cancer–a brief overview. Adv Biol Regul. 2015;57:1–9.
- Li J, Xia L, Zhou Z, et al. MiR-186-5p upregulation inhibits proliferation, metastasis and epithelial-to-mesenchymal transition of colorectal cancer cell by targeting ZEB1. Arch Biochem Biophys. 2018;640:53–60.
- Jiao D, Wu M, Ji L, et al. MicroRNA-186 suppresses cell proliferation and metastasis through targeting sentrin-specific protease 1 in renal cell carcinoma. Oncol Res. 2018;26:249–259.
- Jones DZ, Schmidt ML, Suman S, et al. Micro-RNA-186-5p inhibition attenuates proliferation, anchorage independent growth and invasion in metastatic prostate cancer cells. BMC Cancer. 2018;18:421.
- Bezrukov EA, Rapoport LM, Shpot EV, et al. Prostate cancer of high oncological risk. Current trends in diagnosis and surgical treatment. Urologiia (Moscow, Russia: 1999). 2017;4:129–134.
- Wang S, Xue X, Wang R, et al. CircZNF609 promotes breast cancer cell growth, migration, and invasion by elevating p70S6K1 via sponging miR-145-5p. CMAR.. 2018;10:3881–3890.
- Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8:73271–73281.
- Dai Y, Li D, Chen X, et al. Circular RNA myosin light chain kinase (MYLK) promotes prostate cancer progression through modulating Mir-29a expression. Med Sci Monit. 2018;24:3462–3471.
- Xiong Y, Zhang J, Song C. CircRNA ZNF609 functions as a competitive endogenous RNA to regulate FOXP4 expression by sponging miR-138-5p in renal carcinoma. J Cell Physiol. 2019;234:10646–10654.
- Tian J, Shen R, Yan Y, et al. miR-186 promotes tumor growth in cutaneous squamous cell carcinoma by inhibiting apoptotic protease activating factor-1. Exp Ther Med. 2018;16:4010–4018.
- Hua X, Xiao Y, Pan W, et al. miR-186 inhibits cell proliferation of prostate cancer by targeting GOLPH3. Am J Cancer Res. 2016;6:1650–1660.
- Zhu P, Wang Y, Wu J, et al. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nat Commun. 2016;7:13608.
- Sugiyama M, Takahashi H, Hosono K, et al. Adiponectin inhibits colorectal cancer cell growth through the AMPK/mTOR pathway. Int J Oncol. 2009;34:339–344.
- Sheng X, Li WB, Wang DL, et al. YAP is closely correlated with castration-resistant prostate cancer, and downregulation of YAP reduces proliferation and induces apoptosis of PC-3 cells. Mol Med Rep. 2015;12:4867–4876.
- Chen RJ, Hung CM, Chen YL, et al. Monascuspiloin induces apoptosis and autophagic cell death in human prostate cancer cells via the Akt and AMPK signaling pathways. J Agric Food Chem. 2012;60:7185–7193.
- Zhang J, Zhao X, Zhang J, et al. Circular RNA hsa_circ_0023404 exerts an oncogenic role in cervical cancer through regulating miR-136/TFCP2/YAP pathway. Biochem Biophys Res Commun. 2018;501:428–433.
- Zhou GK, Zhang GY, Yuan ZN, et al. Has_circ_0008274 promotes cell proliferation and invasion involving AMPK/mTOR signaling pathway in papillary thyroid carcinoma. Eur Rev Med Pharm Sci. 2018;22:8772–8780.
- Chen MB, Wei MX, Han JY, et al. MicroRNA-451 regulates AMPK/mTORC1 signaling and fascin1 expression in HT-29 colorectal cancer. Cell Signal. 2014;26:102–109.