Abstract
Diabetes mellitus (DM) is one of the severe metabolic diseases found in all types of people’s lives in lower, middle and high income countries. It is suggested that the prevalence of diabetes is increasing in many countries and most of the cases are type 2 DM, clinical treatments are changing now to manage type 2 DM, however, up-to-date, there is no diagnostic, prognostic, and therapeutic target for type 2 DM. MicroRNAs (miRNAs) is one of a non-protein coding RNAs that have been regulating a wide range cellular processes and induce the development of many diseases. Most of the researchers concluded that miRNAs involvement is an important process in a broad range of signaling pathways such as cell proliferation, stem cell maintenance, migration, apoptosis and gene or protein expressions. Expressed sequence tags (ESTs) are the best source of coding and non-coding sequences for the identification of miRNAs. Although DNA methylation is an important mechanism for miRNAs up-regulation, this has not been highly explored in type 2 DM. The present study is useful to elucidate the molecular mechanism of MiR-1285-5p in type 2 DM and its role in disease progression and we discovered miR-1285 as a novel prognostic, diagnostic and therapeutic target for type 2 DM.
Introduction
Diabetes mellitus (DM) is one of the severe metabolic diseases found in all types of people’s lives in lower, middle and high income countries [Citation1]. It has been suggested that DM is a multi-factor disease which arise mainly because of imbalance in glucose concentration in the blood. Unfortunately, both in type 1 DM and type 2 DM, the beta cells of the pancreatic organ cannot secrete the proper amount of insulin to control blood glucose level in the blood cells [Citation2]. Many reports suggested that there will be an increase of 300 million DM patient by 2025 and 366 million by 2030 and interestingly, the majority of these patients are from developing countries [Citation3–5]. Specifically, Type 2 DM becomes more in the older and urban population [Citation6]. It has been suggested that the prevalence of diabetes is increasing in many countries and most of the cases are type 2 DM, clinical treatments are changing now to manage type 2 DM, however, till date, there is no prognostic, diagnostic and therapeutic target for type 2 DM.
MicroRNAs (miRNAs) are one of the non-protein coding RNAs that have been modulating a broad range of cellular processes and induce the development of many diseases [Citation7–9]. Single-stranded mature miRNAs after processing in cytoplasm induces negative regulation of gene expression in the cells. Most of the researchers concluded that miRNAs involvement is an important process in a broad range of signaling pathways such as cell proliferation, stem cell maintenance, migration, apoptosis and gene or protein expressions [Citation10,Citation11]. Expressed sequence Tags (ESTs) are the best source of coding and non-coding sequences for the identification of miRNAs [Citation2]. On the basis of the above information, the present study focused on the identification of novel miRNAs from DM ESTs using computational approaches and their involvement in type 2 DM. Our results suggested that using type 2 DM ESTs, we identified hsa-Mir-1285-5p by computational approach, and it is involved in diminishing the expression of sterol carrier protein 2 (SCP2) through hypermethylation of the promoter region. The present study is useful to elucidate the molecular mechanism of MiR-1285-5p in type 2 DM and its role in disease progression. We discovered that MiR-1285-5p as a novel prognostic, diagnostic and therapeutic target for type 2 DM and results are discussed in detail.
Materials and methods
Creating local nucleotide database using ESTs and miRNAs
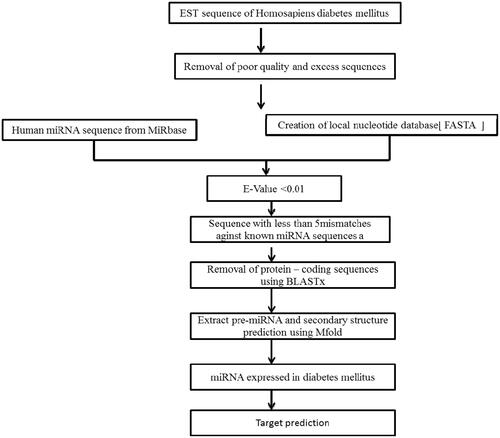
Bioinformatic tools were used in search of miRNA responsible for type 2 DM from the National Center for Biotechnology Information (NCBI) ESTs sequences (6533) (November: 10, 2018) are retrieved by using the search word “type 2 diabetes mellitus homo sapiens”. miRbase (http://www.mirbase.org/) online tool was used to get mature and pre-miRNAs. Human miRNAs were used as a reference sequence and the local nucleotide database were formed for type 2 DM EST sequence after the elimination of unwanted sequences. The nucleotide databases were searched for their cognitive miRNAs. shows the flow chart for the identification of miRNAs from a computational approach.
Extracting pre- miRNAs and their precursor sequences
Novel miRNA has been identified by using the procedure from Priyanka et al 2018 [Citation2]. BLAST, BLASTx and clustal W were used to align the sequences with reference sequences and then the aligned regions were extracted and considered to be a Pre-miRNA sequence.
Validation of pre-miRNAs and identification of target mRNA for mature miRNA
Mfold online tool (http://unafold.rna.albany.edu/) was used for finding pre-miRNAs secondary structure and the criteria for selecting candidate miRNA procedure were adopted from Priyanka et al. 2018 [Citation2]. The incorporated miRNA were evaluated by higher energy MFEs and A + U contents and Target scan software were used for target prediction.
Type 2 DM clinical samples and baseline clinical data
In this study, genetically related 40 type 2 DM were selected and divided into two groups. The Group I contain 20 healthy volunteers who are randomly involved in this study and Group II consisted of 20 type 2 DM. shows that the data from the type 2 DM patient including body mass index (BMI), family history, age, height and weight. With use of automatic biochemistry analyzer, the levels of fasting blood glucose (FBG), post prandial (PP), glycosylated hemoglobin (HbA1c), triglyceride (TG), total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C) and C-reactive protein (CRP) were detected. Totally, 7 ml of blood was collected from each subject and 2 ml of each sample were stored for genomic DNA isolation and the remaining sample was processed for the serum to determine the above parameters. All the parameters were calculated by the Friedewald formula. Each experimental protocol was approved by Institutional ethical commitee and for all human subjects, informed consent was obtained.
Table 1. Shows that the various target mRNA for MiR-1285.
RNA isolation and quantification of MiR-1285-5p and SCP2
The expression patterns of MiR-1285-5p were performed according to the procedure described by Krishnan et al. 2017. The RNA was isolated using an RNeasy Mini Kit (Qiagen, New Delhi, India). Quantification of miR-1285 was performed by PCR using the following primers: Forward: 5′-TTTTTTTTACGTTTTTTTTAAGGTC-3′; reverse: 5′-CTATTACTAAACCCAACTACACGC -3′. MiR-1285-5p levels were normalized to 18 s RNA levels using the 2ΔΔCt model. The quantification of the target gene SCP2 was performed by the same method by using the primers: Forward: 5′- TTTTTTATGTTTTTTTTAAGGTTGT -3′; Reverse: 5′- CCCTATTACTAAACCCAACTACACAC -3′.
Bisulfite conversion and methylation analysis
DNA was extracted from type 2 DM using the Qiagen mini kit and bisulfite-treated DNA was extracted by using EZ DNA Methylation-Gold Kit (Zymo Research, Chennai, India) following the manufacturer's instructions. Hypermethylation experiments were carried out by using the method described by Vogelsang et al. (2014). The primers used in these experiments are MSP-MiR-1285-5p-Fw (GGTATGATTAAGGTAAATCGAGTAGC) and MSP-MiR-1285-5p Rv(AACAATCGAAACTAAAAAACCGA) has been used to amplify CpG rich promoter region (450 bp) from bisulfate treated DNA templates followed by a second round of methylated and unmethylated specific primers amplification. MiRNA-1285-5p Fwd M: TTTGAGTTTTCGGTAGTTTTTGAAC, MirRNA-1285-5p ReverseM: TCCCTAATAATATCAACCTCCGAC, MirRNA-1285-5p FwdU: TGAGTTTTTGGTAGTTTTTGAATGT. MirRNA-1285-5p reverseU: ATCCCTAATAATATCAACCTCCAAC.
Statistical calculation
Statistical analysis of all data was made by SPSS software version 25.0 for Microsoft Windows. Two-tailed student’s t-test was performed and p < .05 was considered to be statistically significant. All data are expressed as the mean ± SD.
Results
Identification of miR-1285 and its targets through computational approach
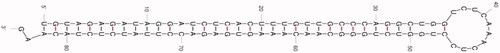
After analyzing all the ESTs from NCBI, MiR-1285 has been identified by using BLAST, BLASTx and secondary structure evaluation procedure. depicts the predicted secondary structure. After the careful analysis of miRNAs sequence, precursor sequence length, minimum folding free energies and A + U content for the predicted miRNA, we have identified miR-1285 and the details are provided in . Sterol Carrier Protein 2 (SCP 2) is the topmost cognate target for miR-1285 and fortunately, it is linked with diabetes and cholesterol metabolism [Citation12]. represents various target mRNA for MiR-1285-5p. From the above results, it has been understandable that MiR-1285-5p directly involved in the regulation of DM.
Table 2. Represents the information about Mir-1285 after computational prediction.
Up-regulation and hypermethylation status of MiR-1285-5p in type 2 DM
In our study, totally 40 subjects were selected and subdivided into 20 for type 2 DM patients and 20 for healthy controls. Interestingly, baseline clinical data shows that () there is no significant difference between type 2 DM and control samples with respect to BMI, total cholesterol, HDL, LDL and triglycerides (p > .05). HbA1C, CRP and blood glucose levels showed high differences between patients and controls (p < .05). represents the baseline clinical data of the study populations.
Table 3. Represents the baseline clinical parameters for the study population.
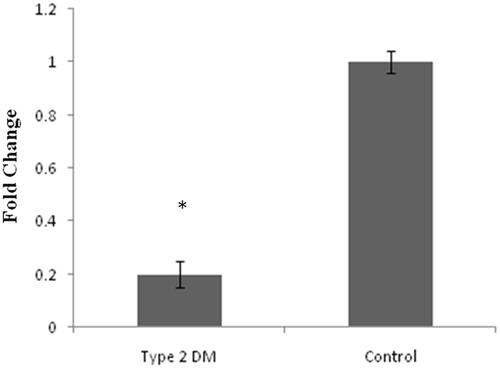
According to our previous publications [Citation2,Citation9], hypermethylation is one of the key molecular mechanisms for the induction of DM and it has been documented that miRNAs follow the similar molecular machinery like protein-coding genes including epigenetic events. Many miRNAs involved in the progression of type 2 DM but the exact mechanisms behind its action are still questionable. So in this study, we intend to analyze and evaluate hypermethylation and the expression status of identified miR-1285 from available type 2 DM ESTs. In order to test this concept, we analyzed the mature MiR-1285-5p expression in type 2 DM and also in healthy samples. Interestingly, our result shows that the MiR-1285-5p in control sample was significantly lower than the type 2 DM patients (. Sterol Carrier Protein 2 (SCP2) is the topmost target for MiR-1285, according to the literature [Citation12], SCP directly involved in diabetes and cholesterol metabolism. We analyzed the expression status of SCP2 in all the above clinical samples and control samples. As we expected, SCP2 gene expression was found to be higher in control samples compared to type 2 DM (.
Figure 3. Represents expression level of circulating miR-1285 type 2 DM by quantitative real time PCR (q PCR). Unpaired t-test data p values <.005*vs. control samples.
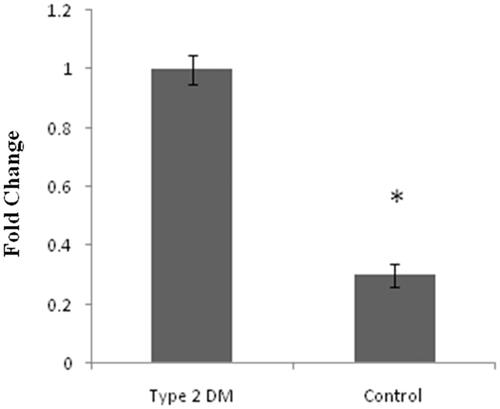
Figure 4. Represents expression level of SCP2 gene in type 2 DM by quantitative real time PCR (q PCR). Unpaired t-test data p values <.005*vs. control samples.
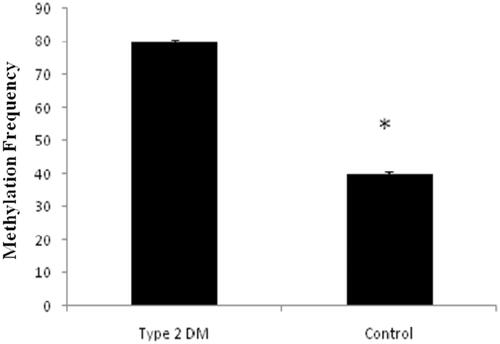
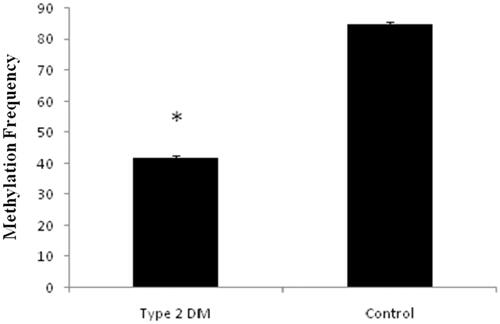
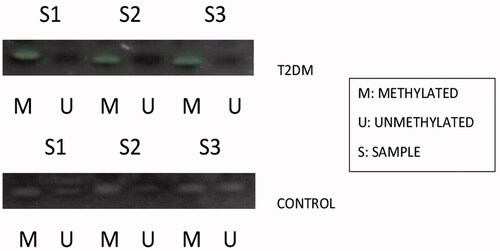
Based on our gene expression data, MiR-1285-5p levels were higher in type 2 DM and it may be relevant to the initiation and development of type 2 DM. In contrary, SCP2 gene expressions are lower in type 2 DM (. We performed the hypermethylation experiments on the same samples since methylation and expressions are inversely proportional as stated by many research articles [Citation13,Citation14]. As per the literature, the epigenetic regulation of miRNAs is a widespread phenomenon [Citation2,Citation13,Citation14] and we evaluated the hypermethylation status of MiR-1285-5p in type 2 DM and control samples by using methylation-specific PCR (MSP) or nested MSP. Our study indicated that the promoter region of MiR-1285-5p was unmethylated in type 2 DM samples, but in contrast, methylated control samples have been seen in the experiments. (. On the other hands, the SCP2 promoter region was methylated in type 2 DM and unmethylated on control samples (. Simultaneously, we analyzed the frequency of methylation in all samples ( and ) and we found that the methylation frequency was statistically significant between these samples (55.0 vs. 85.0%, and 57 vs. 80% p ≤ .05). Overall, our data suggested that the expression nature of MiR-1285-5p corroborate with methylation status. MiR-1285-5p was found to be a candidate miRNA for type 2 DM. In addition, it needs more investigation to prove the above mechanism.
Figure 5. Represents the methylation status of miR-1285 promoter region in type 2 DM samples and the control samples by methylation sensitive PCR (MSP).
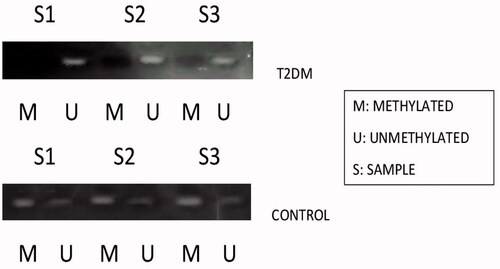
Figure 6. Represents the methylation status of SCP 2 promoter region in type 2 DM samples and the control samples by methylation sensitive PCR (MSP).
Discussion
It has been suggested that the mechanisms in which miRNA repress the targets are diverse, for example, the miRNAs bind to the complementary seed region and may also in the central region causing mRNA cleavage and leads to subsequent degradation of mRNAs. Many articles demonstrated that miRNAs reduce the expression of their cognate target genes by degrading mRNA, inducing decapping, promoting deadenylation, altered cap protein binding, and reduced ribosome occupancy. MiRNAs regulate wide a range of molecular mechanism such as transcriptional activation or inhibition, epigenetic repression, cell proliferation, differentiation and other cellular function. It is considered to be non-invasive biomarkers since it is found in many body fluids. Computational approach is the best method of identifying miRNA through conditional specific ESTs. It has been reported that miRNAs is a key regulator in type 2 DM and till date, there is a scarcity in developing prognostic, diagnostic or therapeutic molecules for type 2 DM.
This computational approach found that SCP2 is a direct target for MiR-1285-5p and interestingly, SCP2 involved in cholesterol and diabetic complications. MiR-1285-5p levels were higher in type 2 DM when compared to SCP2 levels and the promoter region of MiR-1285-5p was unmethylated in type 2 DM samples, thus our results indicated that MiR-1285-5p and SCP2 expression directly depends on their promoter methylation. In addition, methylation frequency was statistically significant between these samples, suggesting that expression analysis corroborates with promoter analysis. Surprisingly, only a few studies reported on the SCP2 expression status in diabetes and importantly, the association with MiR-1285-5p has not been explored, moreover, the molecular mechanism behind their role has not been elucidated. It was found that MiR-1285-5p may act as a candidate target for type 2 DM, but future functional studies with more clinical samples and the molecular action of antimiR-1285 are required to prove MiR-1285-5p used as a biomarker and a treatment option for type 2 DM.
Acknowledgements
All the authors thank Saveetha Dental College and Hospitals, Saveetha Institute of Medical and Technical sciences (SIMATS), Chennai, India for providing moral support for writing this manuscript.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Data availability
The data used to support the findings of this study are included within the article.
References
- Ramachandran A. Know the signs and symptoms of diabetes. Indian J Med Res. 2014;140:579–581.
- Priyanka P, M, Panagal P, Sivakumar V, et al. Identification, expression, and methylation of miR-7110 and its involvement in type 1 diabetes mellitus. Gene Reports. 2018;11:229–234.
- Animaw W, Seyoum Y. Increasing prevalence of diabetes mellitus in a developing country and its related factors. PLoS One. 2017;12:e0187670.
- Campbell R. Type 2 diabetes: where we are today: an overview of disease burden, current treatments, and treatment strategies. J Am Pharm Assoc. 2009;49: S5–S9.
- Levitt NS. Diabetes in Africa: epidemiology, management and healthcare challenges. Heart. 2008;94:1376–1382.
- Hall V, Thomsen R, Henriksen O, et al. Diabetes in Sub Saharan Africa 1999–2011: epidemiology and public health implications. A systematic review. BMC Public Health. 2011;11:564.
- Sekar D, Venugopal B, Sekar P, et al. Role of microRNA 21 in diabetes and associated/related diseases. Gene. 2016;582:14–18.
- Panagal M, Biruntha M, Vidhyavathi RM, et al. Dissecting the role of miR-21 in different types of stroke. Gene. 2019;681:69–72.
- Sekar D. Comment on the potential role of microRNAs in hypertension. J Hum Hypertens. 2018;32:639–640.
- Shincy JS, Panagal M, Jereena J, et al. Computational Identification of microRNA-17-3p in breast cancer cells. MIRNA. 2017;6:208–212.
- Sekar D, Krishnan R, Panagal M, et al. Deciphering the role of microRNA 21 in cancer stem cells (CSCs). Genes Dis. 2016;3:277–281.
- McLean MP, Billheimer JT, Warden KJ, et al. Differential expression of hepatic sterol carrier proteins in the streptozotocin-treated diabetic rat. Endocrinology. 1995;136:3360–3368.
- Lujambio A, Esteller M. How epigenetics can explain human metastasis: a new role for microRNAs. Cell Cycle. 2009;8:377–382.
- Krishnan R, Mani P, Sivakumar P, et al. Expression and methylation of circulating microRNA-510 in essential hypertension. Hypertens Res. 2017;40:361–363.