Abstract
Recent studies showed that long non-coding RNAs (lncRNAs) could play critical roles in tumors progression. However, the performance of LINC01354 is still limited in non-small cell lung cancer (NSCLC). In the current study, our results showed that LINC01354 was significantly increased in NSCLC tissues and cell lines. High LINC01354 expression was associated with advanced TNM stage and poor prognosis in NSCLC patients. Loss-of-function assays revealed that knockdown of LINC01354 reduced lung cancer cells proliferation and invasive ability in vitro. Subsequently, mechanism studies showed that LINC01354 positively regulated the ATF1 expression via competitive binding to miR-340-5p. Therefore, our results illustrated that LINC01354 might act as an oncogenic role by modulating the miR-340-5p/ATF1 axis, providing a novel therapeutic therapy for NSCLC treatment.
Keywords:
Introduction
Lung cancer is a malignant tumor with a high mortality rate, which is originated from the bronchial mucosa or glands [Citation1]. About 85% of lung cancers are categorized as non-small cell lung cancer (NSCLC) and the others are classified as small cell lung cancer [Citation2]. Despite the improvement of clinical treatments (including surgery and chemotherapy et al), the prognosis of NSCLC are still poor due to the metastasis and recurrence [Citation3,Citation4]. Therefore, it is necessary to determine the underlying mechanism and identify novel therapies to improve NSCLC patients’ prognosis.
Long non-coding RNAs (lncRNAs) which are longer than 200 nt, have little or no protein coding capacity [Citation5,Citation6]. Recently, increasing studies showed that aberrant expression of lncRNAs play critical roles in cancers, such as metastasis, proliferation, and apoptosis [Citation7]. For instance, Zhang et al found that in renal cancer, lncRNA MALAT1 was significantly increased and associated with prognosis [Citation8]. Liu et al showed that GAS5 was downregulated in bladder cancer and reduced the proliferation by regulating the expression of CDK6 [Citation9]. Hu et al suggested that in osteosarcoma, lncRNA NEAT1 inhibition restored the availability of miR-34c and improved the sensitivity to cisplatin [Citation10]. Recently, increasing studies showed that LINC01354 play critical roles in tumor progression. However, the roles and underlying mechanisms of LINC01354 in NSCLC remain unclear.
In our study, we showed that LINC01354 was increased in NSCLC tissues. High LINC01354 expression promoted lung cancer cells proliferation and invasive ability in vitro. Furthermore, we suggested that LINC01354 might exert the oncogenic roles partially by regulating the miR-340–5p/ATF1 axis.
Materials and methods
Patients and tissue specimens
A total of 43 paired NSCLC tissues were collected from Ninghai First Hospital and People's Hospital of Bozhou City. All the patients had not undergone any radiotherapy or chemotherapy before operation. Samples were dissected during the surgery and immediately transferred to liquid nitrogen for quick-freeze and then to −80 °C fridge for further study. The research has been approved by the Institutional Research Ethics Committee of our hospital.
Cell culture and transfection
Human lung cancer cell lines (A549, SPC-A1, NCI-H23, PC-9, H1299, C422L, H1975) and normal lung epithelial cell line EBAS-2B were purchased from ATCC (Manassas, VA, USA). All cells were cultured with RPMI-1640 medium containing 10% FBS (Gibco), and cell culture conditions were 37 °C and 5% CO2.
The synthetic miR-340-5p mimics, miR-340-5p inhibitor (anti-miR-340-5p), siRNA against LINC01354 (si-LINC01354), siRNA against ATF1 (si-ATF1) and negative controls were obtained from GenePharma (Shanghai, China). All transfections were performed by Lipofectamine 2000 reagents (Invitrogen, Carlsbad, CA, USA) following the protocols.
Quantitative real-time PCR (qRT-PCR)
TRIzol solution (Invitrogen) was purchased to isolate total RNA. In accordance with the manufacturer’s protocols, cDNA was synthesized using reverse transcription kit (Takara, Japan). Subsequently, real-time PCR was used with SYBR Premix Ex Taq II (Takara) on the 7900HT system. GAPDH functioned as the housekeeping gene to normalize the expression of LINC01354 and ATF1. U6 served as the internal control to normalize miR-340-5p expression [Citation11,Citation12]. The expression level was calculated by 2−ΔΔCt method.
CCK-8 and colony formation assays
Cell Counting Kit-8 (CCK-8, Solarbio, China) were used to determine cells proliferation ability. Cells were seeded in 96-well plates at a density of 5 × 103 per well and incubated for 24 h, 48 h, 72 h, and 96 h. Then 20 μL CCK-8 regents were added and incubated at 37 °C for additional 1 h. The absorbance was tested at 450 nm (BioTek, VT, USA).
Cells were seeded in 6-well plates (500/well) and cultured for 2 weeks to form cell colony. Then cells were fixed with 4% polyoxymethylene and stained with 0.1% crystal violet. Finally, colony numbers were counted.
Transwell assay
Cell invasiveness was determined by transwell invasion assay. The upper transwell inserts were coated with matrigel (BD, CA, USA) and inserted into 24-well plate. 1 × 104/cells were added to the upper chamber and 800 μl medium containing 10% FBS was added to the lower chamber. 48 h later, the transwell chambers were washed with PBS, fixed in 4% paraformaldehyde for 15 min and stained with 0.4% crystal violet for 5 min. The invaded cells were counted under a microscope (Olympus).
Western blot
The lung cancer cells were collected, washed and lysed in RIPA lysis buffer. After that, the equal amounts of protein were separated on a 10% SDS-PAGE and then transferred to a PVDF membrane (Millipore, Schwalbach, Germany). Then, the membrane was maintained with 5% non-fat milk for 1 h and blocked with primary antibodies (Abcam, Cambridge, MA, USA) at 4 °C overnight. Then the membranes were probed with HRP-conjugated secondary antibodies (Cell Signaling, Danvers, MA, USA) for another 1.5 h.
Dual-luciferase assay
PmirGLO plasmid was used to clone the LINC01354 Wt/Mut 3’-UTR and ATF1 Wt/Mut 3’-UTR sequences. After co-transfected with LINC01354 Wt/MUT or ATF1 Wt/MUT and miR-340-5p mimics or negative controls, cells were inoculated into 96-well plates (1 × 104/well) for 48 h. Then, luciferase activity was measured using a Dual-Luciferase Assay Kit (GeneCopoeia, Rockville, MD, USA).
RNA immunoprecipitation (RIP) assay
For the RIP assay for Ago2, RIP was performed using the Magna RIP™ RNA-binding protein immunoprecipitation kit (Millipore, Merck KGaA, Germany) according to the manufacturer’s guidelines and previous reports [Citation13].
Biotin pull-down assay
Pull-down assay was performed as described previously [Citation10]. And and the purified RNA was subjected to real-time PCR analysis.
Statistical analysis
Statistical analysis was conducted using SPSS 18.0 software. Experimental data were denoted as means ± SD. The differences among groups were tested using Student’s t-test or one-way ANOVA. Kaplan-Meier survival method with log-rank test was used to assess the prognosis of NSCLC patients. The correlation was explored by Spearman’s correlation analysis. p < .05 had statistically significance.
Results
LINC01354 was upregulated in NSCLC
To explore the roles of LINC01354 in lung cancer, we firstly determined LINC01354 expression in 43 paired NSCLC tissues. Results showed that LINC01354 levels were significantly increased in NSCLC tissues (. High LINC01354 expression was associated with advanced TNM stage of NSCLC patients (). LINC01354 expression in lung cancer cells was also upregulated compared to EBAS-2B cells (). Subsequently, we showed that LINC01354 expression was significantly reduced in lung tissues by NCBI and UCSC online database (). Kaplan-Meier analysis revealed that high LINC01354 expression was associated with poor overall survival (OS) and disease free survival (DFS) rate compared to low LINC01354 expression group in NSCLC patients ().
Figure 1. LINC01354 was upregulated in lung cancer. (A) qRT-PCR was used to determine analysis for LINC01354 expression in 43 paired lung cancer tissues. (B) High LINC01354 expression was correlated with advanced TNM stage of patients. (C) LINC01354 expression in lung cancer cells by qRT-PCR. (D, E) LINC01354 expression was significantly reduced in lung tissues by NCBI and UCSC online database. (F, G) High LINC01354 expression predicted a poor overall survival (F) and disease free survival (G). *p < .05.
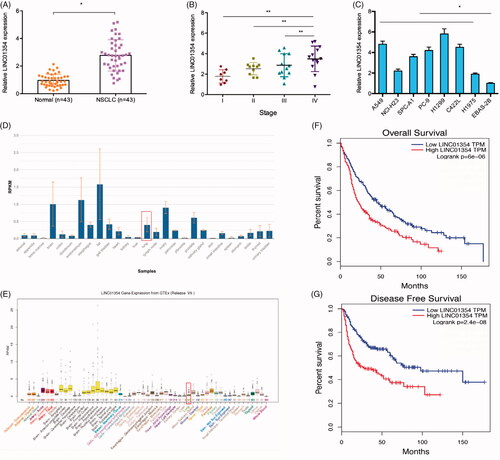
Knockdown of LINC01354 inhibited lung cancer cells proliferation and invasion
To study the specific roles of LINC01354 in lung cancer, si-LINC01354 was used to knockdown LINC01354 expression in lung cancer cells (). CCK-8 and colony formation assays revealed that LINC01354 suppression inhibited A549 and H1299 cells viabilities in vitro (). Transwell assays showed that the invasiveness of si-LINC01354 transfected A549 and H1299 cells was significantly inhibited compared to si-NC group (). These data suggested that LINC01354 might serve as an oncogenic role in lung cancer progression.
Figure 2. LINC01354 suppression inhibited the proliferation and invasion in lung cancer. (A, B) Relative expression of LINC01354 in lung cancer cells transfected with siRNAs or si-NC. (C-F) CCK8 and colony formation assays were conducted to determine the proliferation in lung cancer. (G, H) Transwell assay was utilized to analyze the invasion in lung cancer. *p < .05.
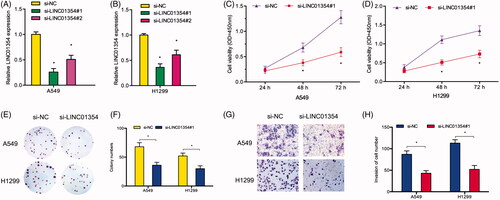
LINC01354 directly interacted with miR-340-5p
To determine the molecular mechanism of LINC01354, we firstly explored the localization of LINC01354. LncLocator prediction results showed that LINC01354 expression was mainly located in the cytoplasm (). And the results were further confirmed by qRT-PCR (). Subsequently, bioinformatics prediction analysis showed that there is a complementary sequences between LINC01354 and miR-340-5p (). QRT-PCR results showed that LINC01354 inhibition significantly increased miR-340-5p expression in A549 and H1299 cells (). Correlation analysis showed that LINC01354 expression was negatively correlated with miR-340-5p expression in NSCLC tissues (). Luciferase assay reported that miR-340-5p overexpression reduced the activity of LINC01354-Wt group (). RNA pull-down analysis showed a significant amount of LINC01354 and miR-340-5p in the LINC01354 pulled down pellet compared with control group (). Moreover, RIP assay further verified the binding sites between miR-340-5p and LINC01354 (). These data suggested that LINC01354 might directly interact with miR-340-5p in lung cancer.
Figure 3. LINC01354 served as a sponge for miR-340-5p. (A, B) LINC01354 location was determined by LncLocator and qRT-PCR. (C-E) Schematic view of miR-340-5p putative binding site in the LINC01354. (F) LINC01354 inhibition reduced miR-340-5p expression in lung cancer cells. (G) LINC01354 expression was negatively correlated with miR-340-5p expression in NSCLC tissues. (H) Dual luciferase reporter assays showed that miR-340-5 bind to LINC01354 and inhibits luciferase activity. (I) RNA pull-down analysis showed a significant amount of LINC01354 and miR-340-5p in the LINC01354 pulled down pellet compared with control group. (J) RIP assay verified the binding sites between miR-340-5p and LINC01354. *p < .05.
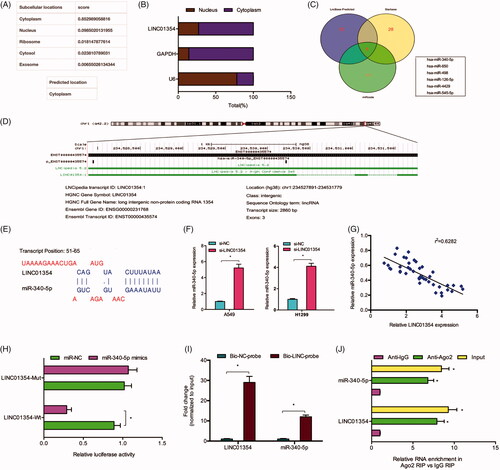
miR-340-5p targeted ATF1
QRT-PCR reported that the expression of miR-340-5p was downregulated in NSCLC (). Kaplan-Meier analysis revealed that low miR-340-5p expression was associated with poor OS rate in NSCLC patients (). Subsequently, TargetScan online website was used to explore the potential targets of miR-340-5p, and ATF1 was selected with its high score (). The predication was further confirmed by luciferase assay (). In addition, western blot assay showed that miR-340-5p mimics significantly inhibited the protein levels of ATF1 in A549 and H1299 cells ().
Figure 4. ATF1 acted as a target of miR-340-5p. (A) miR-340-5p expression in 43 paired lung cancer tissues was determined by qRT-PCR. (B, C) Low miR-340-5p expression predicted a poor overall survival rate of patients. (D. E) The predicted complementary binding sites between miR-340-5p and ATF1. (F) miR-340-5p mimics decreased the luciferase activity of ATF1-Wt group by dual-luciferase assay. (G) miR-340-5p mimics reduced the ATF1 protein expression in lung cancer cells. *p < .05.
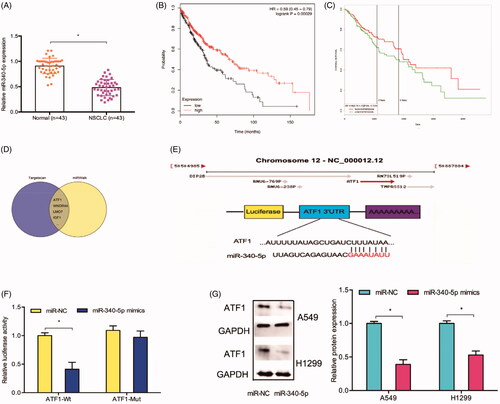
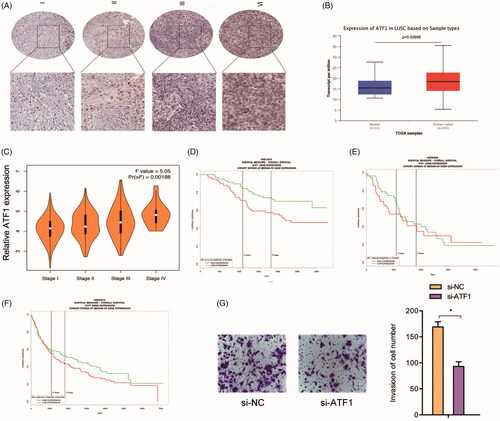
Next, we explored ATF1 expression in NSCLC. IHC data indicated that the expression of ATF1 was upregulated (). TCGA database further confirmed that ATF1 was upregulated and associated with advanced tumor stage in NSCLC patients (). Kaplan-Meier analysis revealed that patients with high ATF1 expression had a poor OS rate (). In addition, we explored the effects of ATF1 on lung cancer cells invasion ability, transwell assays showed that ATF1 inhibition reduced A549 cells invasion ability in vitro ()
Figure 5. The ATF1 roles in NSCLC. (A) ATF1 expression was upregulated and associated advanced TNM stage in patients with lung cancer. (B) TCGA database showed that ATF1 expression was increased in lung cancer tissues. (C) High ATF1 expression was associated with advanced tumor stage in NSCLC patients. (D-F) High ATF1 expression predicted a poor overall survival rate in NSCLC patients. (G) ATF1 inhibition reduced A549 cells invasion ability in vitro. *p < .05.
LINC01354 promotes NSCLC progression via miR-340-5p/ATF1 axis
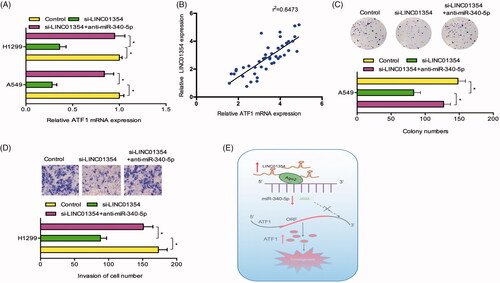
Furthermore, we explore whether LINC01354 regulate ATF1 expression by sponging miR-340-5p. QRT-PCR results revealed that LINC01354 inhibition reduced the expression of ATF1 in lung cancer cells, while miR-340-5p inhibitors rescued the effects (). Correlation analysis revealed that LINC01354 expression was positively correlated with ATF1 expression in NSCLC tissues (). Furthermore, rescue assays were used. Colony formation assay showed that LINC01354 inhibition reduced A549 cells colony numbers, whereas miR-340-5p inhibitors abolished the effects (). Transwell assay showed that miR-340-5p inhibitors abolished the functions of LINC01354 inhibition on H1299 cells invasion ability in vitro (). Thus, we suggested that the LINC01354/miR-340-5p/ATF1 axis might play critical roles in lung cancer progression ().
Figure 6. LINC01354/miR-340-5p/ATF1 axis promoted lung cancer progression. (A) miR-340-5p inhibitors rescued the effects of LINC01354 inhibition on ATF1 mRNA expression in lung cancer cells. (B) LINC01354 expression was positively correlated with ATF1 expression in NSCLC tissues. (C, D) LINC01354 suppression on lung cancer cells colony numbers and invasion ability could be rescued by miR-340-5p inhibitors. (E) Schematic diagram of LINC01354/miR-340-5p/ATF1 axis in lung cancer progression *p < .05.
Discussion
In the past years, numbers lncRNA were identified to play essential roles in lung cancer progression. For instance, Huang et al found that high PVT1 could promote the migration and invasion in NSCLC cells [Citation14]. She et al found that SNHG7 might promote lung cancer cells proliferation and invasion through increasing the expression of FAIM2 [Citation15]. Fang et al suggested that MALAT1 contributed to the cisplatin-resistance in lung cancer through increasing MRP1 and MDR1 by STAT3 activation [Citation16]. In the present study, we explored the functions of LINC01354 in lung cancer development. We found that LINC01354 increased ATF1 expression by regulating miR-340-5p, which provided a potential therapeutic target for the treatment of lung cancer.
LINC01354 is a member of lncRNAs, plays an oncogenic role on tumor development, and prognosis [Citation11]. Recently, LINC01354 was found to play critical roles in tumor progression. For example, Wang et al found that LINC01354 might play critical roles in colorectal cancer [Citation17]. Li et al showed that LINC01354 might interact with hnRNP-D contributed to colorectal cancer cells growth and metastasis by regulating Wnt/β-catenin pathway [Citation12]. Guo et al showed that LINC01354 was downregulated in papillary thyroid carcinoma [Citation18]. However, the roles of LINC01354 in lung cancer progression remain unknown. Our data showed that the expression of LINC01354 was aberrantly upregulated in NSCLC, high LINC01354 expression was associated with advanced TNM stage and poor prognosis of NSCLC patients. In function assays, we found that the inhibition of LINC01354 reduced lung cancer cells proliferation, and invasion in vitro. These data supported the oncogenic roles of LINC01354 in lung cancer.
Increasing evidence reported that lncRNAs could exert their role by constructing a regulatory network lncRNA-miRNA-mRNA [Citation19]. For instance, Cui et al showed that MIR22HG/miR-141-3p/DAPK1 axis might play critical roles in the proliferation of endometrial carcinoma cells [Citation20]. Gao et al suggested that SBF2-AS1 promoted the cervical cancer progression via regulating miR-361-5p/FOXM1 axis [Citation21]. In our research, we found that miR-340-5p was a potential target of LINC01354. Subsequently, we used qRT-PCR, luciferase reporter assay, and RIP assay to further verified the correlation between LINC01354 and miR-361-5p.
Previous studies suggested that miR-340-5p play critical roles in tumor progression. For example, Strong et al suggested that miR-340 might act as an important modulator in the RAS/RAF/MAPK pathway in melanoma [Citation22]. Wu et al showed that miR‐340 reduced the metastasis via regulating c‐Met expression in breast cancer [Citation23]. In addition, Fernandez et al reported that miR-340 inhibited lung cancer cells growth via regulating the expression of p27 [Citation24]. However, miR-340-5p functions in lung cancer remain unclear. In the currently study, we found that miR-340-5p was significantly decreased and associated with poor OS in NSCLC patients. MiR-340-5p inhibitors could promote the proliferation and invasion in lung cancer cells. Those data demonstrated that miR-340-5p acted as a tumor suppressor in NSCLC progression.
Next, we showed that ATF1 could serve as a direct target of miR-340-5p. And high ATF1 expression was associated with lung cancer metastasis, tumor stage and poor prognosis. ATF1 suppression reduced cell invasion in vitro. In addition, we showed that the expression of ATF1 could be regulated by LINC01354/miR-340-5p axis. And the effects of LINC01354 knockdown on lung cancer cells progression could be reversed by miR-340-5p inhibitors. Therefore, our present study suggested that the LINC01354/miR-340-5p/ATF1 axis play critical roles in lung cancer progression.
In conclusion, we showed that LINC01354 was upregulated in NSCLC and contributed to the high proliferation and invasive abilities through sponging miR-340-5p/ATF axis. Thus, we provided a diagnostic biomarker as well as a potential treatment target for lung cancer patients.
Ethics approval and consent to participate
Ethical approval was given by the Ethics Committee of People's Hospital of Bozhou City.
Disclosure statement
All the authors declare that they have no conflict of interest.
Data availability
The dataset supporting the conclusions of this article is included within the article.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
- Cruz CSD, Tanoue LT, Matthay RA. Lung cancer: epidemiology, etiology, and prevention. Clin Chest Med. 2011;32(4):605–644.
- de Groot P, Munden RF. Lung cancer epidemiology, risk factors, and prevention. Radiol Clin. 2012;50(5):863–876.
- Molina JR, Yang P, Cassivi SD, et al. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship[C]//Mayo Clinic Proceedings. Elsevier. 2008;83(5):584–594.
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641.
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol cell. 2011;43(6):904–914.
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361.
- Zhang H, Yang F, Chen SJ, et al. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol. 2015;36(4):2947–2955.
- Liu Z, Wang W, Jiang J, et al. Downregulation of GAS5 promotes bladder cancer cell proliferation, partly by regulating CDK6. PloS one. 2013;8(9):e73991
- Hu Y, Yang Q, Wang L, et al. Knockdown of the oncogene lncRNA NEAT1 restores the availability of miR-34c and improves the sensitivity to cisplatin in osteosarcoma. Biosci Rep. 2018;38(3):BSR20180375.
- Li J, He M, Xu W, et al. LINC01354 interacting with hnRNP-D contributes to the proliferation and metastasis in colorectal cancer through activating Wnt/β-catenin signaling pathway. J Exp Clin Cancer Res. 2019;38(1):161.
- Xu Y, Zhou W, Zhang C, et al. Long non-coding RNA RP11-552M11.4 favors tumorigenesis and development of cervical cancer via modulating miR-3941/ATF1 signaling]. Int J Biol Macromol. 2019;130:24–33.
- Zhao CC, Jiao Y, Zhang YY, et al. Lnc SMAD5-AS1 as ceRNA inhibit proliferation of diffuse large B cell lymphoma via Wnt/β-catenin pathway by sponging miR-135b-5p to elevate expression of APC. Cell death Dis. 2019;10(4):252.
- Huang C, Liu S, Wang H, et al. LncRNA PVT1 overexpression is a poor prognostic biomarker and regulates migration and invasion in small cell lung cancer. Am J Transl Res. 2016;8(11):5025.
- She K, Huang J, Zhou H, et al. lncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep. 2016;36(5):2673–2680.
- Fang Z, Chen W, Yuan Z, et al. LncRNA-MALAT1 contributes to the cisplatin-resistance of lung cancer by upregulating MRP1 and MDR1 via STAT3 activation. Biomed Pharmacother. 2018;101:536–542.
- Wang X, Zhou J, Xu M, et al. A 15-lncRNA signature predicts survival and functions as a ceRNA in patients with colorectal cancer. Cancer Manag Res. 2018;10:5799.
- Guo K, Chen L, Wang Y, et al. Long noncoding RNA RP11‐547D24. 1 regulates proliferation and migration in papillary thyroid carcinoma: Identification and validation of a novel long noncoding RNA through integrated analysis of TCGA database. Cancer Med. 2019;8:3105–3119.
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344.
- Cui Z, An X, Li J, et al. LncRNA MIR22HG negatively regulates miR-141-3p to enhance DAPK1 expression and inhibits endometrial carcinoma cells proliferation. Biomed Pharmacother. 2018;104:223–228.
- Gao F, Feng J, Yao H, et al. LncRNA SBF2-AS1 promotes the progression of cervical cancer by regulating miR-361-5p/FOXM1 axis. Artif Cells Nanomed Biotechnol. 2019;47(1):776–782.
- Strong AMP, Setaluri V, Spiegelman VS. microRNA-340 as a modulator of RAS-RAF-MAPK signaling in melanoma. Arch Biochem Biophys. 2014;563:118–124.
- Wu Z, Wu Q, Wang C, et al. miR-340 inhibition of breast cancer cell migration and invasion through targeting of oncoprotein c-Met. Cancer. 2011;117(13):2842–2852.
- Fernandez S, Risolino M, Mandia N, et al. miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene. 2015;34(25):3240.
